Submitted:
08 July 2025
Posted:
09 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Mathematical Framework
2.1. Stochastic differential equations and the Fokker-Planck equation
2.2. Fisher equation
3. Glioblastoma Infiltration Modeling
- polar coordinate formulation: because of the assumption of symmetry and isotropy of cell movements (already assumed in Pompa et al. [28]) within the tumour mass we can reduce the spatial dependency to only the radial coordinate;
- model outputs reproducing the available data on two different cell lines: U87WT (already considered in [28]) and U87EGFR, which is a common mutation (data reported in Stein et al. [20], see the next section); the proposed models are specified to the different cell lines by means of a suitable parameter estimation.
3.1. Data Collection
- the invasive radius, , of a tumor that represents the maximum distance that a single tumor cell or a small group of tumor cells can migrate from the primary tumor mass into the surrounding tissue. It is a key parameter in cancer invasion studies, reflecting the ability of malignant cells to infiltrate the extracellular matrix (ECM), evade immune responses, and establish metastases. In [20] it is defined as the farthest distance from the center at which the azimuthally averaged gradient magnitude reaches half of its maximum value;
- the core radius, , refers to the central region of the tumor where cell proliferation is significantly reduced or halted due to limited nutrient and oxygen availability. This region is often characterized by hypoxia, necrosis, or quiescence, depending on the severity of the nutrient and waste diffusion limitations. In Stein et al. [20] the region is defined as the collection of pixels exhibiting an intensity level of on a grayscale image—where 0 corresponds to the darkest pixel and 1 to the brightest—centered around the tumor spheroid;
