Submitted:
14 July 2025
Posted:
15 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Results of Amoebae Cultures, Thermal Tolerance Test, PCRs, and Sequencing
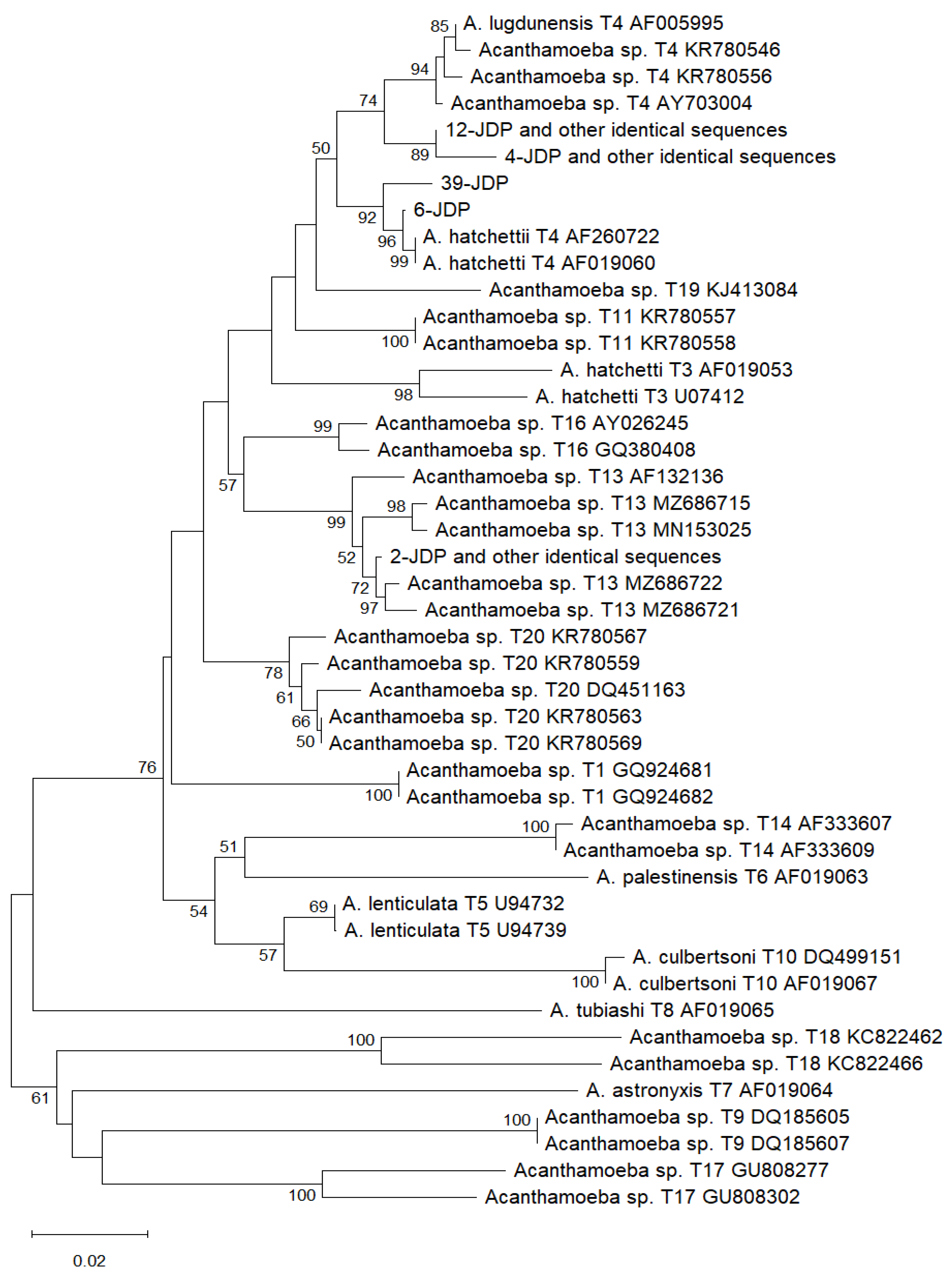
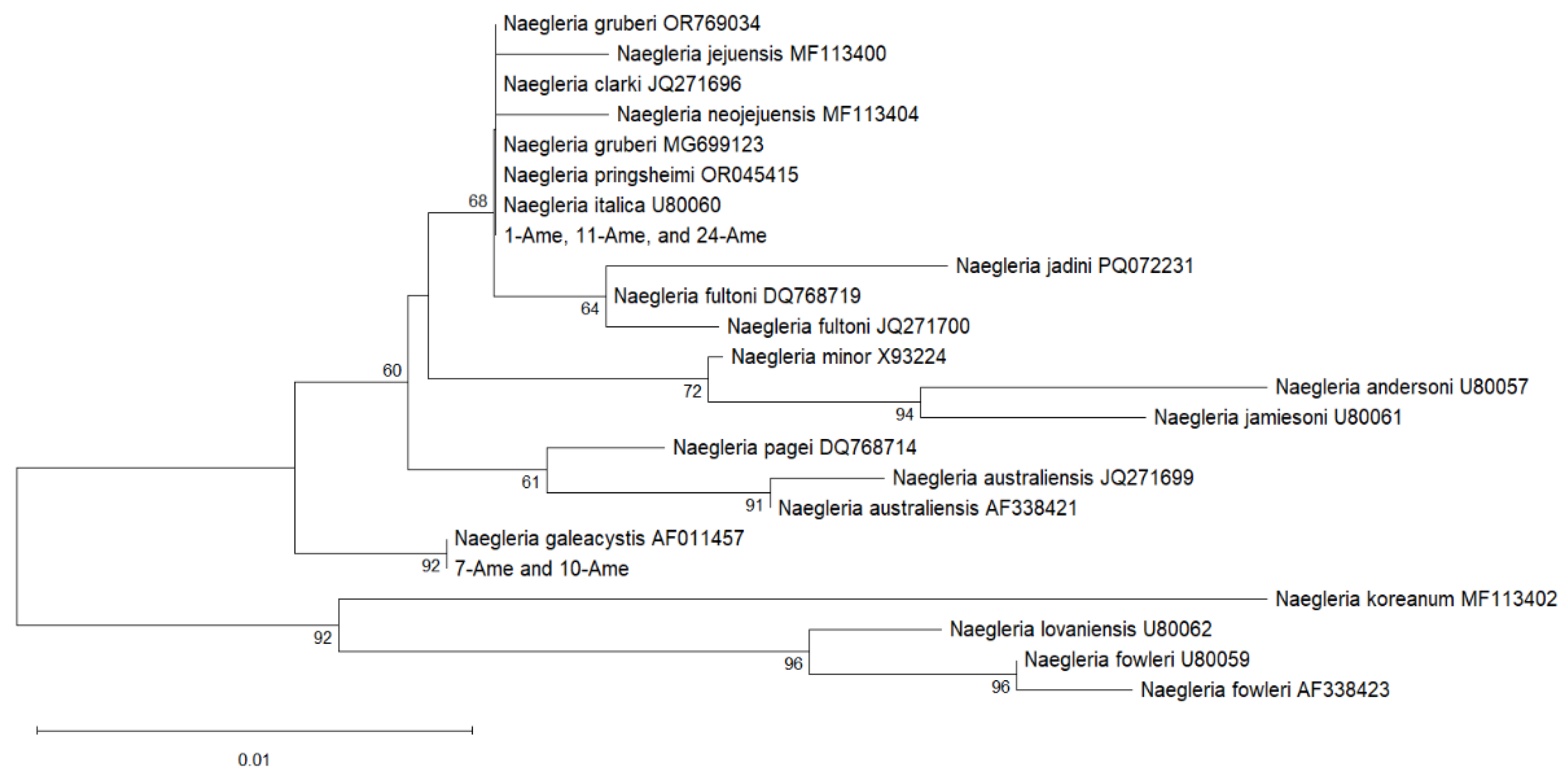
2.2. Results of Sequence Comparison and Phylogenetic Analysis
2.3. Results of Statistical Analysis
3. Discussion
4. Materials and Methods
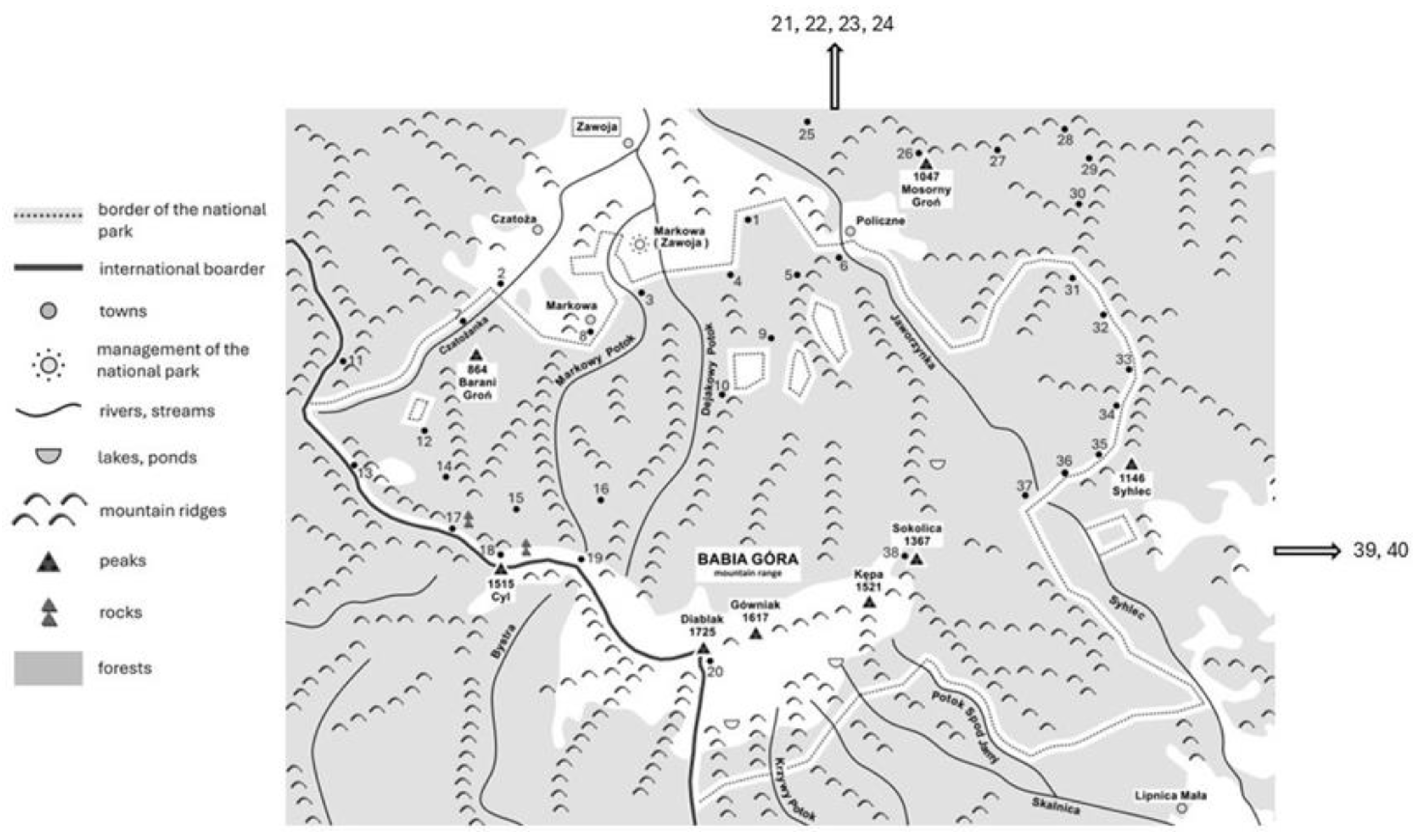
4.1. Study Area and Soil Sampling
4.2. Amoebae Cultures and Thermal Tolerance Test
4.3. DNA Extraction from Soil and Cultures, and PCR Protocols
4.4. Sequencing, Genotyping, and Phylogenetic Analysis
4.5. Statistical Analysis
5. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ali, M.; Rice, C.A.; Byrne, A.W.; Paré, P.E.; Beauvais, W. Modelling dynamics between free-living amoebae and bacteria. Environ. Microbiol. 2024, 26, e16623. [CrossRef]
