1. Introduction
The field of developmental biology seeks to unravel the complex molecular mechanisms governing organismal growth, differentiation, and morphogenesis. Central to these processes is the dynamic regulation of RNA molecules, which serve as key mediators of gene expression, cellular signaling, and functional adaptation during development (Barta and Jantsch, 2017; Morris and Mattick, 2014; Rosa et al., 2021). The ability to track and analyze RNA dynamics in real-time has become increasingly important for understanding how spatiotemporal gene expression patterns shape developmental outcomes. However, traditional approaches for studying RNA function, such as in situ hybridization, RNA sequencing, and immunostaining, often provide only static or endpoint measurements, limiting their utility in capturing the full complexity of RNA-mediated processes. To overcome these challenges, RNA biosensors have emerged as powerful tools with the potential to revolutionize research in developmental biology.
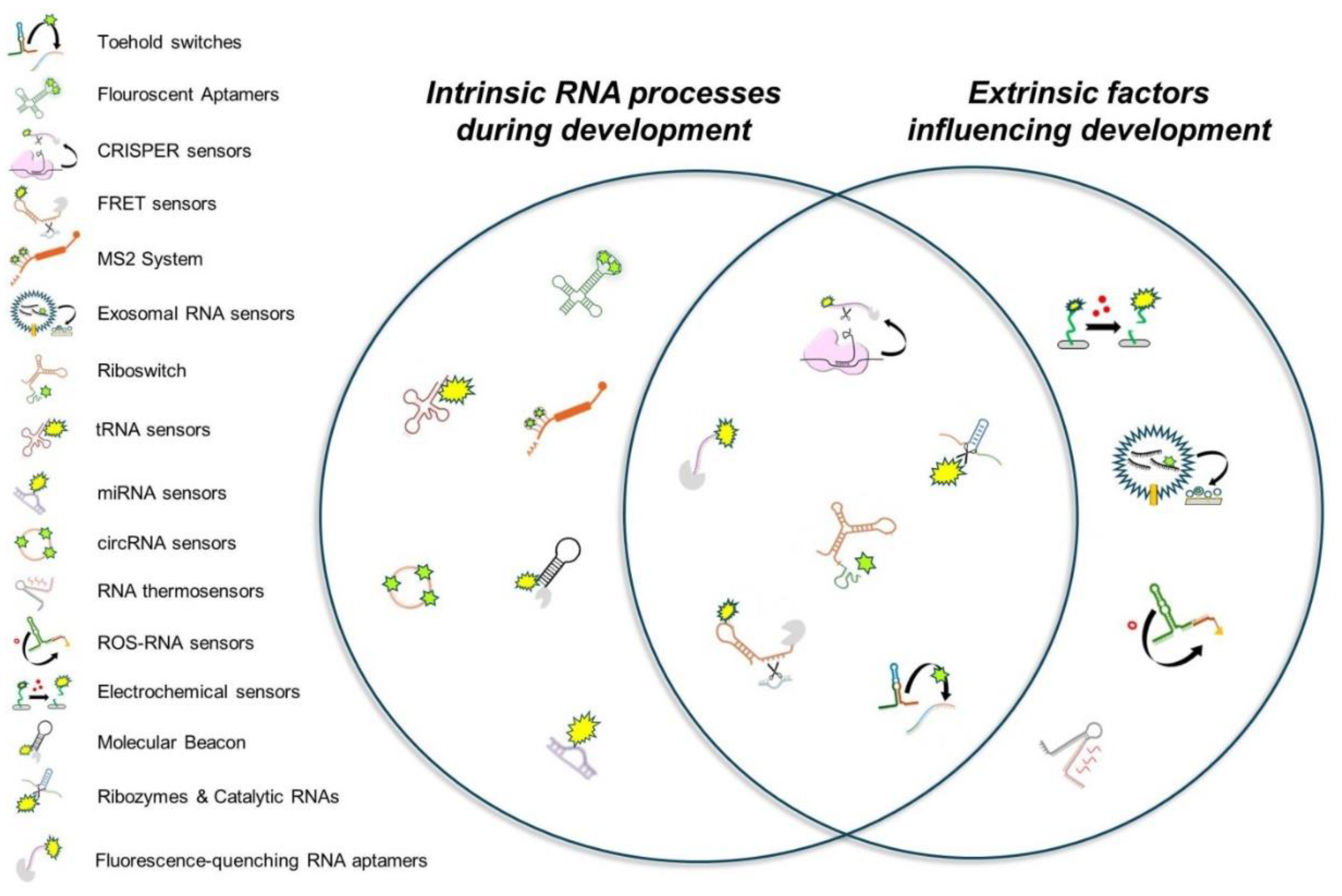
RNA biosensors are highly specialized molecular tools designed to detect, monitor, and analyze RNA in live cells and organisms (Halstead et al., 2015; Su and Hammond, 2020). These sensors employ a range of mechanisms, including fluorescent aptamers, CRISPR-Cas-based systems, riboswitches, and catalytic RNA elements, to provide real-time, sequence-specific, and context-dependent insights into RNA localization, modifications, and interactions. In addition to tracking intrinsic aspects of RNA biology during development (such as expression, localization, splicing and stability), RNA-based biosensors can also be engineered to study extrinsic physical and chemical factors (e.g., temperature, pH, redox state, mechanical stress) that influence RNA behavior and developmental outcomes (
Figure 1). While these technologies have been extensively explored in fields such as synthetic biology, diagnostics, and environmental monitoring (Jones et al., 2019; Mikaeeli Kangarshahi et al., 2024; Palchetti and Mascini, 2008), they have seen comparatively fewer applications in developmental biology to date. This gap is particularly notable given the growing recognition that RNA dynamics play a pivotal role in cellular differentiation, tissue patterning, and morphogenetic signaling.