- the radial cell density at day 3 (expressed in []), function of the radius , that is denoted by with ; it is extracted based on the concentration of darker pixels observed in the digital photomicrograph data.
3.2. Radial Distribution Models of Invasive Cells
3.2.1. Pure Diffusion Model (PD)
3.2.2. Diffusion and Growth Model (DG)
3.2.3. Modulated Diffusion and Growth Model (MDG)
4. Parameter Identification
- i)
- representing the radial distribution of cell density on days , measured at different spatial points with a sample size ;
- ii)
- representing the core radius at different times , with a sample size ;
- iii)
- representing the invasive radius at different times , with a sample size .
5. Simulation Results
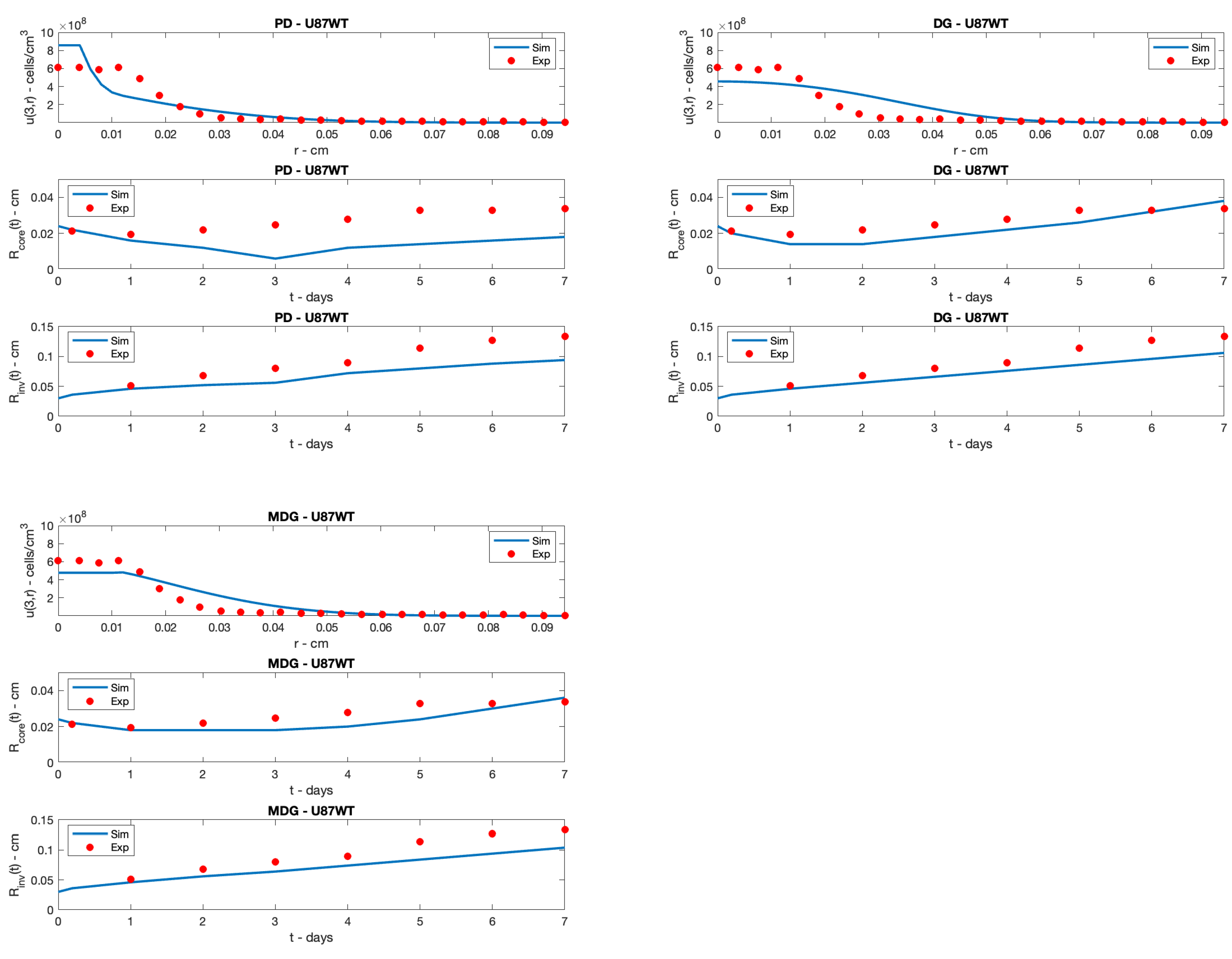
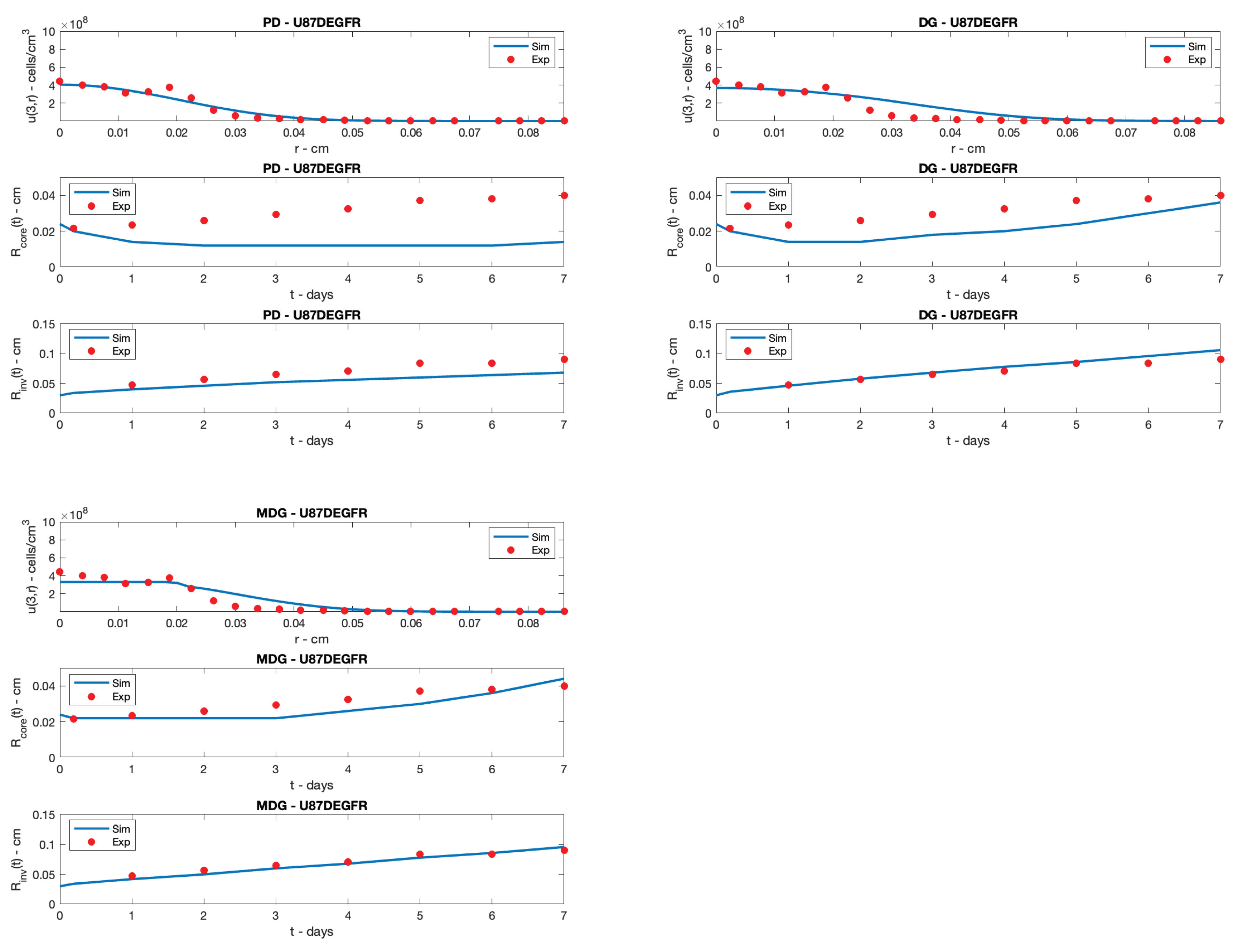
6. Conclusions
Funding
Conflicts of Interest
References
- Louis, D.; Perry, A.; Wesseling, P.; Brat, D.; Cree, I.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol 2021, 23, 1231–51. [Google Scholar] [CrossRef] [PubMed]
- Farina, H.; Muzaffar, K.; Perveen, K.; Malhi, S.; Simjee, S. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac J Cancer Prev 2017, 18, 3–9. [Google Scholar]
- Tan, A.; Ashley, D.; Lopez, G.; Malinzak, M.; Friedman, H.; Khasraw, M. Management of glioblastoma: state of the art and future directions. CA Cancer J Clin. 2020, 299–312. [Google Scholar] [CrossRef]
- Musielak, E.; Krajka-Kuzniak, V. Lipidic and Inorganic Nanoparticles for Targeted Glioblastoma Multiforme Therapy: Advances and Strategies. Micro 2025, 5. [Google Scholar] [CrossRef]
- Wirsching, H.G.; Weller, M. Glioblastoma. Malignant Brain Tumors: State-of-the-Art Treatment 2017, pp. 265–288.
