Submitted:
03 July 2025
Posted:
04 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
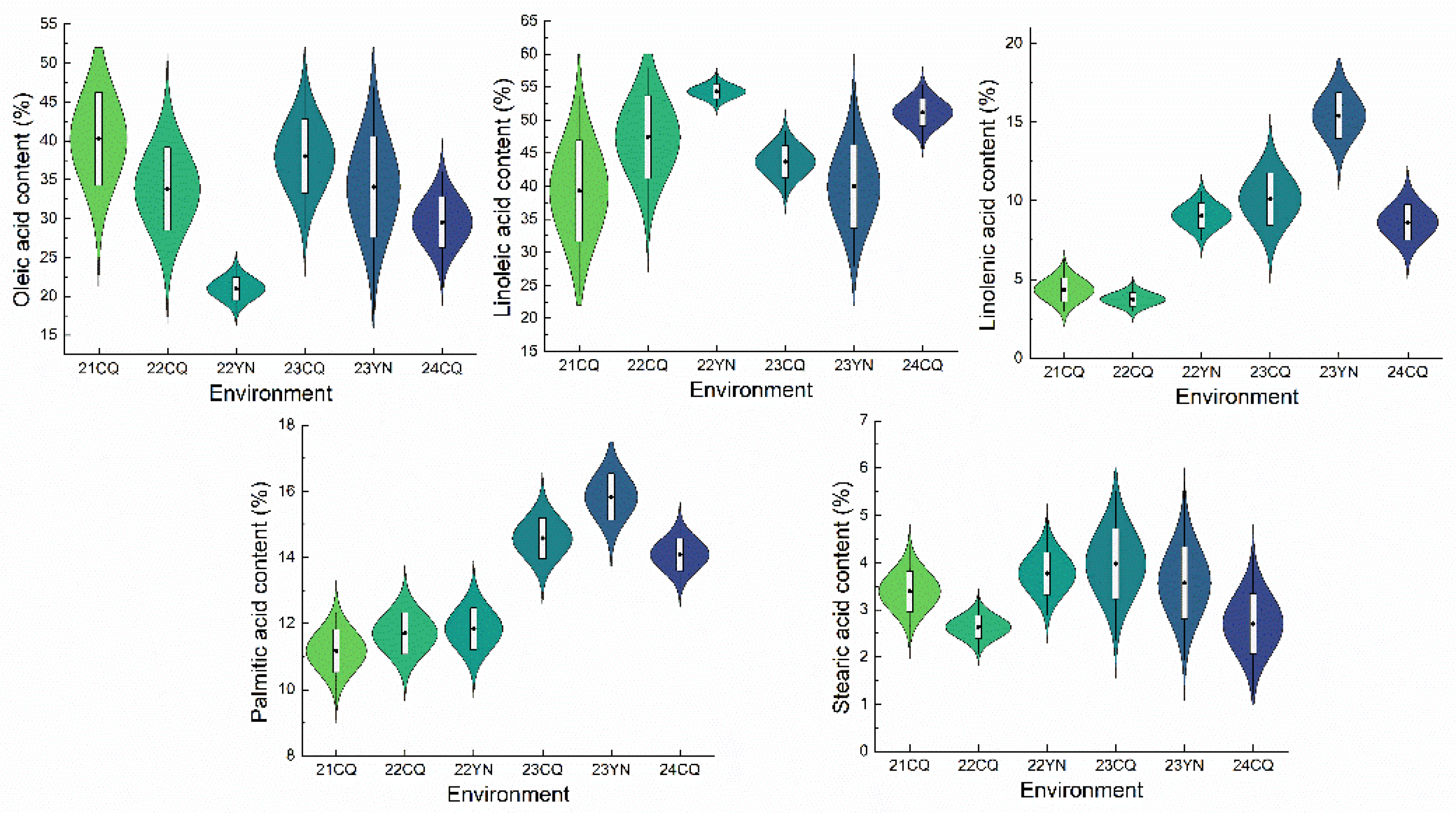
2.1. Trait Phenotype Analysis
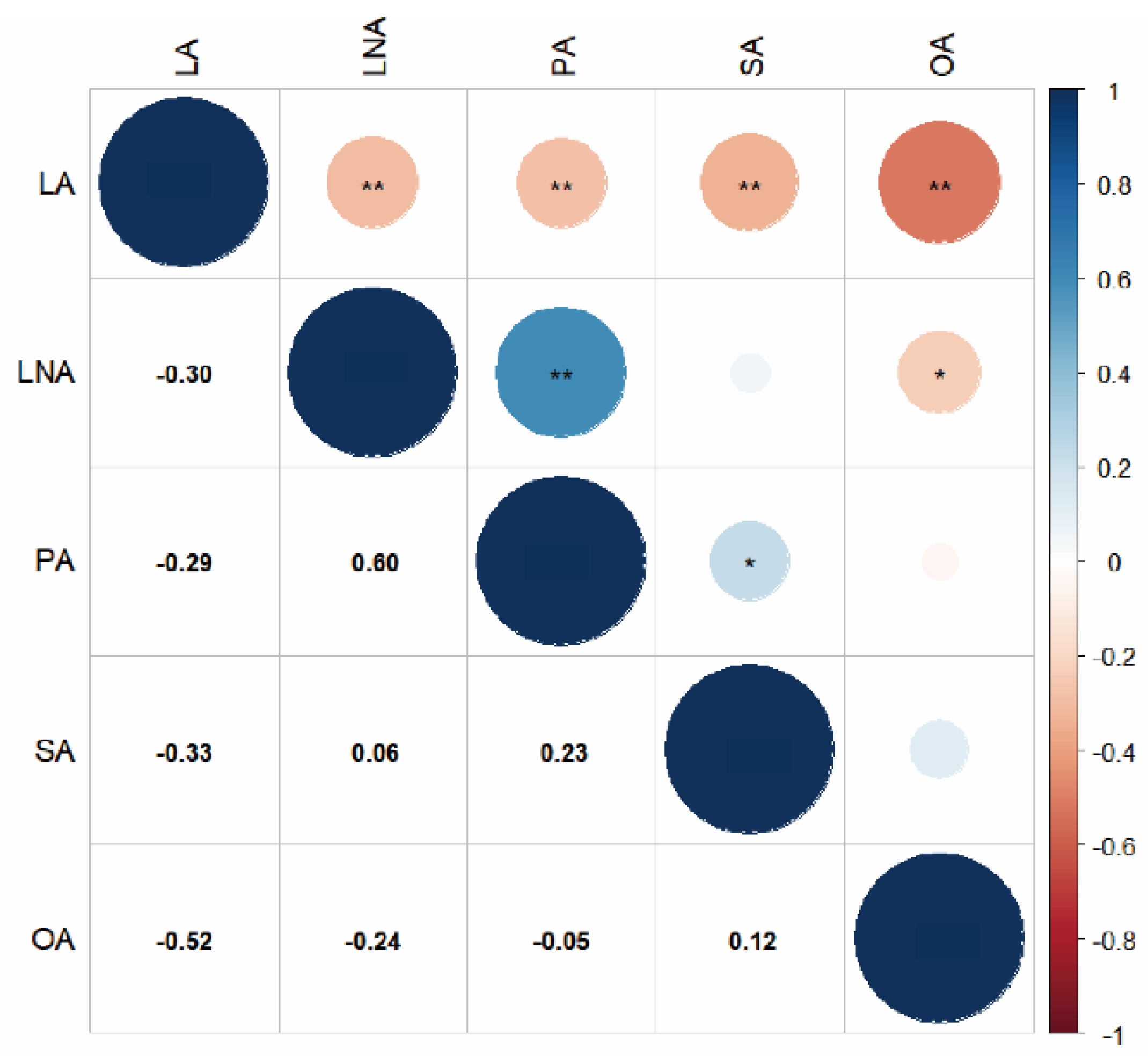
2.2. Correlation Analysis of Five Fatty Acids in Seeds
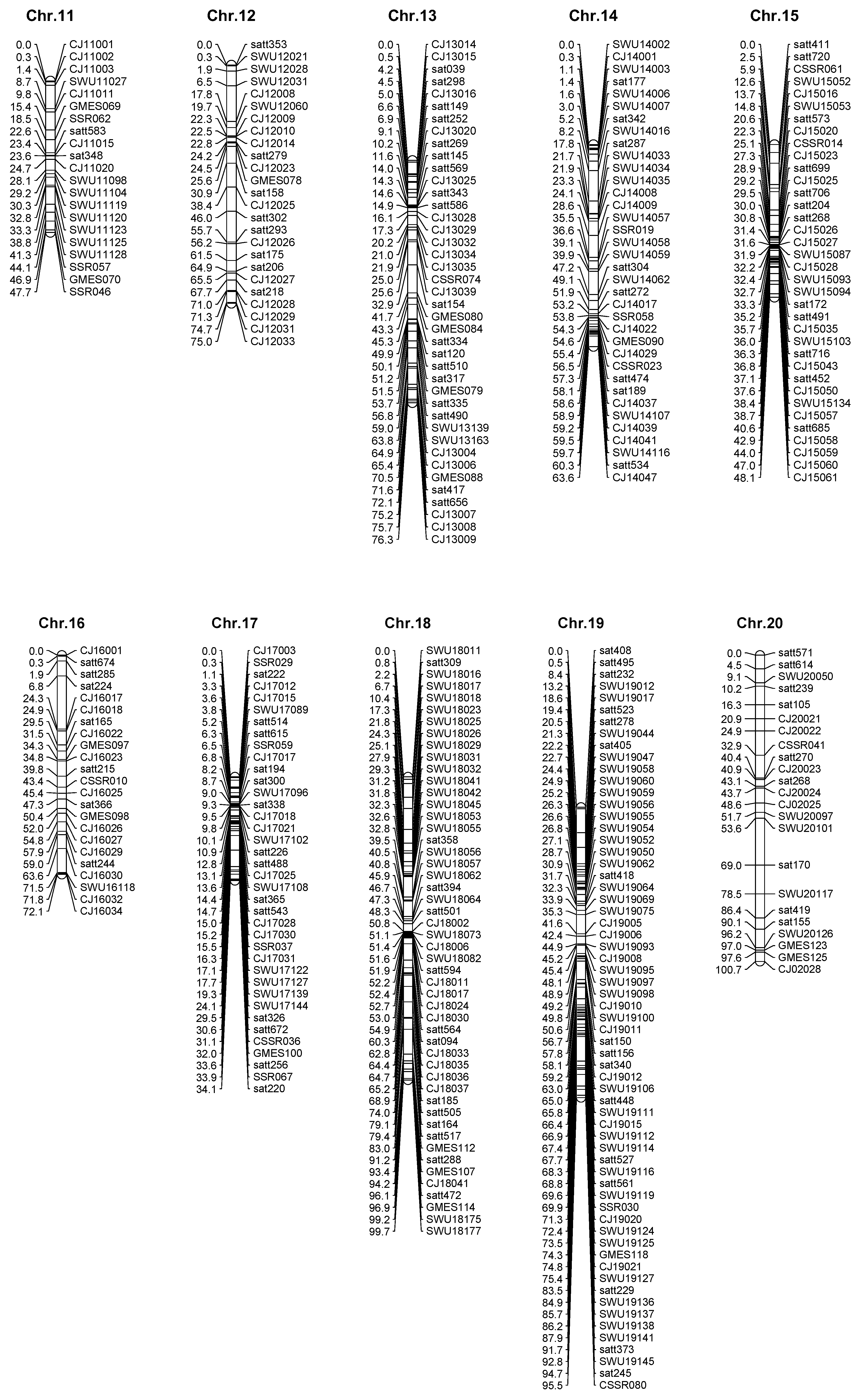
2.3. High Density Genetic Map Construction
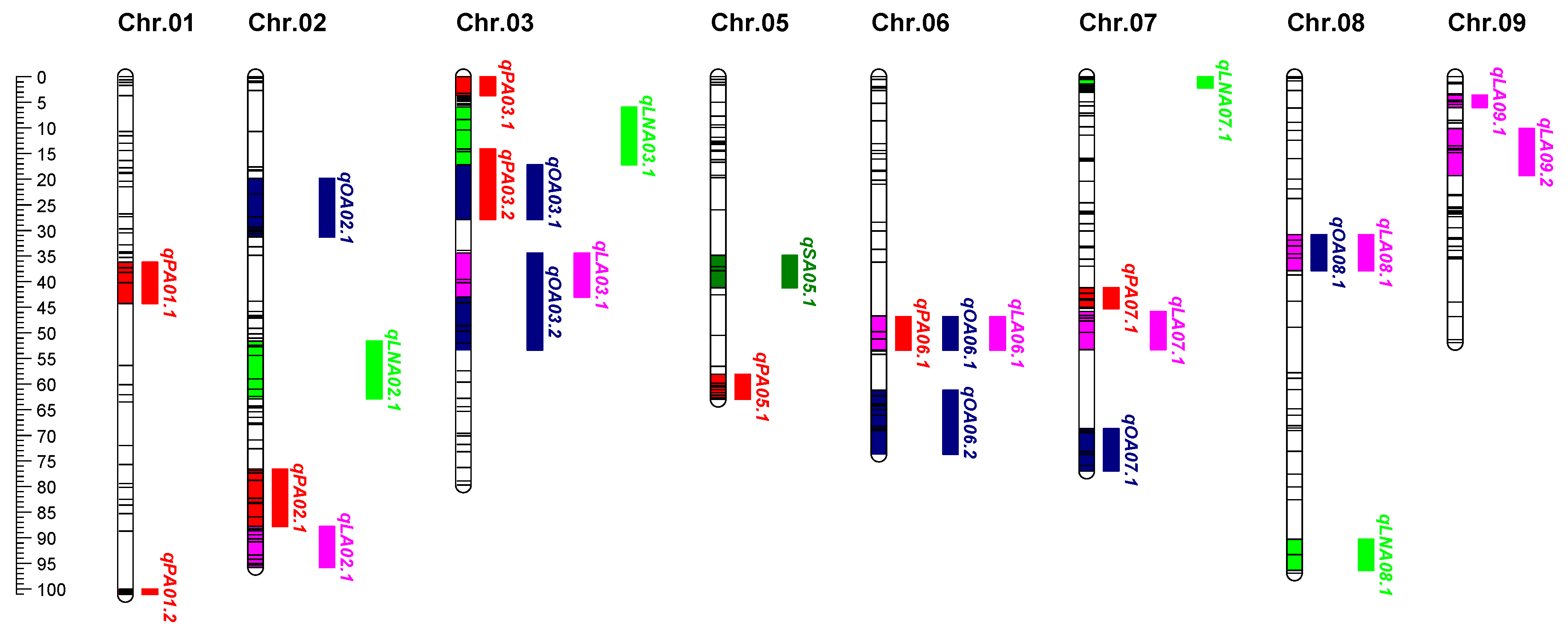
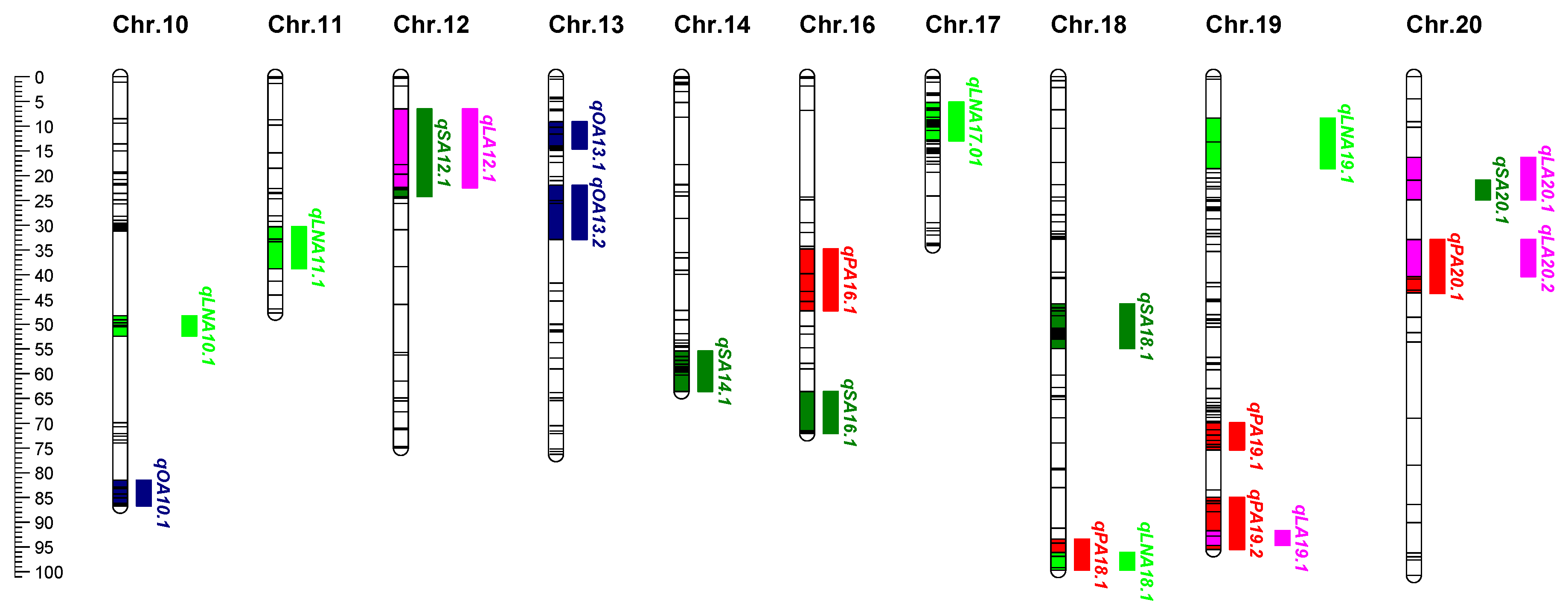
2.4. QTL Mapping for Five Fatty Acids in Seeds
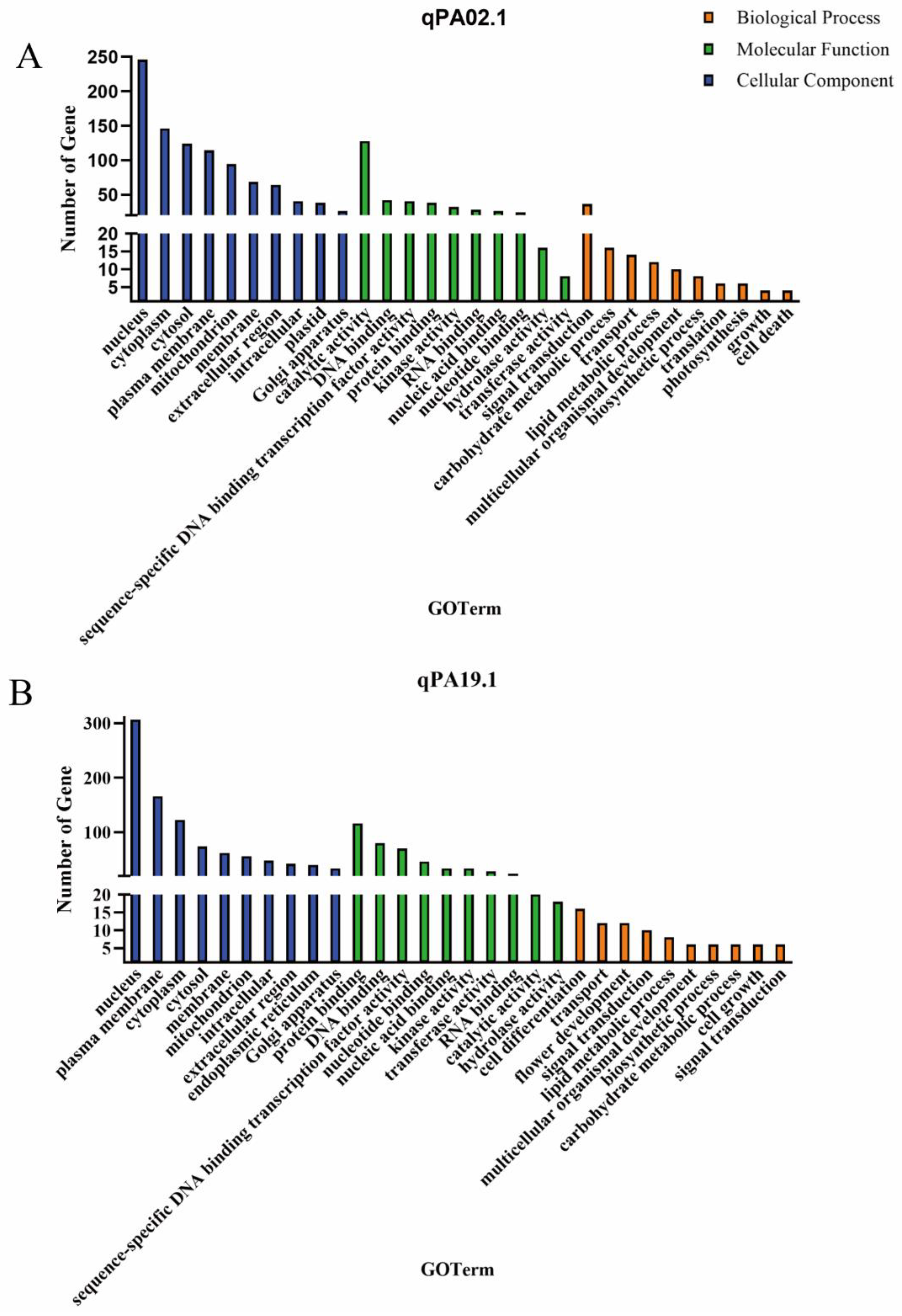
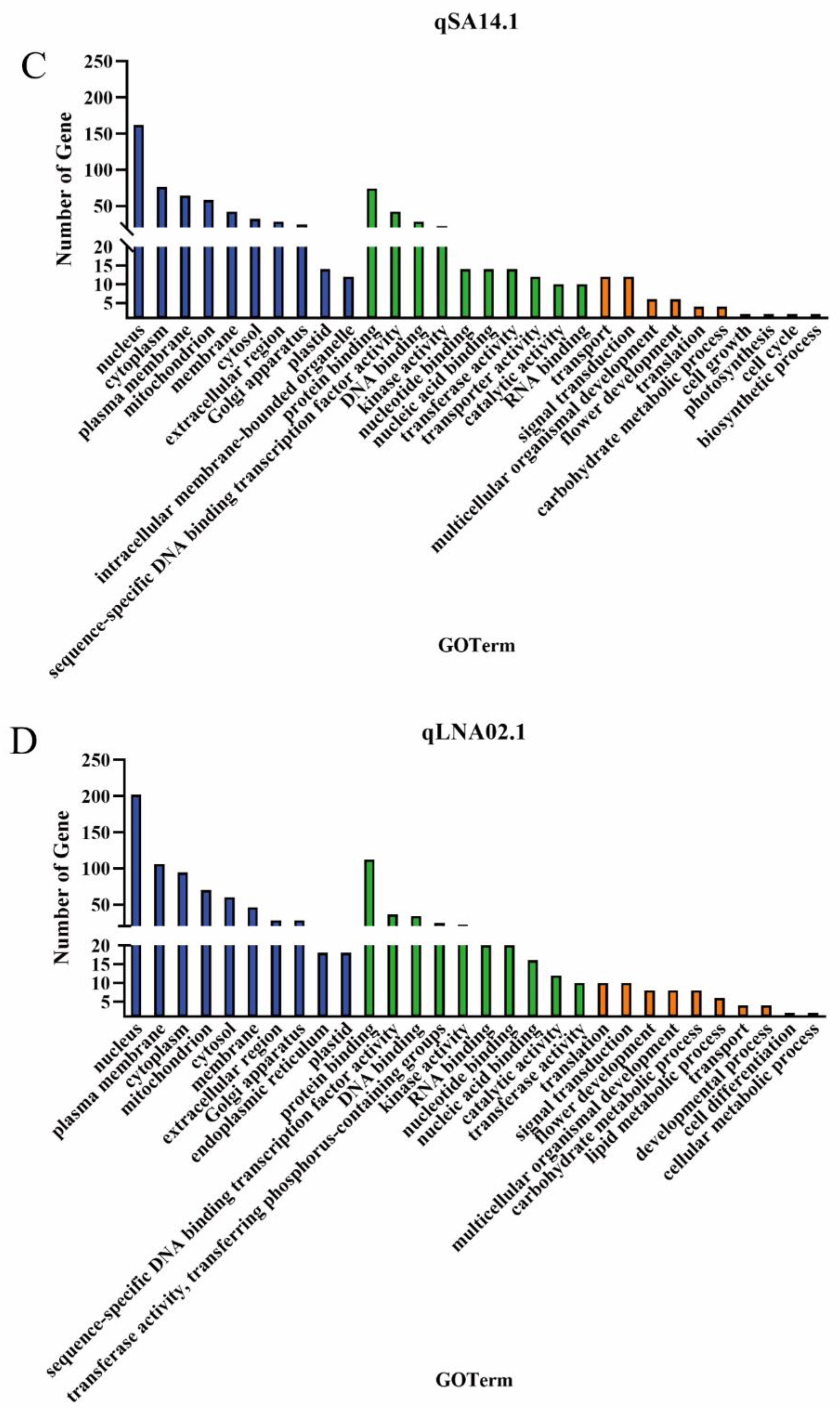
2.5. Candidate Gene Prediction
3. Discussion
4. Materials and methods
4.1. Plant Materials
4.2. DNA Extraction and SSR Genotyping
4.3. Determination of Traits
4.4. Map Construction and QTL Detection
4.5. Candidate Genes Prediction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Han, L.D. , Gai, J. Y., Zhang, W. M. Research status of soybean nutritional components. Seed 2003, 57–59. [Google Scholar]
- Bellaloui, N.; Bruns, H.A.; Abbas, H.K.; Mengistu, A.; Fisher, D.K.; Reddy, K.N. Agricultural practices altered soybean seed protein, oil, fatty acids, sugars, and minerals in the midsouth USA. Frontiers in plant science 2015, 6, 31. [Google Scholar] [CrossRef]
- Clevinger, E.M.; Biyashev, R.; Haak, D.; Song, Q.; Pilot, G.; Saghai Maroof, M.A. Identification of quantitative trait loci controlling soybean seed protein and oil content. PloS one 2023, 18, e0286329. [Google Scholar] [CrossRef] [PubMed]
- Dou, X.; Zhang, L.; Wang, X.; Yang, R.; Wang, X.; Ma, F.; Yu, L.; Mao, J.; Li, H.; Wang, X. , et al. Identification and validation of metabolic markers for adulteration detection of edible oils using metabolic networks. Metabolites 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Z. , Yang, J. Y., Li, X., et al. Nutrition of soybean and human health. New Agricultural Technologies 2004. [Google Scholar]
