Submitted:
02 July 2025
Posted:
03 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
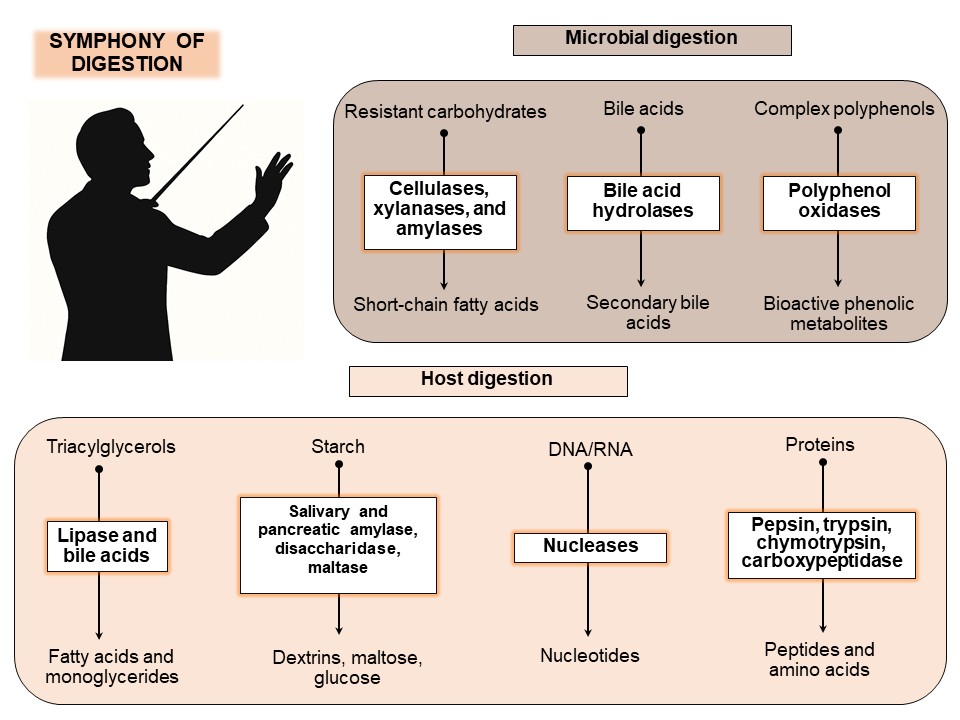
2. The Enzymatic Roles of the Host and the Gut Microbiota
2.1. Host Digestive Enzymes
2.2. Microbial Digestive Enzymes
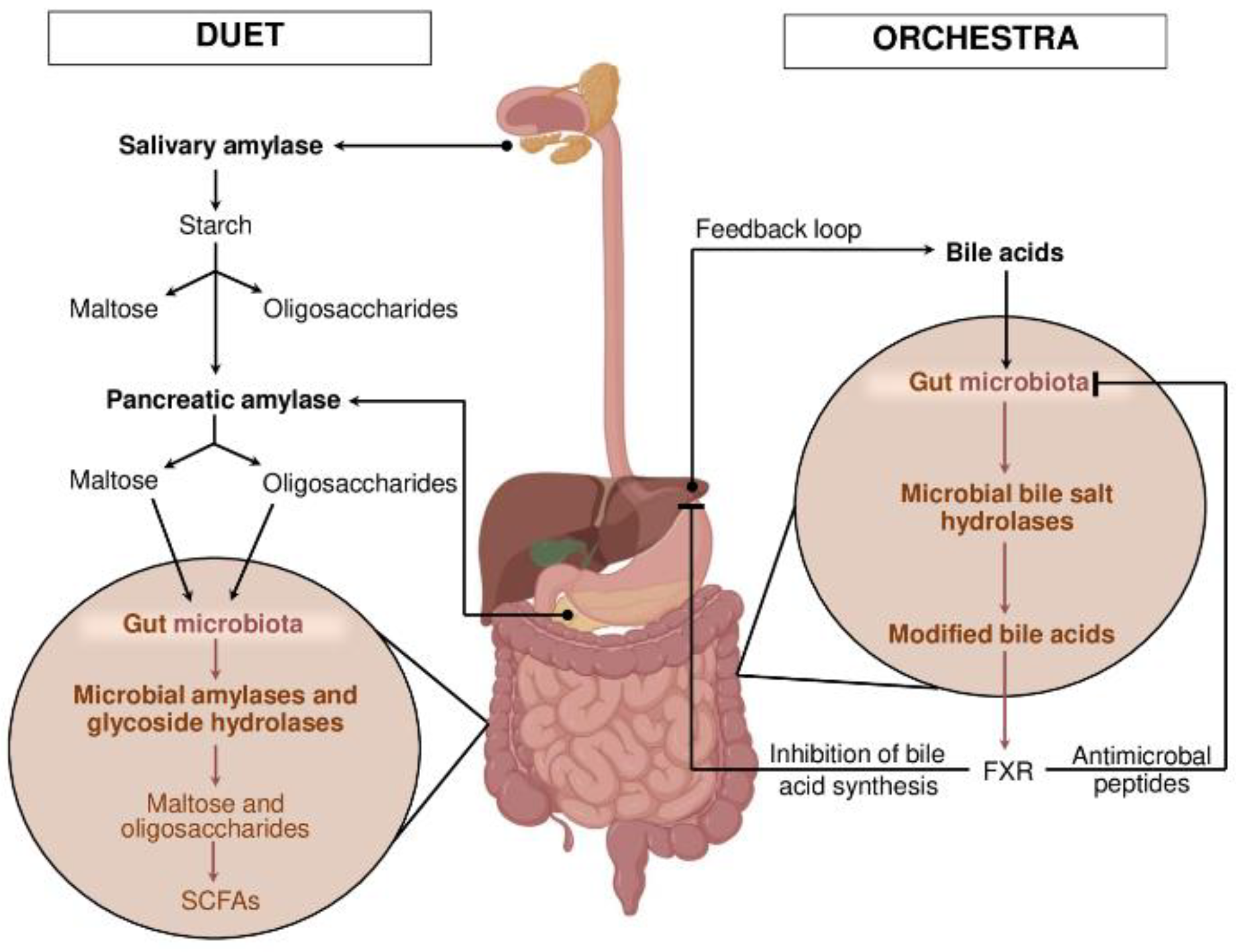
3. Models of Cooperation: Duet or Orchestra?
4. Disruption of Enzymatic Harmony: Dysbiosis and Disease
5. Therapeutic Perspectives and Future Directions
6. Some Poorly Covered or Overlooked Points
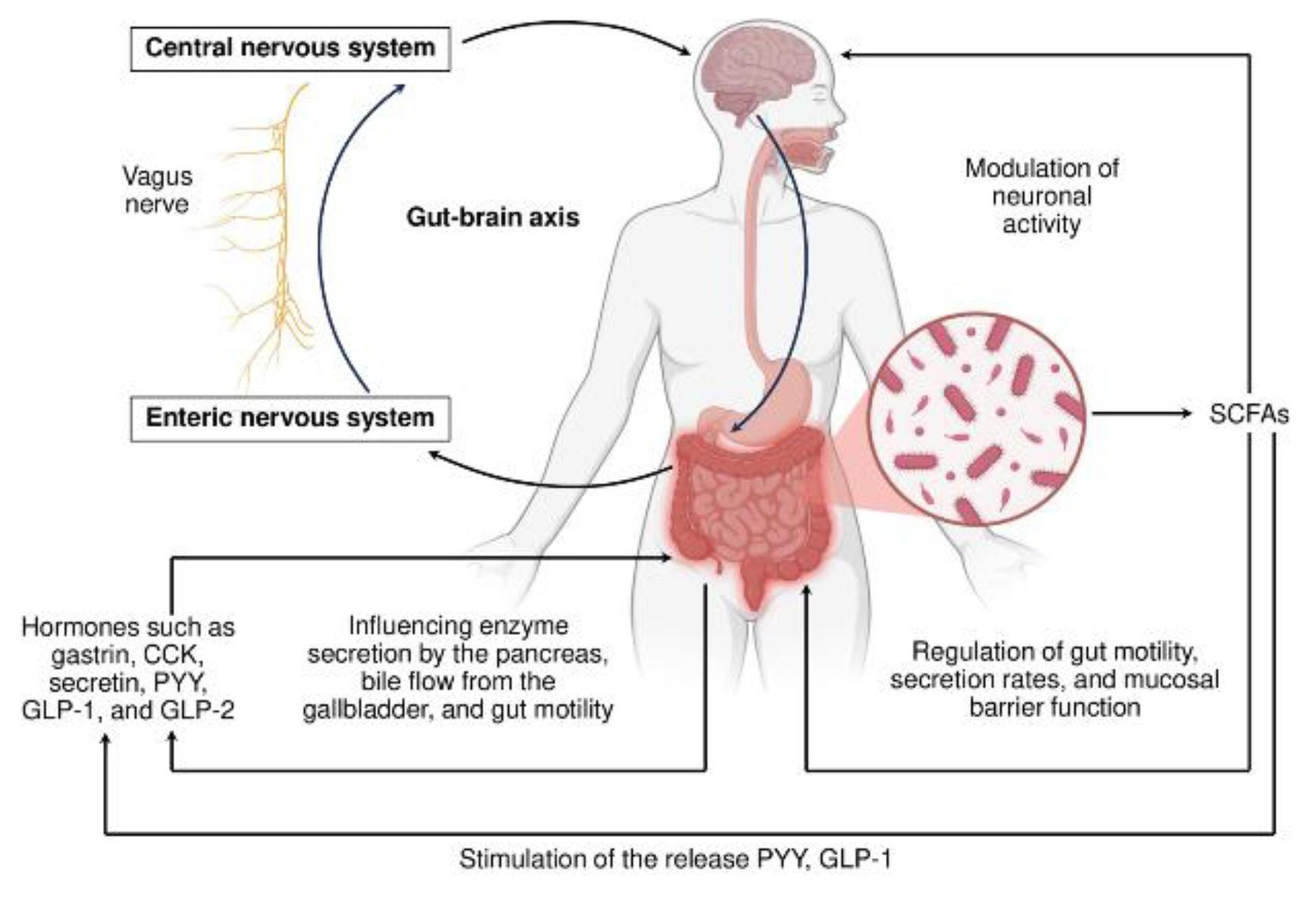
7. Orchestrating Host-Gut microbiota Cooperation: a System Conducting
8. Conclusions and Perspectives
Funding
Acknowledgements
Abbreviations
References
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut Microbiota Functions: Metabolism of Nutrients and other Food Components. Eur J Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, X.; Li, L.; Zhong, C.; Zhang, Y.; Yang, X.; Li, M.; Yang, C. The Role of Gut Microbiota in Intestinal Disease: From an Oxidative Stress Perspective. Front. Microbiol. 2024, 15, 1328324. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Mei, L.; Li, Y.; Guo, Y.; Yang, B.; Huang, Z.; Li, Y. Enzymatic Regulation of the Gut Microbiota: Mechanisms and Implications for Host Health. Biomolecules 2024, 14, 1638. [Google Scholar] [CrossRef] [PubMed]
- Karasov, W. H.; Diamond, J. M. Interplay between Physiology and Ecology in Digestion: Intestinal Nutrient Transporters Vary within and between Species According to Diet. BioScience 1988, 38, 602–611. [Google Scholar] [CrossRef]
- Clark, J. A.; Coopersmith, C. M. Intestinal Crosstalk: a New Paradigm for Understanding the Gut as the "Motor" of Critical Illness. Shock 2007, 28, 384–393. [Google Scholar] [CrossRef]
- Gregor, R.; Probst, M.; Eyal, S.; Aksenov, A.; Sasson, G.; Horovitz, I.; Dorrestein, P. C.; Meijler, M. M.; Mizrahi, I. Mammalian Gut Metabolomes Mirror Microbiome Composition and Host Phylogeny. ISME J. 1262. [Google Scholar] [CrossRef]
- Deehan, E. C.; Duar, R. M.; Armet, A. M.; Perez-Muñoz, M. E.; Jin, M.; Walter, J. Modulation of the Gastrointestinal Microbiome with Nondigestible Fermentable Carbohydrates to Improve Human Health. Microbiol Spectr. 2017, 5, 10.1128. [Google Scholar] [CrossRef]
