Submitted:
11 July 2025
Posted:
11 July 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Phenotypic Adaptations: Pulmonary Elasticity
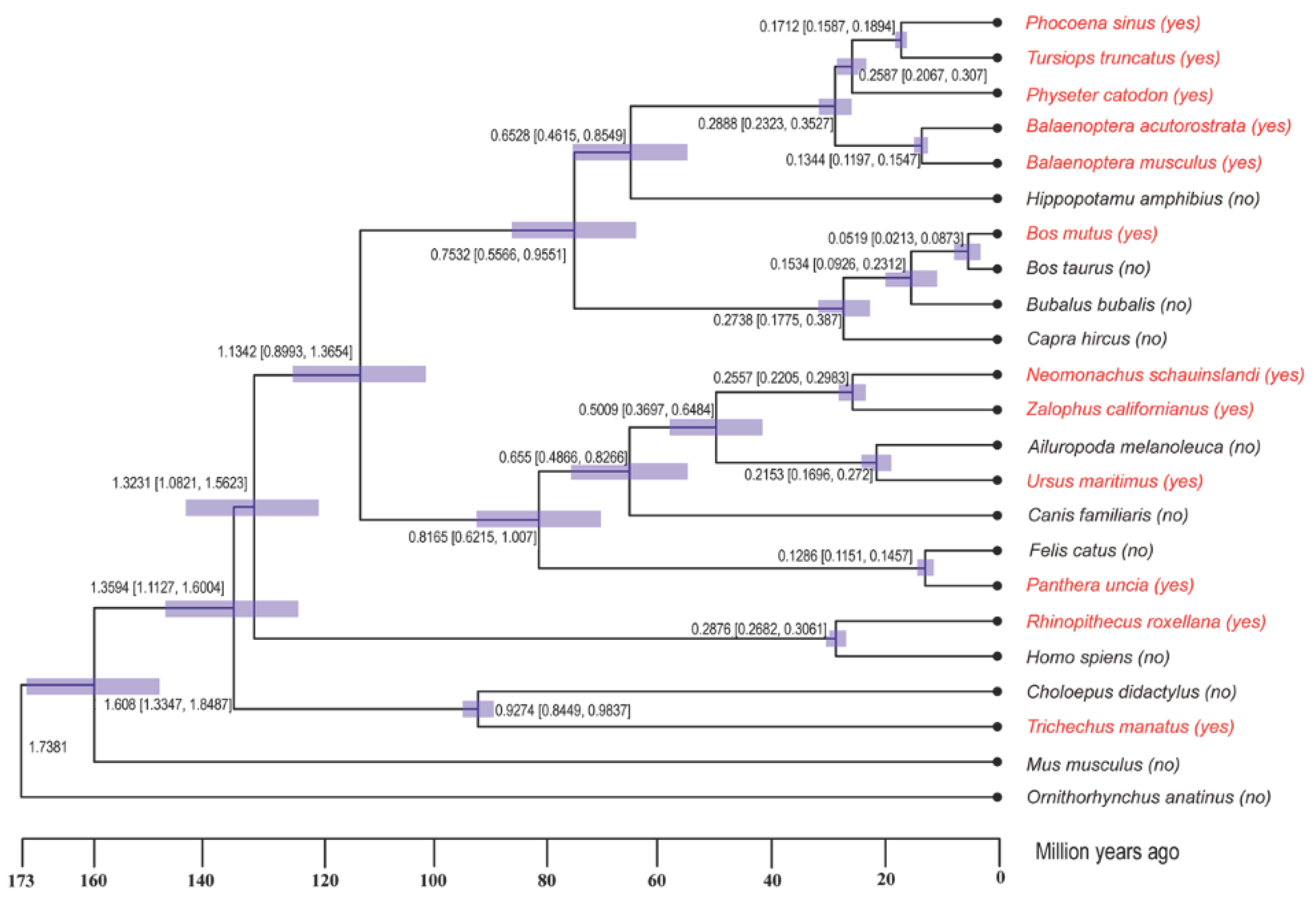
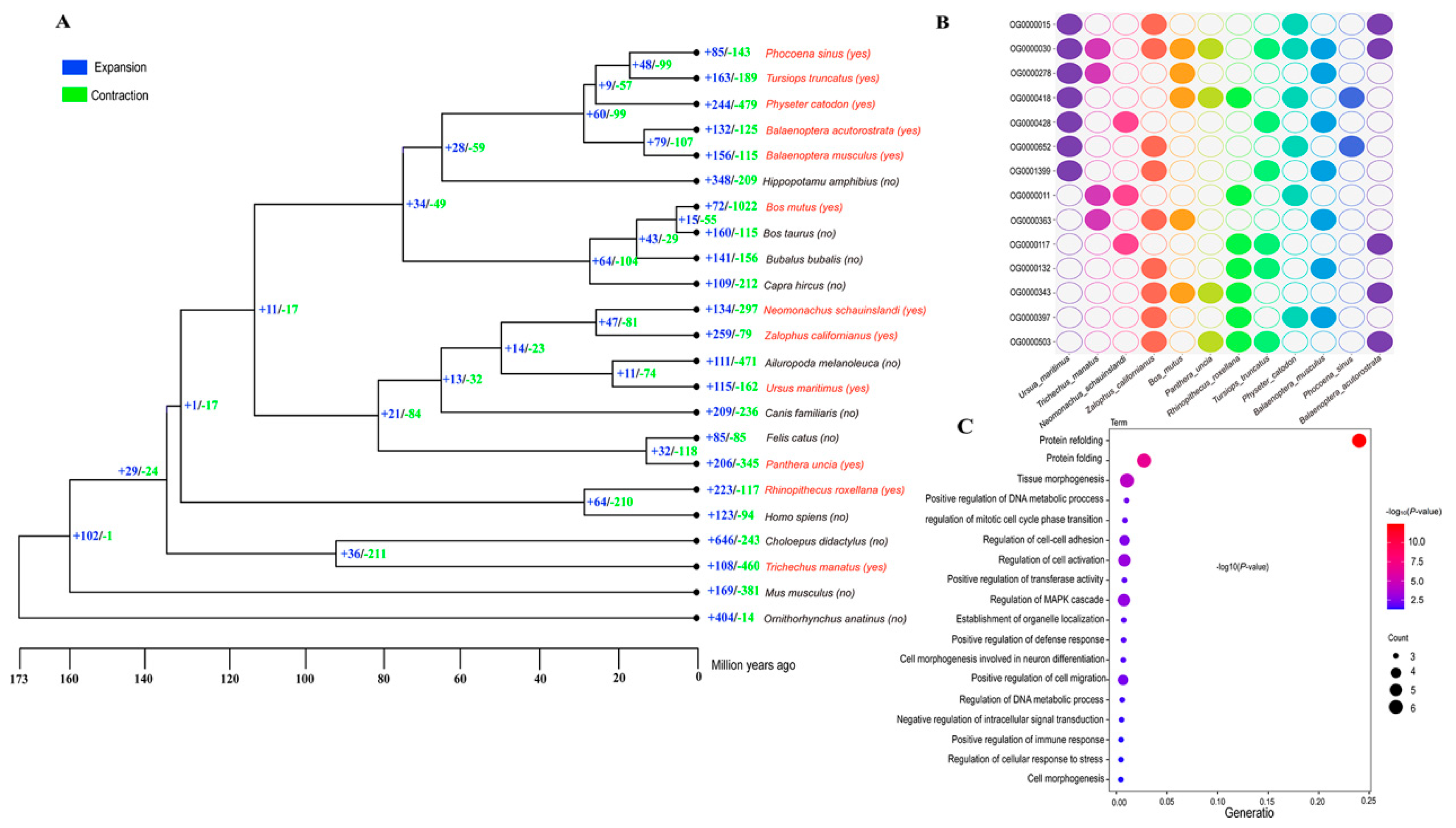
Gene Family Expansion and Contraction
Genes Under Positive Selection and Accelerated Evolution
Convergent Amino Acid Substitutions
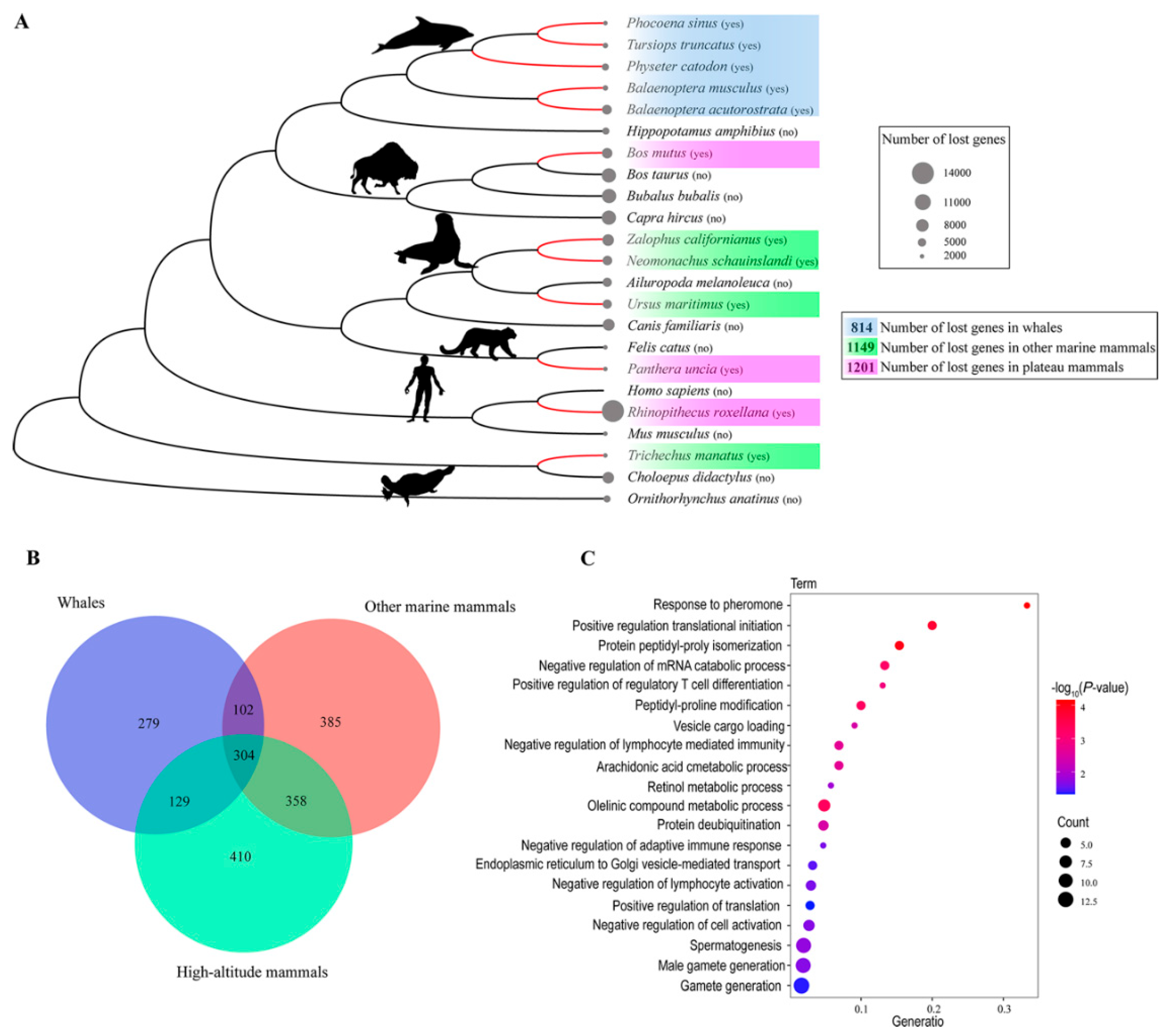
Convergent Gene Loss
Implications for Human Health
Conclusion and Future Directions
Acknowledgments
Conflict of Interest Statement
References
- Mallet RT, Burtscher J, Pialoux V, Pasha Q, Ahmad Y, Millet GP, Burtscher M. Molecular Mechanisms of High-Altitude Acclimatization. Int J Mol Sci. 2023 Jan 15;24(2):1698. [CrossRef] [PubMed] [PubMed Central]
- Nathan J, Ramachandran A. Efficacy of marine biomolecules on angiogenesis by targeting hypoxia inducible factor/vascular endothelial growth factor signaling in zebrafish model. J Biochem Mol Toxicol. 2022;36(2):e22954. [CrossRef]
