Submitted:
08 July 2025
Posted:
11 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Methods
2.1. Patients
2.2. Materials and Methods
2.2.1. Immunohistochemical Assessment of the Hormones and Transcriptions Factors
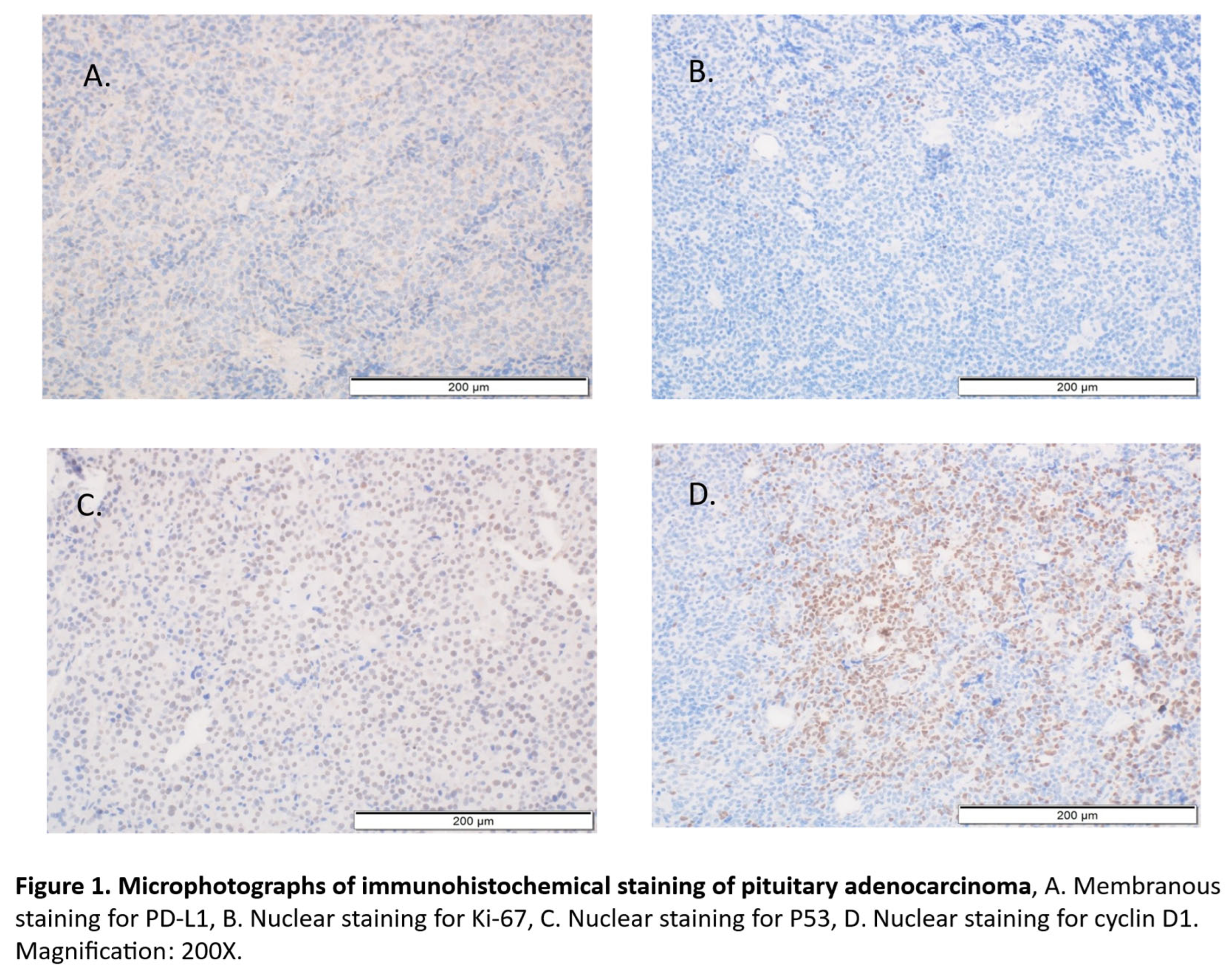
2.2.2. Immunohistochemistry of the PDL1, cyclin D1, Ki-67 and P53
2.2.3. Magnetic Resonance Imaging of the Tumor
3. Statistics
4. Results
4.1. Patients Characteristics
4.2. Immunoexpression of PDL-1, Ki-67, P53 and Cyclin D1 and Their Correlation with Epidemiological, Clinical and Histopathological Features
4.3. Combined Analysis of Ki-67 and P53 Expression or Ki-67 and Cyclin D1
4.3.1. Combination Analysis of Ki-67 and P53 Factors
4.3.2. Combination Analysis of Cyclin D1 and P53 Factors
4.3.3. Analysis of the Effect of the Combination of Ki-67, P53 and Cyclin D1 on the Invasiveness of PitNETs
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data availability
Acknowledgments
Conflicts of interest
References
- Molitch ME (2017) Diagnosis and treatment of pituitary adenomas: a review. JAMA 317:516–524. https ://doi.org/10.1001/Jama.2016.19699. [CrossRef]
- Melmed S. Pathogenesis of pituitary tumors. Nat Rev Endocrinol. 2011 May;7(5):257-66. doi: 10.1038/nrendo.2011.40. Epub 2011 Mar 22. Erratum in: Nat Rev Endocrinol. 2011 May;7(5):following 266. PMID: 21423242. [CrossRef]
- Melmed S, Kaiser UB, Lopes MB, Bertherat J, Syro LV, Raverot G, Reincke M, Johannsson G, Beckers A, Fleseriu M, Giustina A, Wass JAH, Ho KKY. Clinical Biology of the Pituitary Adenoma. Endocr Rev. 2022 Nov 25;43(6):1003-1037. doi: 10.1210/endrev/bnac010. PMID: 35395078; PMCID: PMC9695123. [CrossRef]
- Asa SL, Casar-Borota O, Chanson P, Delgrange E, Earls P, Ezzat S, Grossman A, Ikeda H, Inoshita N, Karavitaki N, Korbonits M, Laws ER Jr, Lopes MB, Maartens N, McCutcheon IE, Mete O, Nishioka H, Raverot G, Roncaroli F, Saeger W, Syro LV, Vasiljevic A, Villa C, Wierinckx A, Trouillas J; attendees of 14th Meeting of the International Pituitary Pathology Club, Annecy, France, November 2016. From pituitary adenoma to pituitary neuroendocrine tumor (PITNET): an International Pituitary Pathology Club proposal. Endocr Relat Cancer. 2017 Apr;24(4):C5-C8. doi: 10.1530/ERC-17-0004. PMID: 28264912. [CrossRef]
- Asa SL, Mete O, Perry A, Osamura RY. Overview of the 2022 WHO Classification of Pituitary Tumors. Endocr Pathol. 2022 Mar;33(1):6-26. doi: 10.1007/s12022-022-09703-7. Epub 2022 Mar 15. PMID: 35291028. [CrossRef]
- Raverot G, Dantony E, Beauvy J, et al. Risk of recurrence in pituitary neuroendocrine tumors: a prospective study using a five-tiered classification. J Clin Endocrinol Metab. 2017;102(9):3368-3374.