- Delaney, B.; Appenzeller, L.M.; Munley, S.M.; Hoban, D.; Sykes, G.P.; Malley, L.A.; Sanders, C. Subchronic feeding study of high oleic acid soybeans (event dp-3Ø5423-1) in sprague-dawley rats. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association 2008, 46, 3808–3817. [Google Scholar] [CrossRef]
- Hyten, D.L.; Pantalone, V.R.; Sams, C.E.; Saxton, A.M.; Landau-Ellis, D.; Stefaniak, T.R.; Schmidt, M.E. Seed quality qtl in a prominent soybean population. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik 2004, 109, 552–561. [Google Scholar] [CrossRef]
- Akond, M.; Liu, S.; Boney, M.; Kantartzi, S.K.; Meksem, K.; Bellaloui, N.; Lightfoot, D.A.; Kassem, M.A. Identification of quantitative trait loci (qtl) underlying protein, oil, and five major fatty acids’ contents in soybean. American Journal of Plant Sciences 2014, 05, 158–167. [Google Scholar] [CrossRef]
- Ha, B.K.; Kim, H.J.; Velusamy, V.; Vuong, T.D.; Nguyen, H.T.; Shannon, J.G.; Lee, J.D. Identification of quantitative trait loci controlling linolenic acid concentration in pi483463 (glycine soja). TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik 2014, 127, 1501–1512. [Google Scholar] [CrossRef]
- Pathan, S.M.; Vuong, T.; Clark, K.; Lee, J.D.; Shannon, J.G.; Roberts, C.A.; Ellersieck, M.R.; Burton, J.W.; Cregan, P.B.; Hyten, D.L. , et al. Genetic mapping and confirmation of quantitative trait loci for seed protein and oil contents and seed weight in soybean. Crop Science 2013, 53, 765–774. [Google Scholar] [CrossRef]
- Priolli, R.H.G.; Campos, J.B.; Stabellini, N.S.; Pinheiro, J.B.; Vello, N.A. Association mapping of oil content and fatty acid components in soybean. Euphytica 2014, 203, 83–96. [Google Scholar] [CrossRef]