- de Vos, W. M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut Microbiome and Health: Mechanistic Insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Shabani, M.; Ghoshehy, A.; Mottaghi, A. M.; Chegini, Z.; Kerami, A.; Shariati, A.; Taati Moghadam, M. The Relationship between Gut Microbiome and Human Diseases: Mechanisms, Predisposing Factors and Potential Intervention. Front Cell Infect Microbiol. 2025, 15, 1516010. [Google Scholar] [CrossRef]
- Sensoy, I. A Review on the Food Digestion in the Digestive Tract and the Used in vitro Models. Curr Res Food Sci. 2021, 4, 308–319. [Google Scholar] [CrossRef]
- Lu, V. B.; Gribble, F. M.; Reimann, F. Nutrient-Induced Cellular Mechanisms of Gut Hormone Secretion. Nutrients. 2021, 13, 883. [Google Scholar] [CrossRef]
- Cho, H.; Lim, J. The Emerging Role of Gut Hormones. Mol Cells. 2024, 47, 100126. [Google Scholar] [CrossRef] [PubMed]
- Flint, H. J.; Scott, K. P.; Duncan, S. H.; Louis, P.; Forano, E. Microbial Degradation of Complex Carbohydrates in the Gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Wardman, J. F.; Bains, R. K.; Rahfeld, P.; Withers, S. G. Carbohydrate-Active Enzymes (CAZymes) in the Gut Microbiome. Nat Rev Microbiol. 2022, 20, 542–556. [Google Scholar] [CrossRef] [PubMed]
- Diether, N. E.; Willing, B. P. Microbial Fermentation of Dietary Protein: An Important Factor in Diet–Microbe–Host Interaction. Microorganisms. [CrossRef]
- Chand, D.; Avinash, V. S.; Yadav, Y.; Pundle, A. V.; Suresh, C.G.; Ramasamy, S. Molecular Features of Bile Salt Hydrolases and Relevance in Human Health. Biochim Biophys Acta Gen Subj. 2017, 1861(1 Pt A), 2981–2991. [Google Scholar] [CrossRef]
- Dong, Z.; Yang, S.; Tang, C.; Li, D.; Kan, Y.; Yao, L. New Insights into Microbial Bile Salt Hydrolases: from Physiological Roles to Potential Applications. Front Microbiol. 2025, 16, 1513541. [Google Scholar] [CrossRef]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D. J.; Bakker, B. M. The Role of Short-Chain Fatty Acids in the Interplay between Diet, Gut Microbiota, and Host Energy Metabolism. J Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Degnan, P. H.; Taga, M. E.; Goodman, A. L. Vitamin B12 as a Modulator of Gut Microbial Ecology Cell Metab. 2014, 20, 769–778. 20. [CrossRef]