- Otero-Ruiz, A.; Gonzalez-Zuñiga, L.D.; Rodriguez-Anaya, L.Z.; Lares-Jiménez, L.F.; Gonzalez-Galaviz, J.R.; Lares-Villa, F. Distribution and current state of molecular genetic characterization in pathogenic free-living amoebae. Pathogens 2022, 11, 1199. [CrossRef]
- Siddiqui, R.; Makhlouf, Z.; Khan, N.A. The increasing importance of Vermamoeba vermiformis. J. Eukaryot. Microbiol. 2021, 68, e12857. [CrossRef]
- Dinda, S.K.; Hazra, S.; De, A.; Datta, A.; Das, L.; Pattanayak, S.; Kumar, K.; Dey, M.D.; Basu, A.; Manna, D. Amoebae: beyond pathogens – exploring their benefits and future potential. Front. Cell. Infect. Microbiol. 2024, 14, 1518925. [CrossRef]
- Fan, S.; Shen, Y.; Qian, L. Social life of free-living amoebae in aquatic environment – comprehensive insights into interactions of free-living amoebae with neighboring microorganisms. Front. Microbiol. 2024, 15, 1382075. [CrossRef]
- Aykur, M.; Selver, O.B.; Dagci, H.; Palamar, M. Vermamoeba vermiformis as the ethiological agent in a patient with suspected non-Acanthamoeba keratitis. Parasitol. Res. 2024, 123, 323. [CrossRef]
- Salazar-Ardiles, C.; Valencia, K.P.; Andrade, D.C. Amoebas: the omnipotent organism and silent assassin. Mol Biol Rep. 2025, 52, 160. [CrossRef]
- Cardoso, I.R.; de Lima, C.S.; dos Reis, R.B.; Pinto, A.C.A.; Pissinatti, T.; Kugelmeier, T.; da Costa Neto, S.F.; da Silva, F.A.; Santos, H.L.C. Occurrence of free-living amoebae in non-human primate gut. Trop. Med. Infect. Dis. 2024, 9, 108. [CrossRef]
- Zurita-Artaloitia, J.M.; Riviera, J; Vinuesa, P. Extensive cryptic diversity and ecological associations uncovered among Mexican and global collections of Naegleria and Vermamoeba species by 18S ribosomal DNA, internal transcribed spacer, and cytochrome oxidase subunit I sequence analysis. Microbiol. Spectr. 2023, 11(2), 03795–22. [CrossRef]
- Kahraman, M.; Polat, Z.A. Are thermotolerant and osmotolerant characteristics of Acanthamoeba species an indicator of pathogenicity? Türkiye Parazitol Derg. 2024, 48(1), 15–20. [CrossRef]
- Lorenzo-Morales, J.; Khan, N.A.; Walochnik, J. An update on Acanthamoeba keratitis: diagnosis, pathogenesis and treatment. Parasite 2015, 22, 10. [CrossRef]
- Chaúque, B.J.M.; da Silva, T.C.B.; dos Santos, D.L.; Benitez, G.B.; Chaúque, L.G.H.; Benetti, A.D; Zanette, R.A.; Rott, M.B. Global prevalence of free-living amoebae in solid matrices – a systematic review with meta-analysis. Acta Trop. 2023, 247, 107006. [CrossRef]
- Bass, D.; Stentiford, G.D.; Littlewood, D.T.J.; Hartikainen, H. Diverse applications of environmental DNA methods in parasitology. Trends Parasitol. 2015, 31(10), 499–513. https://dx.doi.org/10.1016/j.pt.2015.06.013.