- Diers, B.W.; Shoemaker, R.C. Restriction fragment length polymorphism analysis of soybean fatty acid content. Journal of the American Oil Chemists Society 1992, 69, 1242–1244. [Google Scholar] [CrossRef]
- Li, Z.; Wilson, R.F.; Rayford, W.E.; Boerma, H.R. Molecular mapping genes conditioning reduced palmitic acid content in n87-2122-4 soybean. Crop Science 2002, 42, 373–378. [Google Scholar]
- Li, H.; Zhao, T.; Wang, Y.; Yu, D.; Chen, S.; Zhou, R.; Gai, J. Genetic structure composed of additive qtl, epistatic qtl pairs and collective unmapped minor qtl conferring oil content and fatty acid components of soybeans. Euphytica 2011, 182. [Google Scholar] [CrossRef]
- Xie, D.; Han, Y.; Zeng, Y.; Chang, W.; Teng, W.; Li, W. Ssr- and snp-related qtl underlying linolenic acid and other fatty acid contents in soybean seeds across multiple environments. Molecular Breeding 2011, 30, 169–179. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, X.; Wang, S.; Li, S.; Zeng, Y.; Cheng, Y.; Ma, Q.; Wang, Y.; Pang, Y.; Nian, H. , et al. Mapping and identification of qtls for seed fatty acids in soybean (glycine max l.). Journal of Integrative Agriculture 2024, 23, 3966–3982. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y. , et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nature Biotechnology 2015, 33, 408–414. [Google Scholar] [CrossRef]
- Bachlava, E.; Dewey, R.E.; Burton, J.W.; Cardinal, A.J. Mapping and comparison of quantitative trait loci for oleic acid seed content in two segregating soybean populations. Crop Science 2009, 49, 433–442. [Google Scholar] [CrossRef]