- Degnan, P. H.; Barry, N. A.; Mok, K. C.; Taga, M. E.; Goodman, A. L. Human Gut Microbes Use Multiple Transporters to Distinguish Vitamin B₁₂ Analogs and Compete in the Gut. Cell Host Microbe 2014, 15, 47–57. [Google Scholar] [CrossRef]
- Sorboni, S. G.; Moghaddam, H. S.; Jafarzadeh-Esfehani, R.; Soleimanpour, S. A Comprehensive Review on the Role of the Gut Microbiome in Human Neurological Disorders. Clin Microbiol Rev. 2022, 35, e0033820. [Google Scholar] [CrossRef]
- Liu, J.; Tan, Y.; Cheng, H.; Zhang, D.; Feng, W/; Peng, C. Functions of Gut Microbiota Metabolites Current Status and Future Perspectives. Aging Dis. 2022, 13, 1106–1126. [Google Scholar] [CrossRef]
- Dmytriv, T. R.; Lushchak, O.; Lushchak, V. I. Glucoraphanin Conversion into Sulforaphane and Related Compounds by Gut Microbiota. Front Physiol. 2025, 16, 1497566. [Google Scholar] [CrossRef]
- Stevens, J. F.; Maier, C. S. The Chemistry of Gut Microbial Metabolism of Polyphenols. Phytochem Rev. 2016, 15, 425–444. [Google Scholar] [CrossRef] [PubMed]
- Gasaly, N.; Gotteland, M. Interference of Dietary Polyphenols with Potentially Toxic Amino Acid Metabolites Derived from the Colonic Microbiota. Amino Acids. 2022, 54, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Larabi, A.B.; Masson, H. L. P.; Bäumler, A. J. Bile Acids as Modulators of Gut Microbiota Composition and Function. Gut Microbes. 2023, 15, 2172671. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Yang, S.; Tang, C.; Li, D.; Kan, Y.; Yao, L. New Insights into Microbial Bile Salt Hydrolases: from Physiological Roles to Potential Applications. Front Microbiol. 2025, 16, 1513541. [Google Scholar] [CrossRef]
- Brito Rodrigues, P.; de Rezende Rodovalho, V.; Sencio, V.; Benech, N.; Creskey, M.; Silva Angulo, F.; Delval, L.; Robil, C.; Gosset, P.; Machelart, A.; Haas, J.; Descat, A.; Goosens; F. ; Beury, D.; Maurier, F.; Hot, D.; Wolowczuk, I.; Sokol, H.; Zhang, X.; Ramirez Vinolo, M. A.; Trottein, F. Integrative Metagenomics and Metabolomics Reveal Age-Associated Gut Microbiota and Metabolite Alterations in a Hamster Model of COVID-19. Gut Microbes. 2025, 17, 2486511. [Google Scholar] [CrossRef]