- Dong-Dong Wu, Cui-Ping Yang, Ming-Shan Wang, Kun-Zhe Dong, Da-Wei Yan, Zi-Qian Hao, Song-Qing Fan, Shu-Zhou Chu, Qiu-Shuo Shen, Li-Ping Jiang, Yan Li, Lin Zeng, He-Qun Liu, Hai-Bing Xie, Yun-Fei Ma, Xiao-Yan Kong, Shu-Li Yang, Xin-Xing Dong, Ali Esmailizadeh, David M Irwin, Xiao Xiao, Ming Li, Yang Dong, Wen Wang, Peng Shi, Hai-Peng Li, Yue-Hui Ma, Xiao Gou, Yong-Bin Chen, Ya-Ping Zhang, Convergent genomic signatures of high-altitude adaptation among domestic mammals, National Science Review, Volume 7, Issue 6, June 2020, Pages 952–963. [CrossRef]
- Pamenter ME, Hall JE, Tanabe Y, Simonson TS. Cross-Species Insights Into Genomic Adaptations to Hypoxia. Front Genet. 2020;11:743. Published 2020 Jul 22. [CrossRef]
- Li J, Meng X, Wang L, Yu Y, Yu H, Wei Q. Changes in the expression levels of elastic fibres in yak lungs at different growth stages. BMC Dev Biol. 2021 Apr 20;21(1):9. [CrossRef] [PubMed] [PubMed Central]
- Ivy CM, Scott GR. Control of breathing and the circulation in high-altitude mammals and birds. Comp Biochem Physiol A Mol Integr Physiol. 2015;186:66-74. [CrossRef]
- Tong X, Yang Y, Wang W, et al. Expression profiling of abundant genes in pulmonary and cardiac muscle tissues of Tibetan Antelope (Pantholops hodgsonii). Gene. 2013;523(2):187-191. [CrossRef]
- Zhang Y, Lv W, Yan W, Guo B, Yang G, Ren W. Molecular adaptations in MMP genes support lung elasticity and diving adaptations in cetaceans. BMC Genomics. 2025;26(1):562. Published 2025 Jun 5. [CrossRef]
- Lyu T, Zhou S, Fang J, et al. Convergent Genomic Signatures of High-Altitude Adaptation among Six Independently Evolved Mammals. Animals (Basel). 2022;12(24):3572. Published 2022 Dec 16. [CrossRef]
- Haine L, Bravais J, Yegen CH, et al. Sleep Apnea in Idiopathic Pulmonary Fibrosis: A Molecular Investigation in an Experimental Model of Fibrosis and Intermittent Hypoxia. Life (Basel). 2021;11(9):973. Published 2021 Sep 15. [CrossRef]
- Guo B, Sun Y, Wang Y, et al. Evolutionary genetics of pulmonary anatomical adaptations in deep-diving cetaceans. BMC Genomics. 2024;25(1):339. Published 2024 Apr 4. [CrossRef]
- Yang Y, Zhang S, Guo L. Characterization of Cell Cycle-Related Competing Endogenous RNAs Using Robust Rank Aggregation as Prognostic Biomarker in Lung Adenocarcinoma. Front Oncol. 2022;12:807367. Published 2022 Feb 3. [CrossRef]
- Guo L, Dou Y, Yang Y, et al. Protein profiling reveals potential isomiR-associated cross-talks among RNAs in cholangiocarcinoma. Comput Struct Biotechnol J. 2021;19:5722-5734. Published 2021 Oct 14. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