- Jiang, A.; Liu, J.; Gao, W.; Ma, R.; Tan, P.; Liu, F.; Zhang, J. Construction of a genetic map and qtl mapping of seed size traits in soybean. Frontiers in Genetics 2023, 14. [Google Scholar] [CrossRef]
- Jason T., C. Tzen, Y.-z.C., Pascal Laurent, Chandra Ratnayake, and Anthony H. C. Huang. Lipids, proteins, and structure of seed oil bodies from diverse species. Plant physiology 1993, 101, 267–276. [Google Scholar]
- Chen, X.G., Z. H.. Research and application of DNA molecular marker technology. Mol. Plant Breed 2019, 17(6), 1970–1977. [Google Scholar]
- Wu, X.L.; He, C.Y.; Wang, Y.J.; Zhang, Z.Y.; Dongfang, Y.; Zhang, J.S.; Chen, S.Y.; Gai, J.Y. [construction and analysis of a genetic linkage map of soybean]. Yi chuan xue bao = Acta genetica Sinica 2001, 28, 1051–1061. [Google Scholar]
- Do, P.T.; Nguyen, C.X.; Bui, H.T.; Tran, L.T.N.; Stacey, G.; Gillman, J.D.; Zhang, Z.J.; Stacey, M.G. Demonstration of highly efficient dual grna crispr/cas9 editing of the homeologous gmfad2-1a and gmfad2-1b genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC plant biology 2019, 19, 311. [Google Scholar] [CrossRef] [PubMed]
- Cregan, P.B.; Jarvik, T.; Bush, A.L.; Shoemaker, R.C.; Lark, K.G.; Kahler, A.L.; Kaya, N.; VanToai, T.T.; Lohnes, D.G.; Chung, J. , et al. An integrated genetic linkage map of the soybean genome. Crop Science 1999, 39, 1464–1490. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, A.; Ma, R.; Gao, W.; Tan, P.; Li, X.; Du, C.; Zhang, J.; Zhang, X.; Zhang, L. , et al. Qtl mapping for seed quality traits under multiple environments in soybean (glycine max l.). Agronomy 2023, 13. [Google Scholar]
- 26. Qi, H.; Wang, W.; Tang, X.; Xue, Y.; Cao, D.; Liu, X.; Luan, X.; Du, J.; Qiu, L., QTL Mining of Protein and Oil Content in Elite Soybean Germplasm. Journal of Plant Genetic Resources 2023, 24(05), 1435–1447.