- Rui, Z.; Hongmei, H.; Xiande, W. Novel neutrophil targeting platforms in treating Glioblastoma: Latest evidence and therapeutic approaches. International Immunopharmacology 2025, 150, 114173. [Google Scholar]
- Rončević, A.; Nenad, K.; Anamarija, S.K.; Robert, R. Why Do Glioblastoma Treatments Fail? Future Pharmacology 2025, 5. [Google Scholar] [CrossRef]
- Xiaolin, L.; Xiao, L.; Xiaonan, L.; Maorong, Z.; Nannan, L.; Juan, L.; Qi, Z.; Cheng, Z.; Yuxin, W.; Zhengcong, C.; et al. Synergistic strategies for glioblastoma treatment: CRISPR-based multigene editing combined with immune checkpoint blockade. Nanobiotechnol 2025, 23. [Google Scholar]
- Neyran, K.; Gozde, K.; Serkan, A.; Gokcen, C.; Ahmet, I.I.; Gozde, Y. Evaluating Immunotherapy Responses in Neuro-Oncology for Glioblastoma and Brain Metastases: A Brief Review Featuring Three Cases. Cancer Control 2025, 32. [Google Scholar]
- Jackson, G.A.; Adamson, D.C. Similarities in Mechanisms of Ovarian Cancer Metastasis and Brain Glioblastoma Multiforme Invasion Suggest Common Therapeutic Targets. Cells 2025. [Google Scholar] [CrossRef]
- Saketh R., K.; Dunja, G.; Daniele, T.; Lucy J., B.; Ting, G.; Andreas Christ Sølvsten, J.; Ciaran, S.H.; Melanie, C.; Tammy L., K.; Jack A., W.; et al. Detecting glioblastoma infiltration beyond conventional imaging tumour margins using MTE-NODDI. Imaging Neuroscience 2025. [Google Scholar]
- Correia, C.D.; Sofia M., C.; Alexandra, M.; Filipa, E.; Ana Luísa, D.S.C.; Marco A., C.; Mónica T., F. dvancing Glioblastoma Research with Innovative Brain Organoid-Based Models. Cells 2025, 14. [Google Scholar] [CrossRef]
- Odangowei Inetiminebi, O.; Timipa Richard, O.; Elekele Izibeya, A.; Racheal Bubaraye, E.; Marcella Tari, J.; Ebimobotei Mao, B. Chapter 1 - Radiogenomics and genetic diversity of glioblastoma characterization. In Radiomics and Radiogenomics in Neuro-Oncology; Saxena, S.; Suri, J.S., Eds.; Academic Press, 2025; pp. 3–34.