- Trouillas J, Roy P, Sturm N, et al. A new prognostic clinicopathological classification of pituitary adenomas: a multicentric case-control study of 410 patients with 8 years postoperative follow-up. Acta Neuropathol. 2013;126(1):123-135.
- Raverot G, Burman P, McCormack A, et al. European Society of Endocrinology Clinical Practice Guidelines for the management of aggressive pituitary tumours and carcinomas. Eur J Endocrinol. 2018;178(1):G1-G24.
- Dekkers OM, Karavitaki N, Pereira AM. The epidemiology of aggressive pituitary tumors (and its challenges). Rev Endocr Metab Disord. 2020;21(2):209-212.
- Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms Controlling PD-L1 Expression in Cancer. Mol Cell. 2019 Nov 7;76(3):359-370. doi: 10.1016/j.molcel.2019.09.030. Epub 2019 Oct 24. PMID: 31668929; PMCID: PMC6981282. [CrossRef]
- Wang X, Teng F, Kong L, Yu J. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther. 2016 Aug 12;9:5023-39. doi: 10.2147/OTT.S105862. PMID: 27574444; PMCID: PMC4990391. [CrossRef]
- Hou J, Yu Z, Xiang R, et al. Correlation between infiltration of FOXP3+ regulatory T cells and expression of B7-H1 in the tumor tissues of gastric cancer. Exp Mol Pathol. 2014;96(3):284–291.
- Qing Y, Li Q, Ren T, Xia W, Peng Y, Liu GL, Luo H, Yang YX, Dai XY, Zhou SF, Wang D. Upregulation of PD-L1 and APE1 is associated with tumorigenesis and poor prognosis of gastric cancer. Drug Des Devel Ther. 2015 Feb 16;9:901-9. doi: 10.2147/DDDT.S75152. [CrossRef]
- Shi F, Shi M, Zeng Z, Qi RZ, Liu ZW, Zhang JY, Yang YP, Tien P, Wang FS. PD-1 and PD-L1 upregulation promotes CD8(+) T-cell apoptosis and postoperative recurrence in hepatocellular carcinoma patients. Int J Cancer. 2011 Feb 15;128(4):887-96. doi: 10.1002/ijc.25397. [CrossRef]
- Thompson RH, Gillett MD, Cheville JC, Lohse CM, Dong H, Webster WS, Chen L, Zincke H, Blute ML, Leibovich BC, Kwon ED. Costimulatory molecule B7-H1 in primary and metastatic clear cell renal cell carcinoma. Cancer. 2005 Nov 15;104(10):2084-91. doi: 10.1002/cncr.21470. [CrossRef]
- Ohigashi Y, Sho M, Yamada Y, Tsurui Y, Hamada K, Ikeda N, Mizuno T, Yoriki R, Kashizuka H, Yane K, Tsushima F, Otsuki N, Yagita H, Azuma M, Nakajima Y. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin Cancer Res. 2005 Apr 15;11(8):2947-53. doi: 10.1158/1078-0432.CCR-04-1469. [CrossRef]
- Wang L, Ma Q, Chen X, Guo K, Li J, Zhang M. Clinical significance of B7-H1 and B7-1 expressions in pancreatic carcinoma. World J Surg. 2010 May;34(5):1059-65. doi: 10.1007/s00268-010-0448-x. [CrossRef]
- Peng Z, Li M, Li H, Gao Q. PD-1/PD-L1 immune checkpoint blockade in ovarian cancer: Dilemmas and opportunities. Drug Discov Today. 2023 Aug;28(8):103666. doi: 10.1016/j.drudis.2023.103666. [CrossRef]
- Ghosh S, Nataraj NB, Noronha A, Patkar S, Sekar A, Mukherjee S, Winograd-Katz S, Kramarski L, Verma A, Lindzen M, Garcia DD, Green J, Eisenberg G, Gil-Henn H, Basu A, Lender Y, Weiss S, Oren M, Lotem M, Geiger B, Ruppin E, Yarden Y. PD-L1 recruits phospholipase C and enhances tumorigenicity of lung tumors harboring mutant forms of EGFR. Cell Rep. 2021 May 25;35(8):109181. doi: 10.1016/j.celrep.2021.109181. [CrossRef]
- Luo Y, Ma S, Sun Y, Peng S, Zeng Z, Han L, Li S, Sun W, Xu J, Tian X, Wang F, Wu Q, Xiao Y, Zhang J, Gong Y, Xie C. MUC3A induces PD-L1 and reduces tyrosine kinase inhibitors effects in EGFR-mutant non-small cell lung cancer. Int J Biol Sci. 2021 Apr 12;17(7):1671-1681. doi: 10.7150/ijbs.57964. [CrossRef]
- Deng H, Zhao Y, Cai X, Chen H, Cheng B, Zhong R, Li F, Xiong S, Li J, Liu J, He J, Liang W. PD-L1 expression and Tumor mutation burden as Pathological response biomarkers of Neoadjuvant immunotherapy for Early-stage Non-small cell lung cancer: A systematic review and meta-analysis. Crit Rev Oncol Hematol. 2022 Feb;170:103582. doi: 10.1016/j.critrevonc.2022.103582. Epub 2022 Jan 11. [CrossRef]
- Kaunitz GJ, Cottrell TR, Lilo M, Muthappan V, Esandrio J, Berry S, Xu H, Ogurtsova A, Anders RA, Fischer AH, Kraft S, Gerstenblith MR, Thompson CL, Honda K, Cuda JD, Eberhart CG, Handa JT, Lipson EJ, Taube JM. Melanoma subtypes demonstrate distinct PD-L1 expression profiles. Lab Invest. 2017 Sep;97(9):1063-1071. doi: 10.1038/labinvest.2017.64. [CrossRef]
- Dell’Aquila M, Granitto A, Martini M, Capodimonti S, Cocomazzi A, Musarra T, et al. PD-L1 and thyroid cytology: A possible diagnostic and prognostic marker. Cancer Cytopathol.2020; 128:177–89. doi: 10.1002/cncy.22224. [CrossRef]
- Mei Y, Bi WL, Greenwald NF et al. (2016) Increased expression of programmed death ligand 1 (PD-L1) in human pituitary tumors. Oncotarget 7:76565–76576. https ://doi.org/10.18632 /oncot arget .12088. [CrossRef]
- Salomon MP, Wang X, Marzese DM et al. (2018) The epigenomic landscape of pituitary adenomas reveals specific alterations and differentiates among acromegaly, Cushing’s disease and endocrine-inactive subtypes. Clin Cancer Res 24:4126–4136. https:// doi.org/10.1158/1078-0432.CCR-17-2206. [CrossRef]
- Wang P, Wang T, Yang Y et al. (2018) The expression profile of PD-L1 and CD8 + lymphocyte in pituitary adenomas indicating for immunotherapy. J Neurooncol 139:89–95. https://doi. org/10.1007/s11060-018-2844-2.