- Zhu, X.; Leiser, W.L.; Hahn, V.; Würschum, T. Identification of seed protein and oil related qtl in 944 rils from a diallel of early-maturing european soybean. The Crop Journal 2021, 9, 238–247. [Google Scholar] [CrossRef]
- Cardinal, A.J.; Whetten, R.; Wang, S.; Auclair, J.; Hyten, D.; Cregan, P.; Bachlava, E.; Gillman, J.; Ramirez, M.; Dewey, R. , et al. Mapping the low palmitate fap1 mutation and validation of its effects in soybean oil and agronomic traits in three soybean populations. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik 2014, 127, 97–111. [Google Scholar] [CrossRef]
- Reinprecht, Y.; Poysa, V.W.; Yu, K.; Rajcan, I.; Ablett, G.R.; Pauls, K.P. Seed and agronomic qtl in low linolenic acid, lipoxygenase-free soybean (glycine max (l.) merrill) germplasm. Genome 2006, 49, 1510–1527. [Google Scholar] [CrossRef]
- Panthee, D.R.; Pantalone, V.R.; Saxton, A.M. Modifier qtl for fatty acid composition in soybean oil. Euphytica 2006, 152, 67–73. [Google Scholar] [CrossRef]
- Fan, S.; Li, B.; Yu, F.; Han, F.; Yan, S.; Wang, L.; Sun, J. Analysis of additive and epistatic quantitative trait loci underlying fatty acid concentrations in soybean seeds across multiple environments. Euphytica 2015, 206, 689–700. [Google Scholar] [CrossRef]
- Qi, Z.-m.; Wu, Q.; Han, X.; Sun, Y.-n.; Du, X.-y.; Liu, C.-y.; Jiang, H.-w.; Hu, G.-h.; Chen, Q.-s. Soybean oil content qtl mapping and integrating with meta-analysis method for mining genes. Euphytica 2011, 179, 499–514. [Google Scholar] [CrossRef]
- Yao, Y.; You, Q.; Duan, G.; Ren, J.; Chu, S.; Zhao, J.; Li, X.; Zhou, X.; Jiao, Y. Quantitative trait loci analysis of seed oil content and composition of wild and cultivated soybean. BMC plant biology 2020, 20, 51. [Google Scholar] [CrossRef] [PubMed]
- Phansak, P.; Soonsuwon, W.; Hyten, D.L.; Song, Q.; Cregan, P.B.; Graef, G.L.; Specht, J.E. Multi-population selective genotyping to identify soybean [glycine max (l.) merr.] seed protein and oil qtls. G3 (Bethesda, Md.) 2016, 6, 1635–1648. [Google Scholar] [CrossRef]
- Huo, W.; Li, B.; Kuang, J.; He, P.; Xu, Z.; Wang, J. Functional characterization of the steroid reductase genes gmdet2a and gmdet2b form glycine max. International journal of molecular sciences 2018, 19. [Google Scholar] [CrossRef]
- Zheng, H.; Rowland, O.; Kunst, L. Disruptions of the arabidopsis enoyl-coa reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. The Plant cell 2005, 17, 1467–1481. [Google Scholar] [CrossRef]
- Arias, C.L.; Quach, T.; Huynh, T.; Nguyen, H.; Moretti, A.; Shi, Y.; Guo, M.; Rasoul, A.; Van, K.; McHale, L. , et al. Expression of atwri1 and atdgat1 during soybean embryo development influences oil and carbohydrate metabolism. Plant biotechnology journal 2022, 20, 1327–1345. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Q.; Han, W.; Zhao, Q.; Sun, D.; Shen, Z. Qtl mapping and candidate gene screening for enhancing oil content in silage maize. Plants (Basel, Switzerland) 2025, 14. [Google Scholar] [CrossRef]
- Otyama, P.I.; Chamberlin, K.; Ozias-Akins, P.; Graham, M.A.; Cannon, E.K.S.; Cannon, S.B.; MacDonald, G.E.; Anglin, N.L. Genome-wide approaches delineate the additive, epistatic, and pleiotropic nature of variants controlling fatty acid composition in peanut (arachis hypogaea l.). G3 (Bethesda, Md.) 2022, 12. [Google Scholar] [CrossRef]
- Lusk, H.J.; Neumann, N.; Colter, M.; Roth, M.R.; Tamura, P.; Yao, L.; Shiva, S.; Shah, J.; Schrick, K.; Durrett, T.P. , et al. Lipidomic analysis of arabidopsis t-DNA insertion lines leads to identification and characterization of c-terminal alterations in fatty acid desaturase 6. Plant & cell physiology 2022, 63, 1193–1204. [Google Scholar]
- Reinprecht, Y.; Pauls, K.P. Microsomal omega-3 fatty acid desaturase genes in low linolenic acid soybean line rg10 and validation of major linolenic acid qtl. Front Genet 2016, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Reif, J.C.; Ma, Y.S.; Hong, H.L.; Liu, Z.X.; Chang, R.Z.; Qiu, L.J. Targeted association mapping demonstrating the complex molecular genetics of fatty acid formation in soybean. BMC genomics 2015, 16, 841. [Google Scholar] [CrossRef] [PubMed]