- Kundu, P.; Lee, H. U.; Garcia-Perez, I.; Tay, E. X. Y.; Kim, H.; Faylon, L. E.; Martin, K.A.; Purbojati, R.; Drautz-Moses, D. I.; Ghosh, S.; Nicholson, J. K. , Schuster, S., Holmes, E.; Pettersson, S. Neurogenesis and Prolongevity Signaling in Young Germ-Free Mice Transplanted with the Gut Microbiota of Old Mice. Sci Transl Med. 2019, 11, eaau4760. [Google Scholar] [CrossRef]
- Darnaud, M.; De Vadder, F.; Bogeat, P.; Boucinha, L.; Bulteau, A. L.; Bunescu, A.; Couturier, C.; Delgado, A.; Dugua, H.; Elie, C.; Mathieu, A. , Novotná, T.; Ouattara, D. A.; Planel, S.; Saliou, AA; Šrůtková, D.; Yansouni, J.; Stecher, B.; Schwarzer, M.; Leulier, F.; Tamellini, A. A Standardized Gnotobiotic Mouse Model Harboring a Minimal 15-Member Mouse Gut Microbiota Recapitulates SOPF/SPF Phenotypes. Nat Commun. 2021, 12, 6686. [Google Scholar] [CrossRef]
- Jans, M.; Vereecke, L. A Guide to Germ-Free and Gnotobiotic Mouse Technology to Study Health and Disease. FEBS J. 2025, 292, 1228–1251. [Google Scholar] [CrossRef]
- Haran, J.P.; McCormick, B.A. Aging, Frailty, and the Microbiome-How Dysbiosis Influences Human Aging and Disease. Gastroenterology 2021, 160, 507–523. [Google Scholar] [CrossRef]
- Dmytriv, T. R.; Lushchak, V. I. Gut Microbiome as a Target for Anti-Ageing Interventions. Subcell Biochem. 2024, 107, 307–325. [Google Scholar] [CrossRef]
- Di Vincenzo, F.; Del Gaudio, A.; Petito, V.; Lopetuso, L. R.; Scaldaferri, F. Gut Mmicrobiota, Intestinal Permeability, and Systemic Inflammation: a Narrative Review. Intern Emerg Med. 2024, 19, 275–293. [Google Scholar] [CrossRef] [PubMed]
- Cusumano, G.; Flores, G. A.; Venanzoni, R.; Angelini, P. The Impact of Antibiotic Therapy on Intestinal Microbiota: Dysbiosis Antibiotic Resistance, and Restoration Strategies. Antibiotics (Basel) 2025, 14, 371. [Google Scholar] [CrossRef] [PubMed]
- Colquhoun, C.; Duncan, M.; Grant, G. Inflammatory Bowel Diseases: Host-Microbial-Environmental Interactions in Dysbiosis. Diseases 2020, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Cronin, P.; Joyce, S. A.; O'Toole, P. W.; O'Connor, E. M. Dietary Fibre Modulates the Gut Microbiota. Nutrients 2021, 13, 1655. [Google Scholar] [CrossRef]