- Sato M, Tamura R, Tamura H et al. (2019) Analysis of tumor angiogenesis and immune microenvironment in non-functional pituitary endocrine tumors. J Clin Med. https://doi.org/10.3390/ jcm8050695. [CrossRef]
- Harel E, Hewer E, La Rosa S, Brouland JP, Pitteloud N, Santoni F, Brunner M, Daniel RT, Messerer M, Cossu G. PD-L1 expression in PITNETs: Correlations with the 2022 WHO classification. Brain Spine. 2024 Dec 24;5:104171. doi: 10.1016/j.bas.2024.104171. [CrossRef]
- Cossu, G.; La Rosa, S.; Brouland, J.P.; Pitteloud, N.; Harel, E.; Santoni, F.; Brunner, M.; Daniel, R.T.; Messerer, M. PD-L1 Expression in Pituitary Neuroendocrine Tumors/Pituitary Adenomas. Cancers 2023, 15, 4471. https://doi.org/10.3390/cancers15184471. [CrossRef]
- Suteau V, Collin A, Menei P, Rodien P, Rousselet MC, Briet C. Expression of programmed death-ligand 1 (PD-L1) in human pituitary neuroendocrine tumor. Cancer Immunol Immunother. 2020 Oct;69(10):2053-2061. doi: 10.1007/s00262-020-02611-x. Epub 2020 May 22. [CrossRef]
- Luo M, Tang R, Wang H. Tumor immune microenvironment in pituitary neuroendocrine tumors (PITNETs): increased M2 macrophage infiltration and PD-L1 expression in PIT1-lineage subset. J Neurooncol. 2023 Jul;163(3):663-674. doi: 10.1007/s11060-023-04382-8. [CrossRef]
- Guo X, Yang Y, Qian Z, Chang M, Zhao Y, Ma W, Wang Y, Xing B. Immune landscape and progress in immunotherapy for pituitary neuroendocrine tumors. Cancer Lett. 2024 Jun 28;592:216908. doi: 10.1016/j.canlet.2024.216908. [CrossRef]
- Shi M, Song Y, Zhang Y, Li L, Yu J, Hou A and Han S (2023) PD-L1 and tumor-infiltrating CD8+ lymphocytes are correlated with clinical characteristics in pediatric and adolescent pituitary adenomas. Front. Endocrinol. 14:1151714. doi: 10.3389/fendo.2023.1151714. [CrossRef]
- Guo X, Yang Y, Qian Z, Chang M, Zhao Y, Ma W, Wang Y, Xing B. Immune landscape and progress in immunotherapy for pituitary neuroendocrine tumors. Cancer Lett. 2024 Jun 28;592:216908. doi: 10.1016/j.canlet.2024.216908. [CrossRef]
- Xu Y, Song G, Xie S, Jiang W, Chen X, Chu M, Hu X, Wang ZW. The roles of PD-1/PD-L1 in the prognosis and immunotherapy of prostate cancer. Mol Ther. 2021 Jun 2;29(6):1958-1969. doi: 10.1016/j.ymthe.2021.04.029. [CrossRef]
- Mehdizadeh S, Bayatipoor H, Pashangzadeh S, Jafarpour R, Shojaei Z, Motallebnezhad M. Immune checkpoints and cancer development: Therapeutic implications and future directions. Pathol Res Pract. 2021 Jul;223:153485. doi: 10.1016/j.prp.2021.153485. [CrossRef]
- Raverot G, Ilie MD. Immunotherapy in pituitary carcinomas and aggressive pituitary tumors. Best Pract Res Clin Endocrinol Metab. 2022 Dec;36(6):101712. doi: 10.1016/j.beem.2022.101712. [CrossRef]
- Ilie MD, Vasiljevic A, Jouanneau E, Raverot G. Immunotherapy in aggressive pituitary tumors and carcinomas: a systematic review. Endocr Relat Cancer. 2022 May 27;29(7):415-426. doi: 10.1530/ERC-22-0037. [CrossRef]
- Lopes-Pinto M, Lacerda-Nobre E, Silva AL, Marques P. Therapeutical Usefulness of PD-1/PD-L1 Inhibitors in Aggressive or Metastatic Pituitary Tumours. Cancers (Basel). 2024 Aug 30;16(17):3033. doi: 10.3390/cancers16173033. [CrossRef]
- Gruppetta M, Formosa R, Falzon S, Ariff Scicluna S, Falzon E, Degeatano J, Vassallo J. Expression of cell cycle regulators and biomarkers of proliferation and regrowth in human pituitary adenomas. Pituitary. 2017 Jun;20(3):358-371. doi: 10.1007/s11102-017-0803-0. [CrossRef]
- Gejman R, Swearingen B, Hedley-Whyte ET (2008) Role of Ki-67 proliferation index and p53 expression in predicting progression of pituitary adenomas. Hum Pathol 39:758–766. https ://doi. org/10.1016/j.humpath.2007.10.004. [CrossRef]
- Salehi F, Agur A, Scheithauer BW, Kovacs K, Lloyd RV, Cusimano M. Ki-67 in pituitary neoplasms: a review--part I. Neurosurgery. 2009 Sep;65(3):429-37; discussion 437. doi: 10.1227/01.NEU.0000349930.66434.82. [CrossRef]
- Del Basso De Caro M, Solari D, Pagliuca F, Villa A, Guadagno E, Cavallo LM, Colao A, Pettinato G, Cappabianca P. Atypical pituitary adenomas: clinical characteristics and role of ki-67 and p53 in prognostic and therapeutic evaluation. A series of 50 patients. Neurosurg Rev. 2017 Jan;40(1):105-114. doi: 10.1007/s10143-016-0740-9. [CrossRef]
- Honegger J, Prettin C, Feuerhake F, Petrick M, Schulte-Mönting J, Reincke M. Expression of Ki-67 antigen in nonfunctioning pituitary adenomas: correlation with growth velocity and invasiveness. J Neurosurg. 2003 Oct;99(4):674-9. doi: 10.3171/jns.2003.99.4.0674. [CrossRef]
- Hasanov R, Aydoğan Bİ, Kiremitçi S, Erden E, Güllü S. The Prognostic Roles of the Ki-67 Proliferation Index, P53 Expression, Mitotic Index, and Radiological Tumor Invasion in Pituitary Adenomas. Endocr Pathol. 2019 Mar;30(1):49-55. doi: 10.1007/s12022-018-9563-2. [CrossRef]
- Petry C, Poli JHZ, de Azevedo Dossin I, Rech CGSL, Pereira Lima JFS, Ferreira NP, da Costa Oliveira M. Evaluation of the potential of the Ki67 index to predict tumor evolution in patients with pituitary adenoma. Int J Clin Exp Pathol. 2019 Jan 1;12(1):320-326.