- Sengupta, M.E.; Lynggaard, C.; Mukaratriwa, S.; Vennervald, B.J.; Stensgaard, A.S. Environmental DNA in human and veterinary parasitology – current applications and future prospects for monitoring and control. Food and Waterborne Parasitol. 2022, 29, e00183. [CrossRef]
- Albuquerque, P.; Nicola, A.M.; Almeida, D.; Magnabosco, G.; da Silveira Derengowski, L.; Crisóstomo, L.S.; Xavier, L.C.G.; de Oliveira Frazão, S.; Guilhelmelli, F.; de Oliveira, M.A.; do Nascimento Dias, J.; Hurtado, F.A.; de Melo Teixeira, M.; Guimarães, A.J.; Paes, H.C.; Bagagli, E.; Felipe, M.S.S.; Casadevall, A.; Silva-Pereira, I. A hidden battle in dirt: Soil amoebae interactions with Paracoccidioides spp. NTDs 2019, 13(10), e0007742. [CrossRef]
- Hendiger-Rizo, E.B.; Chmielewska-Jeznach, M.; Poreda, K.; Liendo, A.R.; Koryszewska-Bagińska, A.; Olędzka, G.; Padzik, M. Potentially pathogenic free-living amoebae isolated from soil samples from Warsaw parks and squares. Pathogens 2024, 13, 895. [CrossRef]
- Pérez-Pérez, P.; Reyes-Batlle, M.; Morchón, R.; Piñero, J.E.; Lorenzo-Morales, J. Isolation and molecular identification of pathogenic free-living amoebae from environmental samples in Tenerife, Canary Islands. ACS EST Water 2025, 5, 2861–2869. [CrossRef]
- Sousa-Ramos, D.; Reyes-Batlle, M.; Bellini, N.K.; Rodriguez-Expósito, R.; Piñero.; J.E.; Lorenzo-Morales, J. Free-living amoebae in soil samples from Santiago Island, Cape Verde. Parasitol. Res. 2021, 121, 2399–2404. [CrossRef]
- Sousa-Ramos, D.; Reyes-Batlle, M.; Bellini, N.K.; Rodriguez-Expósito, R.; Martin-Real, C.; Piñero, J.E.; Lorenzo-Morales, J. Pathogenic free-living amoebae from water sources in Cape Verde. Microorganisms 2022, 9, 1460. [CrossRef]
- Reyes-Batlle, M.; Wagner, C.; Zamora-Herrera, J.; Vargas-Mesa, A.; Sifaoui, I.; González, A.C.; López-Arencibia, A.; Valladares, B.; Martinez-Carretero, E.; Piñero, J.E., Lorenzo-Morales, J. Isolation and molecular identification of Vermamoeba vermiformis strains from soil sources in El Hierro Island, Canary Islands, Spain. Curr. Microbiol. 2016, 73, 104–107. [CrossRef]
- Reyes-Batlle, M.; Diaz, F.J.; Sifaoui, I.; Rodriguez-Expósito, R.L.; Rizo-Liendo, A.; Piñero, J.E.; Lorenzo-Morales, J. Free living amoebae isolation in irrigation waters and soils of an insular arid agroecosystem. Sci. Total Environ. 2021, 753, 141833. [CrossRef]
- Denet, E.; Coupat-Goutaland, B.; Nazaret, S.; Pélandakis, M.; Favre-Bonté, S. Diversity of free-living amoebae in soils and their associated human opportunistic bacteria. Parasitol. Res. 2017,116, 3151–3162. [CrossRef]
- Aykur, M.; Dagci, H. Molecular identification of Acanthamoeba spp., Balamuthia mandrillaris and Naegleria fowleri in soil samples using quantitative real-time PCR assay in Turkey; Hidden danger in the soil! Acta Trop. 2023, 244, 106956. [CrossRef]
- Pazoki, H.; Niyyati, M.; Javanmard, E.; Iasjerdi, Z.; Spotin, A.; Mirjalali, H.; Behravan, M.R. Isolation and phylogenetic analysis of free-living amoebae (Acanthamoeba, Naegleria, and Vermamoeba) in the farmland soils and recreational places in Iran. Acta Parasitol. 2020, 65, 36–43. [CrossRef]
- Pérez-Pérez, P.; Artigas, P.; Reyes-Batlle, M.; Córdoba-Lanús, E.; Rodriguez-Expósito, R.L.; Cuervo, P.F.; Dominguez-de Barros, A.; Garcia-Pérez, O.; Valero, M.A.; De Elias, A.; Anglés, R.; Mas-Coma, S.; Piñero, J.E.; Bargues, M.D.; Lorenzo-Morales, J. Potentially pathogenic free-living amoebae at very high altitude: Detection by multiplex qPCR in the Northern Altiplano fascioliasis hyperendemic area in Bolivia. OH 2025, 20, 100985. [CrossRef]
- Reynaud, Y.; Ducat, C.; Talarmin, A.; Marcelino, I. Cartography of free-living amoebae in soil in Guadeloupe (French West Indies) using DNA metabarcoding. Pathogens 2020, 9, 440. [CrossRef]
- Tanzifi, A.; Moghaddam, Y.; Dodangeh, S.; Daryani, A.; Shahabeddin, S.; Gholami, S.; Hosseini, S.A.; Chegeni, T.N.; Hosseininejad, Z. Detection and molecular characterization of potentially pathogenic free-living amoebae from recreational and public soils in Mazandaran, Northern Iran. Iran J Parasitol. 2021, 16(2), 295–304.