- Ray Zirui, Z.; Ivan, E.; Michal, B.; Andy, Z.; Benedikt, W.; Bjoern, M.; John S., L. Personalized predictions of Glioblastoma infiltration: Mathematical models, Physics-Informed Neural Networks and multimodal scans. Medical Image Analysis 2025, 101, 103423. [Google Scholar]
- Kim, Y.; Jeon, H.; Othmer, H. The Role of the Tumor Microenvironment in Glioblastoma: A Mathematical Model. IEEE Transactions on Biomedical Engineering 2017, 64, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Kar, N.; Özalp, N. A fractional mathematical model approach on glioblastoma growth: tumor visibility timing and patient survival. Mathematical Modelling and Numerical Simulation with Applications 2024, 4, 66–85. [Google Scholar] [CrossRef]
- Marek, B.; Monika J., P.; Mariusz, B.; Juan, B.B.; Urszula, F. Dual CAR-T cell therapy for glioblastoma: strategies to cure tumour diseases based on a mathematical model. 2025, 113, 1637–1666. [Google Scholar]
- Cacace, F.; Conte, F.; d’Angelo, M.; Germani, A.; Palombo, G. Filtering linear systems with large time-varying measurement delays. Automatica 2022, 136, 110084. [Google Scholar] [CrossRef]
- Hatzikirou, H.; Deutsch, A.; Schaller, C.; Simon, M.; Swanson, K. Mathematical modelling of glioblastoma tumour development: a review. Mathematical Models and Methods in Applied Sciences 2005, 15, 1779–1794. [Google Scholar] [CrossRef]
- Stein, A.M.; Demuth, T.; Mobley, D.; Berens, M.; Sander, L.M. A mathematical model of glioblastoma tumor spheroid invasion in a three-dimensional in vitro experiment. Biophysical journal 2007, 92, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.; Surulescu, C. Mathematical modeling of glioma invasion: acid-and vasculature mediated go-or-grow dichotomy and the influence of tissue anisotropy. Applied Mathematics and Computation 2021, 407, 126305. [Google Scholar] [CrossRef]
- Falco, J.; Agosti, A.; Vetrano, I.G.; Bizzi, A.; Restelli, F.; Broggi, M.; Schiariti, M.; DiMeco, F.; Ferroli, P.; Ciarletta, P.; et al. In silico mathematical modelling for glioblastoma: A critical review and a patient-specific case. Journal of clinical medicine 2021, 10, 2169. [Google Scholar] [CrossRef]
- Engwer, C.; Knappitsch, M.; Surulescu, C. A multiscale model for glioma spread including cell-tissue interactions and proliferation. Mathematical Biosciences & Engineering 2015, 13, 443–460. [Google Scholar]
- Kumar, P.; Li, J.; Surulescu, C. Multiscale modeling of glioma pseudopalisades: contributions from the tumor microenvironment. Journal of Mathematical Biology 2021, 82, 1–45. [Google Scholar] [CrossRef]
- Jørgensen, A.C.S.; Hill, C.S.; Sturrock, M.; Tang, W.; Karamched, S.R.; Gorup, D.; Lythgoe, M.F.; Parrinello, S.; Marguerat, S.; Shahrezaei, V. Data-driven spatio-temporal modelling of glioblastoma. Royal Society Open Science 2023, 10, 221444. [Google Scholar] [CrossRef]
- Borri, A.; d’Angelo, M.; Palumbo, P. Self-regulation in a stochastic model of chemical self-replication. International Journal of Robust and Nonlinear Control 2023, 33, 4908–4922. [Google Scholar] [CrossRef]