- Dmytriv, T. R.; Storey, K. B.; Lushchak, V. I. Intestinal Barrier Permeability: the Influence of Gut Microbiota, Nutrition, and exercise. Front. Physiol. 2024, 15, 1380713. [Google Scholar] [CrossRef]
- Aggarwal, N.; Kitano, S.; Puah, G. R. Y.; Kittelmann, S.; Hwang, I.Y.; Chang, M. W. Microbiome and Human Health: Current Understanding, Engineering, and Enabling Technologies. Chem Rev. 2023, 123, 31–72. [Google Scholar] [CrossRef]
- Abouelela, M. E.; Helmy, Y. A. Next-Generation Probiotics as Novel Therapeutics for Improving Human Health: Current Trends and Future Perspectives. Microorganisms 2024, 12, 430. [Google Scholar] [CrossRef]
- Lalowski, P.; Zielińska, D. The Most Promising Next-Generation Probiotic Candidates—Impact on Human Health and Potential Application in Food Technology. Fermentation 2024, 10, 444. [Google Scholar] [CrossRef]
- Kumar, A.; Green, K. M.; Rawat, M. A Comprehensive Overview of Postbiotics with a Special Focus on Discovery Techniques and Clinical Applications. Foods 2024, 13, 2937. [Google Scholar] [CrossRef]
- Hijová, E. Postbiotics as Metabolites and Their Biotherapeutic Potential. Int J Mol Sci. 2024, 25, 5441. [Google Scholar] [CrossRef]
- Kasahara, K.; Kerby, R. L.; Zhang, Q.; Pradhan, M.; Mehrabian, M.; Lusis, A. J.; Bergström, G.; Bäckhed, F.; Rey, F. E. Gut Bacterial Metabolism Contributes to Host Global Purine Homeostasis. Cell Host Microbe 2023, 31, 1038–1053.e10. [Google Scholar] [CrossRef] [PubMed]
- Grove, A. The Delicate Balance of Bacterial Purine Homeostasis. Discov Bact 2025, 2, 14. [Google Scholar] [CrossRef]
- Morrison, D. J.; Preston, T. Formation of Short Chain Fatty Acids by the Gut Microbiota and Their Impact on Human Metabolism. Gut Microbes. 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Kawai, T.; Akira, S. Microbial Sensing by Toll-like Receptors and Intracellular Nucleic Acid Sensors. Cold Spring Harb Perspect Biol. 2014, 7, a016246. [Google Scholar] [CrossRef]
- Gil, A. Modulation of the Immune Response Mediated by Dietary Nucleotides. Eur J Clin Nutr. 2002, 56 (Suppl 3), S1–4. [Google Scholar] [CrossRef]
- Swidsinski, A.; Ladhoff, A.; Pernthaler, A.; Swidsinski, S.; Loening-Baucke, V.; Ortner, M.; Weber, J. , Hoffmann, U., Schreiber, S., Dietel, M., Lochs, H. Mucosal Flora in Inflammatory Bowel Disease. Gastroenterology 2002, 122, 44–54. [Google Scholar] [CrossRef]