- Noinarin, P.; Chareonsudjai, P.; Wangsomnuk, P.; Wongratanacheewin, S.; Chareonsudjai, S. Environmental free-living amoebae isolated from soil in Khon Kaen, Thailand, antagonize Burkholderia pseudomallei. PLoS ONE 2016, 11(11), e0167355. [CrossRef]
- Watson, P.M.; Sorrell, S.C.; Brown, M.W. Ptolemeba n. gen., a novel genus of Hartmannellid amoebae (Tubulinea, Amoebozoa); with an emphasis on the taxonomy of Saccamoeba. J. Eukaryot. Microbiol. 2014, 61, 611–619. [CrossRef]
- Kudryavtsev, A.; Volkova, E.; Parshukov, A. Ptolemeba bulliensis Watson et al. 2014 (Amoebozoa, Tubulinea) from freshwater NGD-affected rainbow trout (Oncorhynchus mykiss Walbaum, 1792) gills tolerates brackish water conditions. J. Fish Dis. 2025, 0, e14132. [CrossRef]
- Brown, S.; De Jonckheere, J.F. Isolation of a new valkhampfiid amoeba from soil: Paravahlkampfia lenta n. sp. Eur. J. Protistol. 2004, 40, 289–294. [CrossRef]
- McLaughlin, G.L.; Brandt, F.H.; Visvesvara, G.S. Restriction fragment length polymorphisms of the DNA of selected Naegleria and Acanthamoeba amebae. J. Clin. Microbiol. 1988, 26(9), 1655–1658.
- Cholewiński, M.; Solarczyk, P.; Derda, M.; Wojtkowiak-Giera, A.; Hadaś, E. Presence of potential pathogenic genotypes of free-living amoebae isolated from sandboxes in children’s playgrounds. Folia Parasitol. 2015, 62: 064. [CrossRef]
- Orosz, E.; Posta, K. Genotyping of Acanthamoeba spp. from rizosphere in Hungary. Acta Microbiol Immunol Hung. 2020, 67(3), 171–175. [CrossRef]
- Schmitz-Esser, S.; Toenschoff, E.R.; Haider, S.; Heinz, E.; Hoenninger, V.M.; Wagner, M.; Horn, M. Diversity of bacterial endosymbionts of environmental Acanthamoeba isolates. Appl. Environ. Microbiol. 2008, 74(18), 5822–5831. [CrossRef]
- Geisen, S.; Fiore-Donno, A.M.; Walochnik, J.; Bonkowski, M. Acanthamoeba everywhere: high diversity of Acanthamoeba in soils. Parasitol. Res. 2014, 113, 3151–3158. [CrossRef]
- Wang, Y.; Jiang, L.; Zhao, Y.; Ju, X.; Wang, L.; Jin, L.; Fine, R.D.; Li, M. Biological characteristics and pathogenicity of Acanthamoeba. Front. Microbiol. 2023, 14, 11470077. [CrossRef]
- Garcia, A.; Goñi, P.; Clavel, A.; Lobez, S.; Fernandez, M.T.; Ormad, M.P. Potentially pathogenic free-living amoebae (FLA) isolated in Spanish wastewater treatment plants. Environ. Biol. Rep. 2011, 3(5): 622–626. [CrossRef]
- Ozkoc, S.; Tuncay, S.; Delibas, S.B.; Akisu, C.; Ozbek, Z.; Durak, I.; Walochnik, J. Identification of Acanthamoeba genotype T4 and Paravahlkampfia sp. from two clinical isolates. J. Med. Microbiol. 2008, 57. [CrossRef]
- Visvesvara, G.S.; Sriram, R.; Qvarnstrom, Y.; Bandyopadhyay, K.; da Silva, A.J.; Pieniążek, N.J.; Cabral, G.A. Paravahlkampfia francinae n. sp. masquerading as an agent of primary amoebic meningocephalitis. J. Eukaryot. Microbiol. 2009, 56(4), 357–366 https://doi.org/10.1111/j.1550-7408.2009.00410.x.