- Akond, M.; Yuan, J.; Liu, S.; Kantartzi, S.K.; Meksem, K.; Bellaloui, N.; Lightfoot, D.A.; Kassem, M.A.; Willenborg, C. Detection of qtl underlying seed quality components in soybean [glycine max (l.) merr.]. Canadian Journal of Plant Science 2018, 98, 881–888. [Google Scholar] [CrossRef]

| Traits | Env. | Parent | Population | |||||||||
| CJC2 | JY166 | Mean | Min | Max | SD | Variance | CV(%) | Skewness | Kurtosis | |||
| PA | 21CQ | 11.28 | 9.46 | 11.17 | 8.98 | 12.32 | 0.65 | 0.42 | 0.06 | -0.60 | 0.67 | |
| 22CQ | 11.32 | 10.40 | 11.70 | 9.82 | 13.37 | 0.64 | 0.40 | 0.06 | -0.34 | 0.76 | ||
| 22YN | 11.59 | 10.88 | 11.83 | 10.40 | 13.17 | 0.64 | 0.41 | 0.05 | 0.06 | -0.65 | ||
| 23CQ | 14.21 | 12.37 | 14.58 | 12.82 | 16.04 | 0.62 | 0.39 | 0.04 | -0.35 | 0.76 | ||
| 23YN | 15.95 | 14.56 | 15.83 | 13.91 | 16.95 | 0.70 | 0.49 | 0.04 | -0.44 | -0.30 | ||
| 24CQ | 14.89 | 13.42 | 14.08 | 12.78 | 15.06 | 0.49 | 0.24 | 0.03 | -0.18 | 0.26 | ||
| SA | 21CQ | 1.83 | 3.55 | 3.39 | 2.20 | 4.40 | 0.43 | 0.19 | 0.13 | -0.16 | 0.09 | |
| 22CQ | 1.77 | 2.79 | 2.63 | 1.77 | 3.45 | 0.26 | 0.06 | 0.10 | 0.43 | 0.82 | ||
| 22YN | 2.59 | 3.62 | 3.77 | 2.88 | 5.18 | 0.45 | 0.20 | 0.12 | 0.50 | 0.87 | ||
| 23CQ | 2.26 | 2.87 | 3.98 | 2.34 | 5.52 | 0.75 | 0.56 | 0.19 | -0.08 | -0.52 | ||
| 23YN | 2.65 | 3.76 | 3.57 | 1.93 | 5.72 | 0.76 | 0.58 | 0.21 | 0.40 | -0.14 | ||
| 24CQ | 1.96 | 3.47 | 2.70 | 1.18 | 4.23 | 0.64 | 0.41 | 0.24 | 0.31 | -0.19 | ||
| OA | 21CQ | 39.23 | 37.64 | 40.38 | 28.42 | 50.70 | 6.05 | 36.54 | 0.15 | -0.10 | -0.77 | |
| 22CQ | 33.26 | 31.38 | 33.81 | 22.10 | 46.34 | 5.34 | 28.53 | 0.16 | 0.22 | -0.47 | ||
| 22YN | 20.87 | 19.34 | 20.99 | 17.64 | 24.09 | 1.48 | 2.18 | 0.07 | 0.05 | -0.63 | ||
| 23CQ | 37.57 | 35.78 | 38.03 | 28.74 | 50.88 | 4.77 | 22.76 | 0.13 | 0.65 | 0.44 | ||
| 23YN | 40.92 | 34.34 | 34.07 | 17.73 | 46.93 | 6.47 | 41.86 | 0.19 | -0.24 | 0.26 | ||
| 24CQ | 31.54 | 28.66 | 29.51 | 21.22 | 35.94 | 3.30 | 10.89 | 0.11 | -0.17 | -0.32 | ||
| LA | 21CQ | 38.47 | 42.13 | 39.27 | 23.33 | 53.56 | 7.66 | 58.67 | 0.20 | -0.16 | -0.74 | |
| 22CQ | 48.97 | 51.97 | 47.43 | 32.11 | 57.86 | 6.27 | 39.20 | 0.13 | -0.51 | -0.30 | ||
| 22YN | 53.32 | 55.25 | 54.31 | 52.08 | 57.21 | 1.08 | 1.17 | 0.02 | 0.24 | 0.16 | ||
| 23CQ | 49.53 | 50.11 | 43.71 | 38.07 | 48.35 | 2.43 | 5.88 | 0.06 | -0.23 | -0.42 | ||
| 23YN | 36.40 | 40.16 | 39.95 | 27.97 | 54.40 | 6.30 | 39.64 | 0.16 | 0.23 | -0.30 | ||
| 24CQ | 49.73 | 52.34 | 51.17 | 5.73 | 55.32 | 2.11 | 4.45 | 0.04 | -0.24 | -0.20 | ||
| LNA | 21CQ | 5.34 | 4.17 | 4.33 | 3.01 | 6.29 | 0.77 | 0.60 | 0.18 | 0.49 | -0.38 | |
| 22CQ | 4.68 | 3.45 | 3.73 | 2.92 | 5.01 | 0.46 | 0.20 | 0.12 | 0.88 | 0.57 | ||
| 22YN | 9.87 | 8.22 | 9.04 | 7.49 | 10.63 | 0.81 | 0.66 | 0.09 | 0.02 | -0.99 | ||
| 23CQ | 10.31 | 9.15 | 10.11 | 4.74 | 13.42 | 1.66 | 2.76 | 0.16 | -0.39 | 0.52 | ||
| 23YN | 15.46 | 13.72 | 15.41 | 11.50 | 17.95 | 1.46 | 2.14 | 0.09 | -0.66 | 0.17 | ||
| 24CQ | 9.26 | 7.72 | 8.62 | 6.29 | 10.94 | 1.12 | 1.26 | 0.13 | -0.12 | -0.52 | ||
| Chromosome | Loci | Length | Average interval |
| Chr.1 | 45 | 101.1 | 2.25 |
| Chr.2 | 62 | 95.8 | 1.55 |
| Chr.3 | 37 | 79.7 | 2.15 |
| Chr.4 | 28 | 41.7 | 1.49 |
| Chr.5 | 35 | 63.0 | 1.80 |
| Chr.6 | 34 | 69.1 | 2.03 |
| Chr.7 | 49 | 77.0 | 1.57 |
| Chr.8 | 37 | 96.9 | 2.62 |
| Chr.9 | 36 | 51.9 | 1.44 |
| Chr.10 | 43 | 86.7 | 2.02 |
| Chr.11 | 21 | 47.7 | 2.27 |
| Chr.12 | 25 | 75.0 | 3.00 |
| Chr.13 | 40 | 76.3 | 1.91 |
| Chr.14 | 36 | 63.6 | 1.77 |
| Chr.15 | 36 | 48.1 | 1.34 |
| Chr.16 | 23 | 72.1 | 3.13 |
| Chr.17 | 38 | 34.1 | 0.90 |
| Chr.18 | 50 | 99.7 | 1.99 |
| Chr.19 | 63 | 95.5 | 1.52 |
| Chr.20 | 23 | 100.7 | 4.38 |
| total | 761 | 1475.7 | 1.94 |
| QTL | Env.a | Chr. | Nearest Maker | Interval (cM) | LOD | PVE (%) | Additive | Dominance |
| qPA01.1 | 22YN | 1 | SWU01110 | 36.20-44.29 | 5.44 | 14.2 | -0.27 | -0.07 |
| 23CQ | 1 | SWU01107 | 36.20-40.18 | 6.6 | 16.4 | 0.17 | 0.49 | |
| 23YN | 1 | SWU01109 | 38.20-44.29 | 3.62 | 10.5 | 0.12 | 0.49 | |
| qPA01.2 | 22CQ | 1 | CJ01032 | 100.02-101.12 | 5.14 | 12 | -0.07 | 0.51 |
| 24CQ | 1 | CSSR051 | 100.02-101.12 | 4.15 | 10.6 | 0.20 | -0.17 | |
| qPA02.1 | 22CQ | 2 | sat183 | 83.35-85.87 | 3.06 | 7.3 | 0.03 | -0.34 |