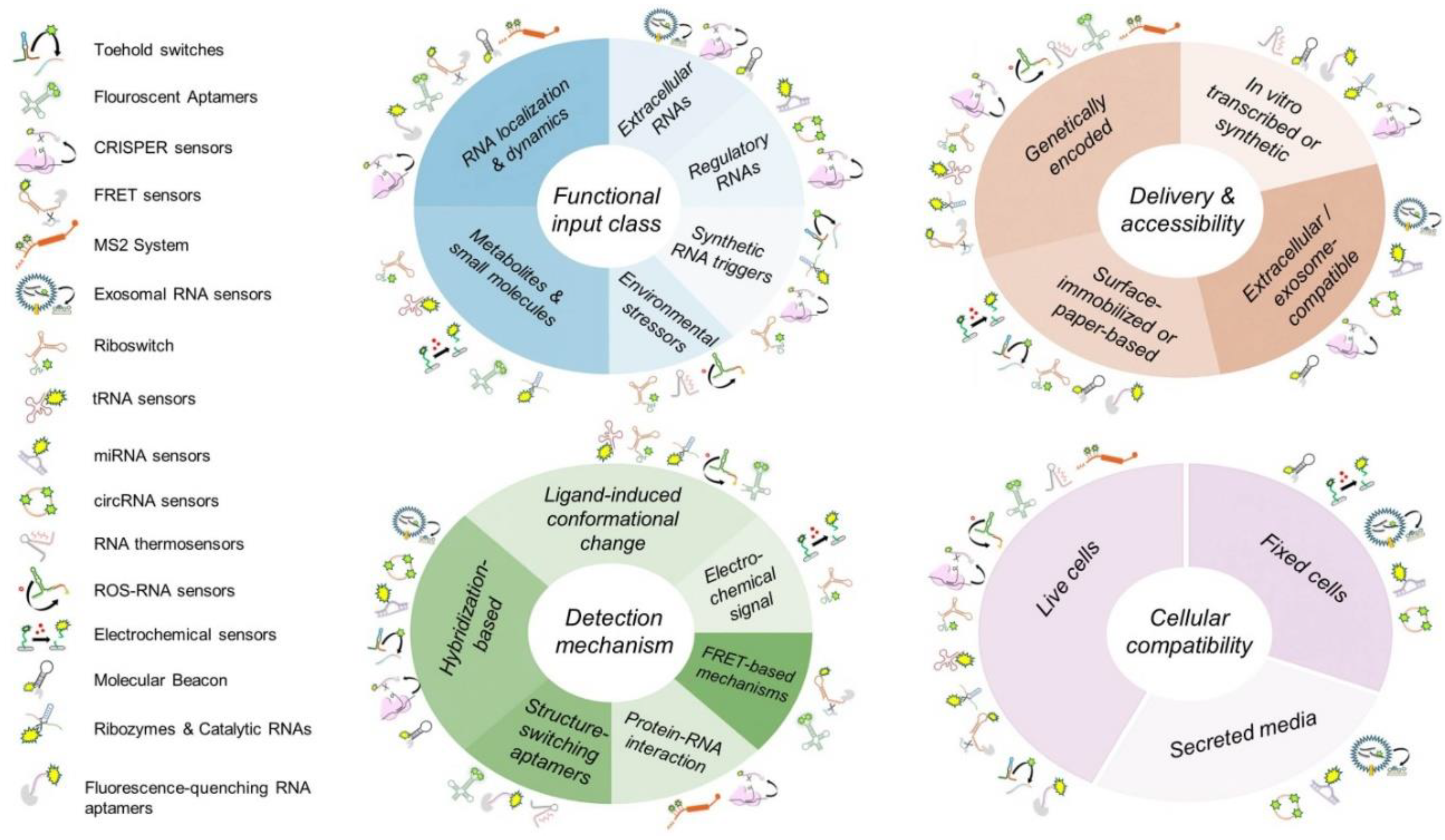
By integrating RNA biosensors into developmental biology research, scientists can gain unprecedented insights into how RNA molecules influence cell fate decisions, regulatory networks, and the orchestration of developmental transitions, as well as how internal and external factors affect RNA behavior during development. The adaptability and specificity of these biosensors make them invaluable tools for addressing fundamental questions about gene regulation, RNA-protein interactions, and cellular signaling cascades during embryogenesis and organogenesis. One of the key challenges is the need for optimized biosensor designs that are compatible with in vivo developmental systems, such as model organisms and organoid cultures. Moreover, considerations regarding biosensor sensitivity, specificity, and minimal perturbation of endogenous biological processes must be carefully addressed to ensure their effective application in developmental research. By highlighting the potential of RNA biosensors and their broad applicability in developmental biology, this review