- Matoušek P, Buzrla P, Reguli Š, Krajča J, Dvořáčková J, Lipina R. Factors That Predict the Growth of Residual Nonfunctional Pituitary Adenomas: Correlations between Relapse and Cell Cycle Markers. Biomed Res Int. 2018 Jul 10;2018:1876290. doi: 10.1155/2018/1876290. PMID: 30112364; PMCID: PMC6077672. [CrossRef]
- Mete O, Cintosun A, Pressman I, Asa SL. Epidemiology and biomarker profile of pituitary adenohypophysial tumors. Mod Pathol. 2018 Jun;31(6):900-909. doi: 10.1038/s41379-018-0016-8. [CrossRef]
- Rak B, Maksymowicz M, Pękul M, Zieliński G. Clinical, Biological, Radiological Pathological and Immediate Post-Operative Remission of Sparsely and Densely Granulated Corticotroph Pituitary Tumors: A Retrospective Study of a Cohort of 277 Patients With Cushing’s Disease. Front Endocrinol (Lausanne). 2021 May 31;12:672178. doi: 10.3389/fendo.2021.672178. [CrossRef]
- George DH, Scheithauer BW, Kovacs K, Horvath E, Young WF Jr, Lloyd RV, Meyer FB. Crooke’s cell adenoma of the pituitary: an aggressive variant of corticotroph adenoma. Am J Surg Pathol. 2003 Oct;27(10):1330-6. doi: 10.1097/00000478-200310000-00005. [CrossRef]
- Osamura RY, Inomoto C, Tahara S, Oyama KI, Matsuno A, Teramoto A. Pathology of Crooke Cells in the Human Pituitaries: A Timely Review. Appl Immunohistochem Mol Morphol. 2023 Aug 1;31(7):485-489. doi: 10.1097/PAI.0000000000001070. Epub 2022 Oct 17. [CrossRef]
- Raymond P, Raverot G, Ilie MD. Outcome and prognostic factors for pituitary carcinomas: lessons from a systematic review. Endocr Relat Cancer. 2023 Mar 31;30(5):e220338. doi: 10.1530/ERC-22-0338. [CrossRef]
- Jordan S, Lidhar K, Korbonits M, Lowe DG, Grossman AB. Cyclin D1 and cyclin E expression in normal and adenomatous pituitary. Eur J Endocrinol. 2000 Jul;143(1):R1-6. doi: 10.1530/eje.0.143r001. PMID: 10870044. [CrossRef]
- Tani Y, Inoshita N, Sugiyama T, Kato M, Yamada S, Shichiri M, Hirata Y. Upregulation of CDKN2A and suppression of cyclin D11 gene expressions in ACTH-secreting pituitary adenomas. Eur J Endocrinol. 2010 Oct;163(4):523-9. doi: 10.1530/EJE-10-0245. Epub 2010 Jul 8. PMID: 20616110. [CrossRef]
- Turner HE, Nagy Z, Sullivan N, Esiri MM, Wass JA. Expression analysis of cyclins in pituitary adenomas and the normal pituitary gland. Clin Endocrinol (Oxf). 2000 Sep;53(3):337-44. doi: 10.1046/j.1365-2265.2000.01088.x. PMID: 10971451. [CrossRef]
- Fedele M, Fusco A. Role of the high mobility group A proteins in the regulation of pituitary cell cycle. J Mol Endocrinol. 2010 Jun;44(6):309-18. doi: 10.1677/JME-09-0178. Epub 2010 Mar 10. PMID: 20219853. [CrossRef]
- Stefanidis P, Kyriakopoulos G, Seretis AM, Korfias S, Theocharis S, Angelousi A. Prognostic Factors for Invasiveness and Recurrence of Pituitary Adenomas: A Series of 94 Patients. Diagnostics (Basel). 2022 Oct 5;12(10):2413. doi: 10.3390/diagnostics12102413. PMID: 36292101; PMCID: PMC9600140. [CrossRef]
- Hibberts NA, Simpson DJ, Bicknell JE, Broome JC, Hoban PR, Clayton RN, Farrell WE. Analysis of cyclin D11 (CCND1) allelic imbalance and overexpression in sporadic human pituitary tumors. Clin Cancer Res. 1999 Aug;5(8):2133-9. PMID: 10473097.
- Hewedi IH, Osman WM, El Mahdy MM. Differential expression of cyclin D11 in human pituitary tumors: relation to MIB-1 and p27/Kip1 labeling indices. J Egypt Natl Canc Inst. 2011 Dec;23(4):171-9. doi: 10.1016/j.jnci.2011.11.003. Epub 2011 Dec 29. PMID: 22776845. [CrossRef]
- Tóth M. Agresszív hypophysisadenoma és hypophysiscarcinoma [Aggressive pituitary adenoma and pituitary carcinoma]. Orv Hetil. 2023 Jul 30;164(30):1167-1175. Hungarian. doi: 10.1556/650.2023.32832. [CrossRef]
- Yagnik G, Jahangiri A, Chen R, Wagner JR, Aghi MK. Role of a p53 polymorphism in the development of nonfunctional pituitary adenomas. Mol Cell Endocrinol. 2017 May 5;446:81-90. doi: 10.1016/j.mce.2017.02.017. Epub 2017 Feb 16. PMID: 28214592; PMCID: PMC5553295. [CrossRef]
- Thapar K, Scheithauer BW, Kovacs K, Pernicone PJ, Laws ER Jr. p53 expression in pituitary adenomas and carcinomas: correlation with invasiveness and tumor growth fractions. Neurosurgery. 1996 Apr;38(4):765-70; discussion 770-1. PMID: 8692397.