| 22YN | 2 | satt703 | 76.64-82.26 | 5.67 | 14.7 | 0.29 | 0.34 | |
| 22CQ | 2 | sat183 | 83.35-87.80 | 3.37 | 8.7 | 0.24 | 0.08 | |
| 24CQ | 2 | satt703 | 76.64-82.26 | 4.51 | 11.5 | 0.25 | 0.06 | |
| qPA03.1 | 22YN | 3 | CJ03002 | 0-3.70 | 5.74 | 14.9 | 0.16 | 0.38 |
| 23CQ | 3 | CJ03002 | 0-3.15 | 3 | 7.8 | 0.24 | -0.22 | |
| 23YN | 3 | CJ03002 | 0-3.15 | 3.34 | 9.8 | 0.27 | -0.33 | |
| qPA03.2 | 22CQ | 3 | satt641 | 17.16-27.92 | 3.23 | 7.7 | 0.08 | 0.35 |
| 23CQ | 3 | SWU03019 | 14.63-27.92 | 3.23 | 8.4 | 0.20 | 0.12 | |
| 24CQ | 3 | SWU03019 | 14.07-17.16 | 5.03 | 12.7 | 0.21 | 0.10 | |
| qPA05.1 | 22YN | 5 | satt599 | 58.13-63.01 | 5.57 | 14.5 | 0.11 | 0.55 |
| 32YN | 5 | CJ05032 | 60.31-63.01 | 4.03 | 11.6 | 0.42 | 0.36 | |
| qPA06.1 | 22YN | 6 | CJ06029 | 46.75-53.38 | 3.48 | 9.3 | -0.12 | 0.34 |
| 23YN | 6 | CJ06029 | 46.75-51.17 | 5.23 | 14.8 | 0.33 | -0.02 | |
| qPA07.1 | 22YN | 7 | SWU07054 | 41.17-45.25 | 6.5 | 16.7 | -0.34 | 0.13 |
| 23YN | 7 | SWU07054 | 41.17-43.34 | 3.58 | 10.4 | 0.21 | -0.34 | |
| qPA16.1 | 22YN | 16 | CSSR010 | 34.84-45.36 | 4.78 | 12.6 | 0.31 | 0.05 |
| 24CQ | 16 | CSSR010 | 39.76-47.29 | 5.01 | 12.7 | 0.21 | -0.19 | |
| qPA18.1 | 21CQ | 18 | SWU18177 | 93.38-99.70 | 5.08 | 12.2 | 0.31 | 0.17 |
| 22CQ | 18 | SWU18177 | 94.19-99.70 | 5.98 | 13.8 | 0.32 | 0.48 | |
| qPA19.1 | 21CQ | 19 | satt527 | 66.36-72.38 | 4 | 9.7 | 0.19 | -0.33 |
| 22CQ | 19 | CJ19020 | 69.92-72.38 | 3.29 | 7.8 | 0.18 | -0.15 | |
| 23CQ | 19 | SSR030 | 69.92-72.48 | 3.74 | 9.6 | 0.19 | -0.21 | |
| 24CQ | 19 | SWU19125 | 71.29-75.38 | 8.16 | 19.8 | 0.28 | -0.06 | |
| qPA19.2 | 22CQ | 19 | sat245 | 92.76-95.53 | 3.59 | 8.5 | -0.20 | 0.20 |
| 22YN | 19 | SWU19138 | 84.86-95.53 | 6.3 | 16.2 | 0.24 | 0.42 | |
| qPA20.1 | 22CQ | 20 | CJ20023 | 40.37-48.60 | 4.45 | 10.4 | 0.27 | 0.31 |
| 22YN | 20 | satt270 | 32.93-40.91 | 4.31 | 12.4 | 0.30 | -0.17 | |
| 24CQ | 20 | satt270 | 32.93-40.91 | 4.1 | 10.5 | 0.20 | -0.12 | |
| qSA05.1 | 21CQ | 5 | GMES021 | 34.89-37.90 | 3.82 | 9.3 | -0.21 | 0.02 |
| 22CQ | 5 | GMES021 | 34.89-41.24 | 6.83 | 15.6 | -0.15 | 0.03 | |
| 22YN | 5 | satt619 | 34.89-41.24 | 4.77 | 12.5 | -0.22 | -0.43 | |
| qSA12.1 | 22CQ | 12 | CJ12008 | 6.50-19.75 | 5.61 | 13 | -0.11 | 0.10 |
| 23CQ | 12 | CJ12009 | 17.81-24.19 | 7.01 | 17.3 | -0.40 | -0.22 | |
| qSA14.1 | 21CQ | 14 | satt534 | 59.18-63.64 | 5.65 | 13.5 | -0.24 | -0.13 |
| 22CQ | 14 | SWU14116 | 58.64-63.64 | 10.64 | 23.2 | -0.18 | 0.07 | |
| 22YN | 14 | satt474 | 55.39-59.72 | 3.92 | 10.4 | -0.22 | -0.17 | |
| 23CQ | 14 | CJ14037 | 51.86-60.26 | 4.08 | 10.5 | -0.36 | 0.00 | |
| qSA16.1 | 21CQ | 16 | SWU16118 | 63.62-72.07 | 5.12 | 12.3 | -0.17 | -0.26 |
| 22CQ | 16 | SWU16118 | 63.62-71.54 | 2.85 | 6.8 | -0.10 | 0.01 | |
| 23CQ | 16 | SWU16118 | 63.62-72.07 | 5.06 | 12.8 | -0.38 | 0.13 | |
| qSA18.1 | 22CQ | 18 | CJ18006 | 48.34-52.98 | 4.71 | 11 | -0.13 | 0.01 |
| 22YN | 18 | CJ18030 | 50.82-54.9 | 7.35 | 18.7 | -0.25 | -0.19 | |
| 23CQ | 18 | SWU18062 | 45.9-48.34 | 3.04 | 7.9 | -0.23 | -0.17 | |
| qSA20.1 | 21CQ | 20 | CJ20022 | 20.88-24.89 | 3.56 | 8.7 | -0.06 | -0.31 |
| 22YN | 20 | CJ20021 | 20.88-24.89 | 7.24 | 18.4 | -0.36 | -0.38 | |
| qOA02.1 | 23CQ | 2 | satt558 | 30.70-43.80 | 3.58 | 9.20 | 0.93 | 2.62 |
| 24CQ | 2 | SWU02044 | 19.81-31.25 | 4.84 | 12.30 | 1.46 | 0.49 | |
| qOA03.1 | 22CQ | 3 | satt641 | 17.16-27.92 | 3.97 | 9.40 | -0.94 | 3.80 |
| 23YN | 3 | SWU03071 | 17.16-27.92 | 3.19 | 9.30 | 0.42 | 0.73 | |
| qOA03.2 | 22YN | 3 | CJ03030 | 34.41-43.01 | 3.88 | 10.30 | 0.40 | -0.62 |
| 23YN | 3 | SWU03115 | 39.60-44.13 | 3.91 | 11.30 | -3.33 | -1.49 | |
| qOA06.1 | 22YN | 6 | satt557 | 46.75-53.38 | 3.84 | 10.20 | 0.69 | 0.19 |
| 23YN | 6 | CJ06029 | 46.75-49.81 | 3.06 | 9.00 | -2.43 | 0.26 | |
| qOA06.2 | 22YN | 6 | CSSR071 | 65.00-69.10 | 3.93 | 10.50 | 0.64 | -0.01 |
| 23CQ | 6 | sat312 | 61.19-66.09 | 3.38 | 8.70 | 1.91 | -0.04 | |
| qOA07.1 | 22CQ | 7 | CJ07044 | 68.69-73.72 | 5.31 | 12.30 | 2.45 | 1.49 |
| 22YN | 7 | CJ07052 | 73.72-77.03 | 3.5 | 9.40 | 0.66 | -0.30 | |
| 24CQ | 7 | CJ07052 | 75.94-77.03 | 3.36 | 8.70 | 1.31 | -1.11 | |
| qOA08.1 | 22YN | 8 | CJ08013 | 30.78-37.90 | 7.02 | 17.90 | 0.88 | 0.01 |
| 23YN | 8 | CJ08009 | 30.78-32.96 | 3.61 | 10.50 | -3.03 | -1.14 | |