- Gandolfi, A.; Franciscis, S.; d’Onofrio, A.; Fasano, A.; Sinisgalli, C. Angiogenesis and vessel co-option in a mathematical model of diffusive tumor growth: The role of chemotaxis. Journal of Theoretical Biology 2021, 512, 110526. [Google Scholar] [CrossRef]
- Pompa, M.; Panunzi, S.; Borri, A.; De Gaetano, A. An Agent-Based Model of Glioblastoma Infiltration. In Proceedings of the 2023 IEEE 23rd International Symposium on Computational Intelligence and Informatics (CINTI). IEEE; 2023; pp. 383–390. [Google Scholar]
- Borri, A.; d’Angelo, M.; D’Orsi, L.; Pompa, M.; Panunzi, S.; De Gaetano, A. Stochastic modeling of glioblastoma spread: a numerical simulation study. In Proceedings of the 13th International Workshop on Innovative Simulation for Healthcare and 21st International Multidisciplinary Modeling & Simulation Multiconference (IWISH 2024), Tenerife, Spain, September 18–20 2024. [Google Scholar]
- Jordan, R.; Kinderlehrer, D.; Otto, F. The variational formulation of the Fokker–Planck equation. SIAM journal on mathematical analysis 1998, 29, 1–17. [Google Scholar] [CrossRef]
- Risken, H. The Fokker-Planck Equation, 1996.
- Tikhomirov, V.M. A Study of the Diffusion Equation with Increase in the Amount of Substance, and its Application to a Biological Problem. 1991.
- Swanson, K.R.; Bridge, C.; Murray, J.; Alvord Jr, E.C. Virtual and real brain tumors: using mathematical modeling to quantify glioma growth and invasion. Journal of the neurological sciences 2003, 216, 1–10. [Google Scholar] [CrossRef]
- Abramowitz, M.; Stegun, I.A. Handbook of mathematical functions with formulas, graphs, and mathematical tables; Vol. 55, US Government printing office, 1968.
- Swanson, K.R.; Rockne, R.C.; Claridge, J.; Chaplain, M.A.; Jr, E.C.A.; Anderson, A. Quantifying the role of angiogenesis in malignant progression of gliomas: in silico modeling integrates imaging and histology. Cancer Research 2011, 71, 7366–75. [Google Scholar] [CrossRef]
- Hegedus, B.; Zàch, J.; Cziròk, A.; Lôvey, J.; Vicsek, T. Irradiation and Taxol treatment result in non-monotonous, dose-dependent changes in the motility of glioblastoma cells. J Neurooncol 2004, 67, 147–157. [Google Scholar] [CrossRef]
- Demuth, T.; Hopf, N.J.; Kempski, O.; Sauner, D.; Herr, M.; Giese, A.; Perneczky, A. Migratory activity of human glioma cell lines in vitro assessed by continuous single cell observation. Clin Exp Metastasis 2000, 18, 589–597. [Google Scholar] [CrossRef]
| U87WT | U87EGFR | ||||||
|---|---|---|---|---|---|---|---|
| Parameter | m.u. | PD | DG | MDG | PD | DG | MDG |
| D | · | 1.84 | 2.28 | 1.25 | 2.08 | 1.11 | |
| cells · | 8.57 | 3.61 | 4.71 | 6.05 | 4.37 | 3.30 | |
| g | - | 3.50 | 2.77 | - | 3.03 | 4.60 | |
| cells · | - | 5.65 | 4.77 | - | 4.56 | 3.18 | |
| - | - | - | 0.7 | - | - | 1.1 | |
| - | - | - | 1.0 | - | - | 1.4 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