- Wall, R.; Ross, R.P.; Ryan, C.A.; Hussey, S.; Murphy, B.; Fitzgerald, G. F.; Stanton, C. Role of Gut Microbiota in Early Infant Development. Clin Med Pediatr. 2009, 3, 45–54. [Google Scholar] [CrossRef]
- Asano, K.; Yoshimura, S.; Nakane, A. Alteration of Intestinal Microbiota in Mice Orally Administered with Salmon Cartilage Proteoglycan, a Prophylactic Agent. PLoS One 2013, 8, e75008. [Google Scholar] [CrossRef]
- Yang, B.; Guo, X.; Shi, C.; Liu, G.; Qin, X.; Chen, S.; Gan, L.; Liang, D.; Shao, K.; Xu, R.; Zhong, J.; Mo, Y.; Li, H.; Luo, D. Alterations in Purine and Pyrimidine Metabolism Associated with Latent Tuberculosis Infection: Insights from Gut Microbiome and Metabolomics Analyses. mSystems 2024, 9, e0081224. [Google Scholar] [CrossRef]
- Semchyshyn, H.; Lushchak, V.; Storey, K. Possible Reasons for Difference in Sensitivity to Oxygen of Two Escherichia coli Strains. Biochemistry (Mosc) 2005, 70, 424–431. [Google Scholar] [CrossRef]
- Vasylkovska, R.; Petriv, N.; Semchyshyn, H. Carbon Sources for Yeast Growth as a Precondition of Hydrogen Peroxide Induced Hormetic Phenotype. Int J Microbiol. 2015, 2015, 697813. [Google Scholar] [CrossRef] [PubMed]
- Bayliak, M.; Semchyshyn, H.; Lushchak, V. Possible Accumulation of Non-Active Molecules of Catalase and Superoxide Dismutase in S. cerevisiae Cells under Hydrogen Peroxide Induced Stress. Cent. Eur. J. Biol. 2007, 2, 326–336. [Google Scholar] [CrossRef]
- Mayer, E. A. Gut Feelings: the Emerging Biology of Gut-Brain Communication. Nat Rev Neurosci. 2011, 12, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Furness, J.B. The Enteric Nervous System and Neurogastroenterology. Nat Rev Gastroenterol Hepatol. 2012, 9, 286–294. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-Generated Metabolites Promote Metabolic Benefits via Gut-Brain Neural Circuits. Cell. [CrossRef]
- Nwako, J. G.; Patel, S. D.; Roach, T. J.; Gupte, S. R.; Williams, S. G.; Riedman, A. M.; McCauley, H. A. Enteroendocrine Cells Regulate Intestinal Barrier Permeability. Am J Physiol Cell Physiol. 2025, 328, C1501–C1508. [Google Scholar] [CrossRef]