- Saeger W, Mawrin C, Meinhardt M, Wefers AK, Jacobsen F. Two Pituitary Neuroendocrine Tumors (PITNETs) with Very High Proliferation and TP53 Mutation - High-Grade PITNET or PitNEC? Endocr Pathol. 2022 Jun;33(2):257-262. doi: 10.1007/s12022-021-09693-y. Epub 2021 Oct 20. Erratum in: Endocr Pathol. 2022 Jun;33(2):263. doi: 10.1007/s12022-021-09699-6. PMID: 34669159; PMCID: PMC9135791. [CrossRef]
- Liu, X., 2012. Classification accuracy and cut point selection. Stat. Med. 31 (23), 2676–2686. https://doi.org/10.1002/sim.4509. [CrossRef]
- Vela-Patiño S, Salazar MI, Taniguchi-Ponciano K, Vadillo E, Gomez-Apo E, Escobar-España A, Perez-Koldenkova V, Bonifaz L, Aguilar-Flores C, Marrero-Rodríguez D, Mercado M. The Immune Microenvironment Landscape of Pituitary NeuroEndocrine Tumors, a Transcriptomic Approach. Genes (Basel). 2024 Apr 24;15(5):531. doi: 10.3390/genes15050531. PMID: 38790160; PMCID: PMC11120841. [CrossRef]
- Yi M, Niu M, Xu L, Luo S, Wu K. Regulation of PD-L1 expression in the tumor microenvironment. J Hematol Oncol. 2021 Jan 7;14(1):10. doi: 10.1186/s13045-020-01027-5. PMID: 33413496; PMCID: PMC7792099. [CrossRef]
- Uraki S, Ariyasu H, Doi A, Takeshima K, Morita S, Inaba H, Furuta H, Fukuhara N, Inoshita N, Nishioka H, Nakao N, Yamada S, Akamizu T. MSH6/2 and PD-L1 Expressions Are Associated with Tumor Growth and Invasiveness in Silent Pituitary Adenoma Subtypes. Int J Mol Sci. 2020 Apr 18;21(8):2831. doi: 10.3390/ijms21082831. PMID: 32325698; PMCID: PMC7215962. [CrossRef]
- Xi Z, Jones PS, Mikamoto M, Jiang X, Faje AT, Nie C, Labelle KE, Zhou Y, Miller KK, Soberman RJ, Zhang X. The Upregulation of Molecules Related to Tumor Immune Escape in Human Pituitary Adenomas. Front Endocrinol (Lausanne). 2021 Oct 21;12:726448. doi: 10.3389/fendo.2021.726448. PMID: 34745002; PMCID: PMC8566912. [CrossRef]
- Mei Y, Bi WL, Agolia J, Hu C, Giantini Larsen AM, Meredith DM, Al Abdulmohsen S, Bale T, Dunn GP, Abedalthagafi M, Dunn IF. Immune profiling of pituitary tumors reveals variations in immune infiltration and checkpoint molecule expression. Pituitary. 2021 Jun;24(3):359-373. doi: 10.1007/s11102-020-01114-3. Epub 2021 Jan 25. PMID: 33492612. [CrossRef]
- Wang X, Li M, Jiang X, Wang F, Ling S, Niu C. Prediction of Higher Ki-67 Index in Pituitary Adenomas by Pre- and Intra-Operative Clinical Characteristics. Brain Sci. 2022 Jul 28;12(8):1002. doi: 10.3390/brainsci12081002. PMID: 36009065; PMCID: PMC9405805. [CrossRef]
- Grimm F, Maurus R, Beschorner R, Naros G, Stanojevic M, Gugel I, Giese S, Bier G, Bender B, Honegger J. Ki-67 labeling index and expression of p53 are non-predictive for invasiveness and tumor size in functional and nonfunctional pituitary adenomas. Acta Neurochir (Wien). 2019 Jun;161(6):1149-1156. doi: 10.1007/s00701-019-03879-4. Epub 2019 Apr 30. PMID: 31037500. [CrossRef]
- Hibberts, N.A.; Simpson, D.J.; Bicknell, J.E.; Broome, J.C.; Hoban, P.R.; Clayton, R.N.; Farrell, W.E. Analysis of cyclin D11 (CCND1) allelic imbalance and overexpression in sporadic human pituitary tumors. Clin. Cancer Res. 1999, 5, 2133–2139. [PubMed].
- Flores L, Sleightholm R, Neilsen B, Baine M, Drincic A, Thorell W, Shonka N, Oupicky D, Zhang C. Highly Aggressive and Radiation-Resistant, “Atypical” and Silent Pituitary Corticotrophic Carcinoma: A Case Report and Review of the Literature. Case Rep Oncol. 2019 Feb 8;12(1):139-146. doi: 10.1159/000496019. PMID: 31043952; PMCID: PMC6477470. [CrossRef]
- Lasolle H, Vasiljevic A, Jouanneau E, Ilie MD, Raverot G. Aggressive corticotroph tumors and carcinomas. J Neuroendocrinol. 2022 Aug;34(8):e13169. doi: 10.1111/jne.13169. Epub 2022 Aug 18. PMID: 35979732; PMCID: PMC9542524. [CrossRef]
- Kovacs K, Diep CC, Horvath E, Cusimano M, Smyth H, Lombardero CC, Scheithauer BW, Lloyd RV. Prognostic indicators in an aggressive pituitary Crooke’s cell adenoma. Can J Neurol Sci. 2005 Nov;32(4):540-5. doi: 10.1017/s0317167100004583. PMID: 16408589. [CrossRef]
- Ilie MD, Raverot G. Treatment Options for Gonadotroph Tumors: Current State and Perspectives. J Clin Endocrinol Metab. 2020 Oct 1;105(10):dgaa497. doi: 10.1210/clinem/dgaa497. PMID: 32735647. [CrossRef]