- Adamska, M.; Leońska-Duniec, A.; Łanocha, N.; Skotarczak, B. Thermophilic, potentially pathogenic amoebae isolated from natural water bodies in Poland and their molecular characterization. Acta Parasitol. 2014, 59(3), 433–441. [CrossRef]
- Schroeder, J.M.; Booton, G.C.; Hay, J.; Niszl, I.A.; Seal, D.V.; Markus, M.B.; Fuerst, P.A.; Byers, T.J. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification and Acanthamoebae from humans with keratitis and from sewage sludge. J. Clin. Microbiol. 2001, 39(5), 1903–1911. [CrossRef]
- Scheikl, U.; Sommer, R.; Kirschner, A.; Rameder, A.; Schrammel, B.; Zweimüller, I.; Wesner, W.; Hinker, M.; Walochnik, J. Free-living amoebae (FLA) co-occurring with legionellae in industrial waters. Eur. J. Protistol. 2014, 50, 422–429. https://dx.doi.org/10.1016/j.ejop.2014.04.002.
- Tsvetkova, N.; Schild, M.; Panaiotov, S.; Kurdova-Mintcheva, R.; Gottstein, B.; Walochnik, J.; Aspӧck, H.; Lucas, M.S.; Müller, N. The identification of free-living environmental isolates of amoebae from Bulgaria. Parasitol. Res. 2004, 92, 405–413. [CrossRef]
- Thomas, V.; Herrera-Rimann, K.; Blanc, D.S.; Greub, G. Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl. Environ. Microbiol. 2006, 72(4), 2428–2438. [CrossRef]
- Liang, S.-Y.; Ji, D.-R.; Hsia, K.-T.; Hung, C.-C.; Sheng, W.-H.; Hsu, B.-M.; Chen, J.-S.; Wu, M.-H.; Lai, C.-H.; Ji, D.-D. Isolation and identification of Acanthamoeba species related to amoebic encephalitis and nonpathogenic free-living amoeba species from the rice field. J. Appl. Microbiol. 2010, 109, 1422–1429. [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, 1–9. [CrossRef]


| Sample number | Culture at 42°C |
PCR and sequencing results with individual primer pairs | ||||
|---|---|---|---|---|---|---|
| JDP1/JDP2 | FLA-F/FLA-R | Ami6F1/Ami9R | AmeF977/ AmeR1534 |
HARTfor/ HARTrev |
||
| 1. | Naegleria sp. | |||||
| 2. | + | Acanthamoeba T13 | V. vermiformis | |||
| 4. | Acanthamoeba T4 | Paravahlkampfia sp. | ||||
| 6. | + | Acanthamoeba T4 | ||||
| 7. | N. galeacystis | N. galeacystis | ||||
| 8. | + | N. gruberi | N. gruberi | |||
| 10. | Acanthamoeba T13 | N. galeacystis | N. galeacystis | |||
| 11. | N. gruberi | N. gruberi | Naegleria sp. | |||
| 12. | Acanthamoeba T4 | |||||
| 15. | + | Acanthamoeba T4 | ||||
| 19. | + | Acanthamoeba T4 | V. vermiformis | V. vermiformis | V. vermiformis | |
| 21. | + | Acanthamoeba T4 | ||||
| 23. | Acanthamoeba T13 | |||||
| 24. | + | N. gruberi | N. gruberi | Naegleria sp. | ||
| 27. | Acanthamoeba T4 | |||||
| 28. | Acanthamoeba T13 | P. bulliensis | ||||
| 29. | + | Acanthamoeba T4 | ||||
| 31. | + | Acanthamoeba T4 | ||||
| 32. | + | Acanthamoeba T13 | V. vermiformis | |||
| 33. | + | Acanthamoeba T13 | ||||
| 35. | + | Acanthamoeba T13 | ||||
| 37. | Acanthamoeba T4 | |||||
| 39. | Acanthamoeba T4 | |||||
| 40. | Acanthamoeba T4 | |||||
| The sequences obtained in this study (sample number and primer set) | The sequences from the GenBank database – accession number, isolation source, and country | Identity |
|---|---|---|
| Identical sequences: 12-JDP, 15-JDP, 19-JDP, 27-JDP, 29-JDP, 31-JDP | ON408415 Acanthamoeba T4 isolated from soil (USA) | 100% |
| 6-JDP | KT985967 Acanthamoeba T4 isolated from soil (Iran) KU894800 Acanthamoeba sp. isolated from water (Uganda) MZ404332 Acanthamoeba sp. isolated from a patient with keratitis (Brazil) |