- Jyoti & Dey, P. Mechanisms and Implications of the Gut Microbial Modulation of Intestinal Metabolic Processes. npj Metab Health Dis. 2025, 3. [Google Scholar] [CrossRef]
- Cho, H.; Lim, J. The Emerging Role of Gut Hormones. Mol Cells 2024, 47, 100126. [Google Scholar] [CrossRef]
- Ramadan, Y. N.; Alqifari, S. F.; Alshehri, K.; Alhowiti, A.; Mirghani, H.; Alrasheed, T.; Aljohani, F.; Alghamdi, A.; Hetta, H. F. Microbiome Gut-Brain-Axis: Impact on Brain Development and Mental Health. Mol Neurobiol. [CrossRef]
- Kolodziejczyk, A. A.; Zheng, D.; Elinav, E. Diet–Microbiota Interactions and Personalized Nutrition. Nat. Rev. Microbiol. 2019, 17, 742–753. [Google Scholar] [CrossRef]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef]
| Scheme | Host-Derived Enzymes | Microbial Enzymes | Primary Function |
|---|---|---|---|
| Starches and simple carbohydrates | Salivary amylase, pancreatic α-amylase | Amylases, pullulanases, maltases, isomaltases (CAZymes) | Hydrolysis of starch and oligosaccharides into monosaccharides |
| Non-digestible polysaccharides | None | Cellulases, hemicellulases, xylanases, pectinases | Fermentation of dietary fibers; SCFA production |
| Disaccharides | Sucrase, maltase, lactase (brush-border enzymes) | β-Galactosidase, β-fructofuranosidase | Breakdown of sucrose, maltose, and lactose into monosaccharides |
| Proteins and peptides | Pepsin, trypsin, chymotrypsin, peptidases | Microbial proteases, peptidases | Peptide hydrolysis; bioactive amine generation |
| Fats and lipids | Gastric and pancreatic lipases, phospholipases | Lipases, esterases | Hydrolysis of triglycerides and other lipids; production of microbial lipid metabolites |
| Bile acids | Liver secretion and reabsorption control | Bile salt hydrolases, hydroxysteroid dehydrogenases | Deconjugation and transformation of bile acids; host–microbiome signaling |
| Polyphenols and phytochemicals | Limited phase I/II metabolism (e.g., glucuronidation) | Glycosidases, esterases, decarboxylases | Microbial conversion into bioavailable phenolic compounds |
| Xenobiotics and drugs | Cytochrome P450s, conjugating enzymes (UGT, SULT, etc.) | Reductases, azoreductases, β-glucuronidases | Biotransformation, reactivation, or detoxification of xenobiotics |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