- Gandhi C, Koumna S, Chik C. Treatment of an Aggressive Gonadotroph Pituitary Neuroendocrine Tumor With 177Lutetium DOTATATE Radionuclide Therapy. JCEM Case Rep. 2024 Jul 15;2(7):luae123. doi: 10.1210/jcemcr/luae123. PMID: 39011402; PMCID: PMC11247163. [CrossRef]
| Characteristic | N (%) |
|---|---|
| Age (years) | |
| Mean (SD); Me (Q1-Q3) | 57.4 (14.0); 60.5 (47.0–69.0) |
| Gender | |
| F/M | 29 (39.2%)/45 (60.8%) |
| Tumor size | |
| Microadenoma | 1 (1.4%) |
| Macroadenoma | 72 (97.3%) |
| Missing | 1 (1.4%) |
| Gigant adenoma (>40mm) | |
| No | 62 (83.8%) |
| Yes | 11 (14.9%) |
| Missing | 1 (1.4%) |
| Tumor volume (cm³) | |
| Mean (SD); Me (Q1-Q3) | 8.6 (8.9); 5.0 (3.1–10.2) |
| Max size of tumor (mm) | |
| Mean (SD); Me (Q1-Q3) | 28.6 (10.2); 25.0 (21.5–34.0) |
| Transcriptions factors | |
| PIT-1 | 9 (12.2%) |
| SF1 | 43 (58.1%) |
| TPIT | 8 (10.8%) |
| PitNETs with no distinct cel lineage | 3 (4.1%) |
| Multiple PitNETs | 11 (14.9%) |
| Hormonal activity | |
| Non - active | 59 (79.7%) |
| Active | 15 (20.3%) |
| Type of PitNETs | |
| Gonadotroph | 42 (56.8%) |
| Gonadotroph/lactotroph | 2 (2.7%) |
| Corticotroph | 8 (10.8%) |
| Lactotroph | 4 (5.4%) |
| Somatotroph | 1 (1.4%) |
| Tyrotroph | 1 (1.4%) |
| Null cell adenoma | 3 (4.1%) |
| Multiple synchronous PITNET | 4 (5.4%) |
| Immature PIT-lineage tumor | 6 (8.1%) |
| Mature PIT-1 lineage tumor | 3 (4.1%) |
| PDL-1 (TPS) | |
| 0% | 31 (41.9%) |
| ≥1% | 35 (47.3%) |
| Missing | 8 (10.8%) |
| Proliferative factors | |
| P53 | |
| <10% | 34 (45.9%) |
| ≥10% | 33 (44.6%) |
| Missing | 7 (9.5%) |
| Ki-67 | |
| <3% | 65 (87.8%) |
| ≥3% | 9 (12.2%) |
| Cyclin D1 | |
| <10% | 41 (55.4%) |
| ≥10% | 29 (39.2%) |
| Missing | 4 (5.4%) |
| Knosp scale | |
| Invasive | 38 (51.4%) |
| Non-invasive | 34 (45.9%) |
| Missing | 2 (2.7%) |
| Hardy scale | |
| Invasive | 57 (77.0%) |
| Non-invasive | 15 (20.3%) |
| Missing | 2 (2.7%) |
| Overall invasiveness | |
| No | 14 (18.9%) |
| Yes | 58 (78.4%) |
| Missing | 2 (2.7%) |
| Feature | PD-L1 | Ki-67 | P 53 | Cyclin D1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <1% (n=31) N (%) |
≥1% (n=35) N (%) |
p- value |
<3% (n=65) N (%) |
≥3 (n=9) N (%) |
p- value |
<10% (n=34) N (%) |
≥10% (n=33) N (%) |
p-value | <10% (n=41) N (%) |
≥10% (n=29) N (%) |
p-value | |
| Age (years) | 0. 59ᵠ | 0.03ᵠ | 0.57ᵠ | 0.38ᵠ | ||||||||
| Me (Q1-Q3) | 59.0 (43.0-70.0) |
60.0 (52.0–68.0) |
62.0 (50.0–69.0) |
41.0 (36.0–57.0) |
62.0 (53.0-70.0) |
61.0 (45.0-67.0) |
57.0 (49.0-68.0) |
62.0 (47.0-68.0) | ||||
| Gender | 0.91ᵡ | 0.08ᵟ | 0.73ᵡ | 0.77ᵡ | ||||||||
| Female | 12 (46.2) | 14 (53.8) | 28 (96.5) | 1 (3.45) | 13 (48.1) | 14 (51.8) | 17 (60.71) | 11 (39.29) | ||||
| Male | 19 (47.5) | 21 (52.5) | 37 (82.2) | 8 (17.78) | 21 (52.5) | 19 (47.5) | 24 (57.14) | 18 (42.86) | ||||
|
Tumor volume [cm3] |
0.55ᵠ | 0.96ᵠ | 0.41ᵠ | 0.31ᵠ | ||||||||
| Me (Q1-Q3) | 4.6 (2.1-10.0) |
6.2 (3.2–10.0) |
4.8 (3.1–10.2) |
7.3 (2.8–11.0) |
5.2 (2.8-10.0) |
7.3 (3.8-11.5) |
4.3 (2.8–9.0) |
8.1 (3.1–12.3) |
||||
| Max size (mm) | 0.59ᵠ | 0.94ᵠ | 0.59ᵠ | 0.86ᵠ | ||||||||
| Me (Q1-Q3) | 23.0 (22.5-34.0) |
30.0 (22.0–34.0) |
25.0 (22.0–33.0) |
29.0 (20.0–36.0) |
28.5 (22.5-34.0) |
25.5 (20.8-33.0) |
25.0 (21.0-34.0) |
26.8 (22.3-33.0) |
||||
|
Gigant adenoma (>40mm) |
1.00ᵡ | 0.62ᵟ | 0.92ᵡ | 0.76ᵡ | ||||||||
| No | 27 (47.4) | 30 (52.6) | 55 (52.1) | 7 (47.9) | 29 (51.0) | 27 (49.0) | 34 (58.6) | 24 (41.4) | ||||
| Yes | 4 (50.0) | 4 (50.0) | 9 (38.3) | 2 (61.7) | 5 (49.2) | 5 (50.8) | 7 (63.6) | 4 (36.4) | ||||
| Missing | 0 (0.0) | 1 (100.0) | 1 (100.0) | 0 (0.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1(100) | ||||
| PIT-1 overexpression | <0.05ᵟ | 1.00ᵟ | 1.00ᵟ | 0.73ᵟ | ||||||||
| No | 25 (42.4) | 34 (57.6) | 57 (87.7) | 8 (12.3) | 30 (50.0) | 30 (50.0) | 35 (57.38) | 26 (42.62) | ||||
| Yes | 6 (85.7) | 1 (14.3) | 8 (88.9) | 1 (11.1) | 4 (57.1) | 3 (42.9) | 6 (66.67) | 3 (33.33) | ||||
| SF1 overexpression | 0.34ᵡ | 0.48ᵟ | 0.36ᵡ | 0.09ᵡ | ||||||||