100% |
| 39-JDP | KC164232 Acanthamoeba sp. isolated from compost (Switzerland) PQ431522, PQ431528 Acanthamoeba sp. isolated from water (France) AF019060 A. hatchetti T4 |
100% 99.48% |
| Identical sequences: 4-JDP, 21-JDP, 37-JDP, 40-JDP | JQ669659 Acanthamoeba T4 isolated from soil (USA) KT892868 Acanthamoeba T4 isolated from soil (Chile) FJ807650 Acanthamoeba sp. isolated from marsh sediment (Austria) LR813621 A. castellani isolated from coprolite (Belgium) KC164234 A. castellani isolated from compost (Switzerland) MN700280 Acanthamoeba T4 isolated from water (Malaysia) MT378239 Acanthamoeba T4 isolated from water (Iran) |
100% |
| Identical sequences: 2-JDP, 10-JDP, 23-JDP, 28-JDP, 32-JDP, 33-JDP, 35-JDP | MZ686722 Acanthamoeba T13 isolated from soil (Malaysia) | 99.48% |
| The sequences obtained in this study (sample number and primer set*) | The sequences from the GenBank database – accession number, isolation source, and country | Identity |
|---|---|---|
| Identical sequences: 7-FLA and 10-FLA | DQ768717 Naegleria sp. isolated from fish (Czech Republic) PP174311 Naegleria sp. |
100% |
| Identical sequences: 8-FLA and 24-FLA; Identical sequences: 8-Ami and 24-Ami |
OR769034 N. gruberi isolated from soil (USA) MG699123, AB298288 – N. gruberi |
100% |
| 11-FLA/11-Ami | 98.64%/ 99.17% |
|
| Identical sequences: 7-Ame and 10-Ame | AF011457 N. galeacystis DQ768717, DQ768718 – Naegleria sp. isolated from fishes |
100% |
| Identical sequences: 1-Ame, 11-Ame, 24-Ame |
OR769034 N. gruberi isolated from soil (USA) MG699123, AB298288 – N. gruberi AF338417, AF338419, JQ271691, JQ271692, JQ271697, JQ271705 – N. clarki isolated from fishes; JQ271704 – N. clarki isolated from sediment (Czech Republic) OR045415 – N. pringsheimi |
100% |
| The sequences obtained in this study (sample number and primer set*) |
The sequences from the GenBank database – accession number, isolation source, and country | Identity |
|---|---|---|
| 19-FLA | KU746978 V. vermiformis isolated from soil (Spain) KP792383, KP792388 V. vermiformis isolated from water (Italy) MF112024 V. vermiformis isolated from water (Pakistan) JQ271688 H. vermiformis isolated from a tapeworm (Croatia) HM363627 Hartmannella sp. isolated from a fish (Germany) |
100% |
| 19-AME | LC764480 V. vermiformis isolated from soil (Japan) LN650671 V. vermiformis isolated from water (Italy) JQ271687 V. vermiformis isolated from a fish (Czech Republic) |
100% |
| Identical sequences: 2-HART, 19-HART, 32-HART | PP732395, PP732396 V. vermiformis isolated from water (Philippines) | 100% |
| The primer set | Detecting FLA | The amplifying fragment of the 18S rRNA gene and the product size | Hybridization temperature | References |
|---|---|---|---|---|
| JDP1/JDP2 | Acanthamoeba spp. | 897-1358 bp of A. castellani sequence (U07400); 462 bp | 55°C | [42] |
| HARTfor/HARTrev | Former genus Hartmannella | 562-1095 bp of V. vermiformis sequence (EU137741); 534 bp | 58°C | [43] |
| FLA-F/FLA-R | All FLA except Balamuthia and Sappinia | 631-1614 bp of A. castellani sequence (U07400); 984 bp | 55°C | [44] |
| Ami6F1/Ami9R | All amoebae | 641-1468 bp of A. castellani sequence (U07400); 828 bp | 55°C | [45] |
| AmeF977/AmeR1534 | All amoebae | 1179-1829 bp of A. castellani sequence (U07400); 651 bp | 60°C | [46] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