| qOA10.1 | 22CQ | 10 | CJ10033 | 81.53-86.72 | 4.06 | 9.60 | 1.78 | 4.01 |
| 24CQ | 10 | CJ10030 | 81.53-86.72 | 3.26 | 8.40 | 1.82 | 0.39 | |
| qOA13.1 | 23CQ | 13 | satt569 | 9.11-14.59 | 4.71 | 12.00 | 2.70 | -1.62 |
| 24CQ | 13 | CJ13025 | 11.56-14.59 | 3.51 | 9.10 | 1.27 | -1.75 | |
| qOA13.2 | 22CQ | 13 | sat154 | 25.59-32.93 | 3.59 | 8.50 | -0.74 | 3.21 |
| 23CQ | 13 | CSSR074 | 21.87-25.59 | 3.31 | 8.60 | 2.05 | 0.13 | |
| qLA02.1 | 22CQ | 2 | CJ02012 | 88.61-93.37 | 4.89 | 11.4 | 2.30 | 3.05 |
| 23CQ | 2 | sat192 | 87.80-95.82 | 6.62 | 16.4 | -1.30 | -0.56 | |
| qLA03.1 | 22CQ | 3 | CJ03030 | 34.41-43.01 | 3.21 | 7.6 | 1.84 | -1.74 |
| 22YN | 3 | CJ03030 | 34.41-43.01 | 4.76 | 12.5 | -0.34 | 0.45 | |
| 23YN | 3 | CJ03030 | 34.41-43.01 | 4.68 | 13.4 | 3.71 | 2.97 | |
| qLA06.1 | 22YN | 6 | CJ06031 | 46.75-51.17 | 3.4 | 9.1 | -0.44 | 0.03 |
| 23YN | 6 | CJ06031 | 46.75-53.38 | 5.08 | 14.4 | 3.40 | -0.52 | |
| qLA07.1 | 22CQ | 7 | CJ07044 | 53.34-69.50 | 4.55 | 10.7 | -2.62 | -1.67 |
| 22YN | 7 | CJ07026 | 49.91-53.34 | 4.59 | 12.1 | -0.51 | 0.29 | |
| 23YN | 7 | satt536 | 45.79-53.34 | 3.96 | 11.4 | 2.91 | -0.97 | |
| qLA08.1 | 22YN | 8 | CJ08013 | 30.78-37.90 | 6.22 | 16 | -0.59 | -0.15 |
| 23YN | 8 | CJ08009 | 30.78-32.96 | 3.85 | 11.1 | 3.16 | 0.68 | |
| qLA09.1 | 23CQ | 9 | satt167 | 3.55-5.99 | 3.67 | 9.5 | -1.02 | 0.02 |
| 24CQ | 9 | CJ09022 | 3.55-5.44 | 3.42 | 8.9 | 0.28 | 1.17 | |
| qLA09.2 | 22CQ | 9 | satt417 | 14.01-19.33 | 3.46 | 8.2 | 0.92 | -3.45 |
| 23YN | 9 | sat281 | 10.10-14.82 | 4.91 | 14 | 2.17 | 4.22 | |
| qLA12.1 | 22YN | 12 | CJ12008 | 6.50-22.26 | 4.49 | 11.8 | -0.47 | 0.14 |
| 23CQ | 12 | CJ12009 | 19.75-22.54 | 3.15 | 8.2 | 0.94 | 0.27 | |
| qLA19.1 | 22YN | 19 | sat245 | 91.67-94.71 | 3.36 | 9 | -0.13 | 0.60 |
| 23CQ | 19 | SWU19145 | 91.67-94.71 | 3.48 | 9 | -0.71 | -1.09 | |
| qLA20.1 | 22CQ | 20 | CJ20021 | 16.34-24.89 | 3.63 | 8.6 | -1.62 | -4.40 |
| 24CQ | 20 | CJ20021 | 16.34-20.88 | 2.59 | 6.8 | -0.72 | 0.33 | |
| qLA20.2 | 22YN | 20 | CSSR041 | 32.93-40.37 | 3.07 | 8.3 | 0.05 | 0.64 |
| 24CQ | 20 | CSSR041 | 32.93-40.37 | 3.93 | 10.1 | -0.78 | 0.51 | |
| qLNA02.1 | 22CQ | 2 | CJ02052 | 51.56-54.36 | 4.14 | 9.7 | -0.16 | -0.24 |
| 22YN | 2 | SWU02115 | 51.56-59.01 | 8.28 | 20.8 | -0.14 | -0.71 | |
| 23CQ | 2 | SWU02120 | 52.66-62.94 | 6.53 | 16.2 | -0.82 | -0.47 | |
| 24CQ | 2 | sat169 | 59.01-62.94 | 5.71 | 14.3 | -0.41 | 0.56 | |
| qLNA03.1 | 22CQ | 3 | SWU03017 | 10.38-17.16 | 3.87 | 9.1 | -0.13 | 0.32 |
| 24CQ | 3 | SSR066 | 5.89-14.07 | 3.55 | 9.2 | -0.35 | -0.50 | |
| qLNA07.1 | 23CQ | 7 | SWU07001 | 0-1.89 | 5.16 | 13.1 | 0.17 | -1.17 |
| 23YN | 7 | CJ07002 | 0.54-2.16 | 3.62 | 10.5 | 0.54 | -0.60 | |
| qLNA08.1 | 23CQ | 8 | satt378 | 90.26-96.39 | 3.97 | 10.2 | -0.68 | -0.14 |
| 23YN | 8 | CJ08041 | 90.26-96.39 | 3.17 | 9.3 | -0.59 | -0.35 | |
| qLNA10.1 | 23CQ | 10 | satt477 | 48.33-52.42 | 6.46 | 16.1 | -0.87 | -0.60 |
| 23YN | 10 | satt262 | 48.33-50.49 | 3.46 | 10.1 | -0.29 | -0.87 | |
| qLNA11.1 | 22CQ | 11 | SWU11120 | 30.27-38.80 | 4.05 | 9.5 | -0.11 | 0.29 |
| 22YN | 11 | SWU11123 | 33.33-38.80 | 3.1 | 8.3 | 0.20 | 0.36 | |
| 24CQ | 11 | SWU11123 | 32.78-38.80 | 3.21 | 8.3 | -0.39 | 0.35 | |
| qLNA17.1 | 22CQ | 17 | sat220 | 30.60-34.14 | 5.1 | 11.9 | -0.24 | 0.11 |
| 24CQ | 17 | sat220 | 30.60-34.14 | 5.16 | 13 | -0.61 | 0.02 | |
| qLNA18.1 | 22CQ | 18 | SWU18175 | 96.12-99.70 | 4.78 | 11.2 | 0.30 | 0.16 |
| 22YN | 18 | SWU18175 | 96.93-99.70 | 3.75 | 10 | 0.20 | -0.37 | |
| 24CQ | 18 | SWU18175 | 96.93-99.70 | 3.22 | 8.3 | 0.47 | -0.09 | |
| qLNA19.1 | 23CQ | 19 | SWU19012 | 8.41-18.61 | 4.01 | 10.3 | -0.60 | -0.39 |
| 24CQ | 19 | SWU19017 | 13.16-18.61 | 3.24 | 8.4 | -0.35 | -0.38 |
| QTL | Candidate gene | Homolog in Arabidopsis | Gene description |
| qPA02.1 | Glyma.02G273300 | AT3G55360 | 3-oxo-5-alpha-steroid 4-dehydrogenase family protein |
| qPA19.1 | Glyma.19G170100 | AT5G43940 | GroES-like zinc-binding dehydrogenase family protein |
| qSA14.1 | Glyma.14G173500 | AT1G55020 | lipoxygenase 1 |
| qLNA02.1 | Glyma.02G203300 | AT4G30950 | fatty acid desaturase 6 |
| Glyma.02G227200 | AT5G05580 | fatty acid desaturase 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