| No | 16 (53.3) | 14 (46.7) | 26 (83.87) | 5 (16.13) | 15 (57.7) | 11 (42.3) | 21 (67.74) | 9 (29.03) | ||||
| Yes | 15 (41.7) | 21 (58.3) | 39 (90.70) | 4 (9.30) | 19 (46.3) | 22 (53.7) | 20 (50.00) | 20 (50.00) | ||||
| TPIT oveexpression | 0.71ᵟ | 0.25ᵟ | 1.00ᵟ | 0.54ᵡ | ||||||||
| No | 28 (48.3) | 30 (51.7) | 59 (89.39) | 7 (10.61) | 30 (50.8) | 29 (49.2) | 35 (56.45) | 27 (43.55) | ||||
| Yes | 3 (37.5) | 5 (62.5) | 6 (75.00) | 2 (25.00) | 4 (50.0) | 4 (50.0) | 6 (75.00) | 2 (25.00) | ||||
| PitNETs with no distinct cell lineage | 0.24ᵟ | 1.00ᵟ | 0.49ᵟ | 0.51ᵟ | ||||||||
| No | 30 (49.23) | 31 (50.8) | 62 (87.35) | 9 (12.65) | 32 (49.2) | 33 (50.8) | 39 (57.35) | 29 (42.65) | ||||
| Yes | 1 (20.0) | 4 (80.0) | 3 (100.00) | 0 (0.00) | 2 (100.0) | 0 (0.0) | 2 (100.00) | 0 (0.00) | ||||
| Multiple PitNETs | 0.72ᵟ | 0.62ᵟ | 0.24ᵟ | 1.00ᵟ | ||||||||
| No | 25 (44.6) | 31 (55.4) | 56 (86.15) | 7 (13.85) | 29 (50.0) | 29 (50.0) | 34 (57.6) | 25 (42.4) | ||||
| Yes | 6 (60.0) | 4 (40.0) | 9 (81.82) | 2 (18.18) | 5 (55.6) | 4 (44.4) | 7 (63.6) | 4 (36.4) | ||||
| Type of PitNETs | - | - | - | - | ||||||||
| Gonadotroph | 17 (44.7) | 21 (55.3) | 39 (92.86) | 3 (7.14) | 19 (47.5) | 21 (52.5) | 20 (51.3) | 19 (48.7) | ||||
| Gonadotroph/Lactotroph | 0 (0.0) | 2 (100.0) | 2 (100.00) | 0 (0.00) | 1 (100.0) | 0 (0.0) | 2 (100.0) | 0 (0.0) | ||||
| Corticotroph | 3 (37.5) | 5 (62.5) | 6 (75.00) | 2 (25.00) | 4 (50.0) | 4 (50.0) | 6 (75.0) | 2 (25.0) | ||||
| Lactotroph | 3 (100.0) | 0 (0.0) | 3 (75.00) | 1 (25.00) | 2 (50.0) | 2 (50.0) | 3 (75.0) | 1 (25.0) | ||||
| Somatroph | 1 (100.0) | 0 (0.0) | 1 (100.00) | 0 (0.00) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (100.0) | ||||
| Tyrotroph | 1 (100.0) | 0 (0.0) | 1 (100.00) | 0 (0.00) | 1 (100.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | ||||
| Null cell adenoma | 0 (0.0) | 3 (100.0) | 3 (100.00) | 0 (0.00) | 2 (100.0) | 0 (0.0) | 2 (100.0) | 0 (0.0) | ||||
| Multiple synchronous PitNET | 1 (33.3) | 2 (66.7) | 3 (75.00) | 1 (25.00) | 2 (50.0) | 2 (50.0) | 2 (50.0) | 2 (50.0) | ||||
| Immature PIT-lineage tumour | 4 (80.0) | 1 (20.0) | 5 (83.33) | 1 (16.67) | 2 (40.00) |
3 (60.0) |
4 (66.7 | 2 (33.3) | ||||
| Mature PIT-lineage tumor | 1 (50.0) | 1 (50.0) | 2 (66.67) | 1 (33.33) | 1 (50. 0) | 1 (50.0) | 1 (33.3) | 2 (66.7) | ||||
| Hormonal activity, | 0.24ᵡ | 0.38ᵟ | 0.14ᵡ | 0.19ᵡ | ||||||||
| Non-active | 23 (43.4) | 30 (56.6) | 53 (89.8) | 6 (10.2) | 25 (46.3) | 29 (53.7) | 30 (54.5) | 25 (45.5) | ||||
| Active | 8 (61.5) | 5 (38.5) | 12 (80.00) | 3 (20.0) | 9 (69.2) | 4 (30.8) | 11 (73.2) | 4 (26.67) | ||||
| Knosp scale | 0.31ᵡ | 0.73ᵟ | 0.88ᵡ | 0.91ᵡ | ||||||||
| Invasive | 18 (51.4) | 15 (45.5) | 34 (89.5) | 4 (10.5) | 18 (51.4) | 17 (48.6) | 22 (59.46) | 15 (40.54) | ||||
| Non-invasive | 16 (53.3) | 18 (58.1) | 29 (85.3) | 5 (14.7) | 16 (53.3) | 14 (46.7) | 18 (58.1) | 13 (41.9) | ||||
| Missing | 0 (0.00) |
2 (100.0) | 2 (100.0) | 0 (0.0) | 0 (0.0) (0.00) |
2 (100.0) | 1 (50.0) | 1 (50.0) | ||||
| Hardy scale, n (%) | 0.29ᵡ | 0.38ᵟ | 0.62ᵡ | 0.92ᵡ | ||||||||
| Invasive | 23 (45.1) | 28 (54.9) | 51 (89.5) | 6 (10.5) | 28 (53.8) | 24 (46.2) | 31 (58.5) | 22 (41.5) | ||||
| Non-invasive | 8 (61.5) | 5 (38.5) | 12 (80.0) | 3 (20.0) | 6 (46.2) | 7 (53.8) | 9 (60.0) | 6 (40.0) | ||||
| Missing | 0 (0.0) | 2 (100.0) | 2 (100.0) | 0 (0.0) | 0 (0.0) | 2 (100.0) | 1 (50.0) | 1 (50.0) | ||||
| Overall invasiveness | 0.16ᵡ | 0.36ᵟ | 0.86ᵡ | 0.64ᵡ | ||||||||
| Non-invasive in Knosp or Hardy scale | 6 (50.0) | 4 (33.3) | 11 (78.6) | 3 (21.4) | 6 (50.0) | 6 (50.0) | 9 (64.3) | 5 (35.7) | ||||
| Invasive in Knosp or Hardy scale | 28 (52.8) | 29 (55.8) | 52 (89.7) | 6 (10.3) | 28 (52.8) | 25 (47.2) | 31 (57.4) | 23 (42.6) | ||||
| Missing | 0 (0.0) | 2 (100.0) | 2 (100.0) | 0 (0.0) | 0 (0.0) | 2 (100.0) | 1 (50.0) | 1 (50.0) | ||||
| Parameters | PD-L1 | p-value | |
| TPS 0% | TPS ≥1% | ||
| Ki-67 overexpression, n (%)ᵟ | 0.71 | ||
| <3% | 28 (48.3) | 30 (51.7) | |
| ≥3% | 3 (37.5) | 5 (62.5) | |
| Missing | |||
| P53 overexpression, n (%)ᵡ | 0.10 | ||
| <10% | 19 (57.6) | 14 (42.4) | |
| ≥10% | 11 (36.7) | 19 (63.3) | |
| Missing | 1 (33.3) | 2 (66.7) | |
| Cyclin D1 overexpression, n (%)ᵡ | 0.27 | ||
| <10% | 14 (41.2) | 20 (58.8) | |
| ≥10% | 16 (55.2) | 13 (44.8) | |
| Missing | 1 (33.3) | 2 (66.7) | |
| Outcome | Optimal TPS cut-point | Sensitivity | Specificity | AUC |
| Transcriptions factors | ||||
| PIT-1 | 0.00 | 0.86 | 0.58 | 0.68 |
| SF 1 | ≥ 1.3% | 0.56 | 0.52 | 0.57 |
| TPIT | ≥ 5% | 0.63 | 0.59 | 0.50 |
| Null cell adenoma | ≥ 5% | 1.00 | 0.59 | 0.75 |
| Multi PITNETs | ≤ 3% | 0.78 | 0.49 | 0.59 |
| Type of PitNETs | ||||
| Gonadotroph | ≥ 1.3% | 0.55 | 0.50 | 0.54 |
| Corticotroph | ≥ 5% | 0.63 | 0.59 | 0.50 |
| Lactotroph | 0.00 | 1.00 | 0.56 | 0.78 |
| Immature PIT1 lineage tumor | 0.00 | 0.80 | 0.56 | 0.67 |
| Mature PIT-1 lineage tumor | ≥ 85% | 0.50 | 0.98 | 0.61 |
| Predictor | OR (95% CI) Crude | P - value | OR (95% CI) Adjusted | P - value |
| PDL 1 | ||||
| ≥1% | 2.52 (0.67 – 9.43) | 0.17 | 2.35 (0.56 – 9.90) | 0.25 |
| 0% (ref) | 1.00 | – | 1.00 | – |
| P53 | ||||
| ≥10 | 0.89 (0.22 – 3.13) | 0.86 | 0.74 (0.16 – 3.39) | 0.70 |
| <10 (ref) | 1.00 | – | 1.00 | – |
| Ki-67 | ||||
| ≥3 | 0.42 (0.09 – 1.96) | 0.27 | 0.76 (0.11 – 5.16) | 0.78 |
| <3 (ref) | 1.00 | – | 1.00 | – |
| Cyclin D1 | ||||
| ≥10 | 1.34 (0.39 – 4.52) | 0.64 | 1.93 (0.42 – 8.84) | 0.40 |
| <10 (ref) | 1.00 | – | 1.00 | – |
| Type of PitNETs | Ki>=3&P53>=10 (1) |
Ki>=3&P53<10 (2) |
Ki<3&P53>=10 (3) |
Ki<3&P53<10 (4) |
| N=6 | N=2 | N=27 | N=32 | |
| Gonadotroph | 3 (7.7%) | 0 (0.0%) | 18 (46.2%) | 18 (46.2%) |
| Gonadotroph/Lactotroph | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) |
| Corticotroph | 2 (25.0%) | 0 (0.0%) | 2 (25.0%) | 4 (50.0%) |
| Lactotroph | 0 (0.0%) | 1 (25.0%) | 2 (50.0%) | 1 (25.0%) |
| Somatotroph | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Tyrotroph | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) |
| Null cell adenoma | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 2 (100.0%) |
| Multiple synchronous PitNET | 1 (25.0%) | 0 (0.0%) | 1 (25.0%) | 2 (50.0%) |
| Immature PIT-1 lineage tumour | 0 (0.0%) | 1 (25.0%) | 3 (75.0%) | 0 (0.0%) |
| Mature PIT-1 lineage tumor | 0 (0.0%) | 0 (0.0%) | 1 (50.0%) | 1 (50.0%) |
| Type of PitNETs | Cyk>=10&P53>=10 (1) |
Cyk>=10&P53<10 (2) |
Cyk<10&P53>=10 (3) |
Cyk<10&P53<10 (4) |
| N=18 | N=10 | N=14 | N=22 | |
| Gonadotroph | 10 (26.3%) | 9 (23.7%) | 10 (26.3%) | 9 (23.7%) |
| Gonadotroph/Lactotroph | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) |
| Corticotroph | 2 (25.0%) | 0 (0.0%) | 2 (25.0%) | 4 (50.0%) |
| Lactotroph | 1 (25.0%) | 0 (0.0%) | 1 (25.0%) | 2 (50.0%) |
| Somatroph | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Tyreotroph | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) |
| Null cell adenoma | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (100.0%) |
| Multiple synchronous PitNET | 2 (50.0%) | 0 (0.0%) | 0 (0.0%) | 2 (50.0%) |
| Immature PIT-lineage tumor | 2 (40.0%) | 0 (0.0%) | 1 (20.0%) | 2 (40.0%) |
| Mature PIT-1 lineage tumor | 1 (50.0%) | 1 (50.0%) | 0 (0.0%) | 0 (0.0%) |
| Features | Knosp scale | Hardy scale | ||
| Non-invasive n (%) |
Invasive n (%) | Non-invasive n (%) |
Invasive n (%) |
|
| Combined analysis of Ki-67 and P53 | ||||
| Ki-67≥3% & P53≥10% | 3 (50.0) | 3 (50.0) | 1 (16.7) | 5 (83.3) |
| Ki-67≥3% & P53<10% | 1 (50.0) | 1 (50.0) | 1 (50.0) | 1 (50.0) |
| Ki-67<3% & P53≥10% | 11 (44.0) | 14 (56.0) | 6 (24.0) | 19 (76.0) |
| Ki-67<3% & P53<10% | 15 (46.9) | 17 (53.1) | 5 (15.6) | 27 (84.4) |
| p valueᵡ | 0.992 | 0.615 | ||
| Combined analysis of Cyclin D1 and P53 | ||||
| Cyclin D1≥10% & P53≥10% | 8 (47.1) | 9 (52.9) | 5 (29.4) | 12 (70.6) |
| Cyclin D1≥10% & P53<10% | 4 (40.0) | 6 (60.0) | 0 (0.0) | 10 (100.0) |
| Cyclin D1<10% & P53≥10% | 5 (38.5) | 8 (61.5) | 2 (15.4) | 11 (84.6) |
| Cyclin D1<10% & P53<10% | 11 (50.0) | 11 (50.0) | 6 (27.3) | 16 (72.7) |
| p valueᵡ | 0.902 | 0.245 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