aims to bridge the gap between these emerging technologies and their transformative impact on studying RNA dynamics in living systems. In the following sections, we provide an overview of commonly used RNA biosensors, categorizing them by their mechanisms of action and applications (summarized in
Figure 2). We discuss their strengths and limitations, as well as their potential for advancing developmental biology research (also summarized in
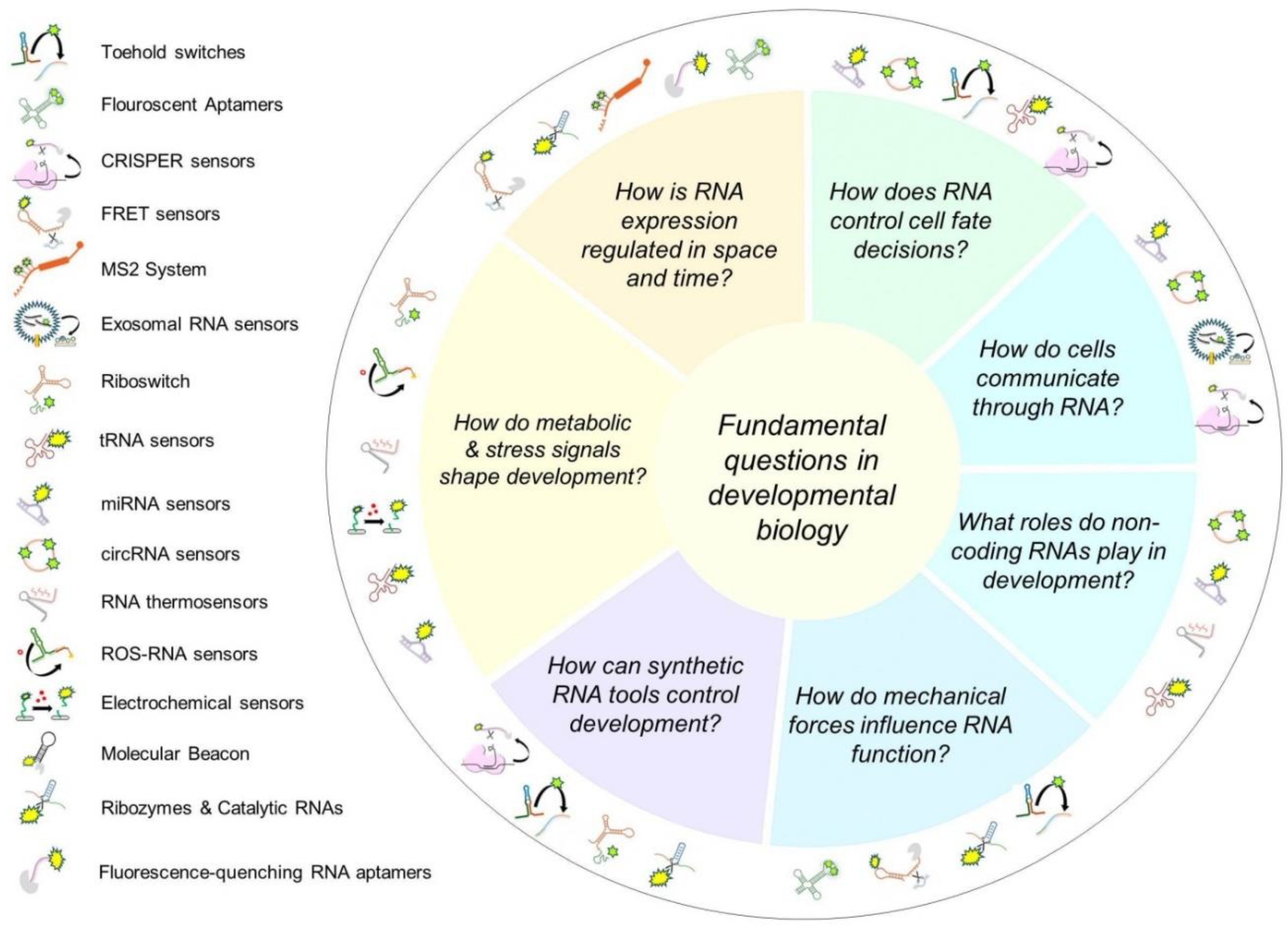
Figure 3 and
Table 1). By revisiting and adapting these tools, we emphasize the necessity of incorporating RNA biosensors into developmental biology studies to gain deeper insights into the molecular and cellular mechanisms that drive organismal development.
2. Fluorescent-Based Sensors
2.1. Fluorescent RNA Aptamers
The real-time monitoring of intracellular RNA dynamics in living cells has been challenging. To address this matter, scientists have used specific aptamers, which are known as synthetic nucleic acid oligos (typically less than 100 nucleotides) selected during a process so called systematic evolution of ligands by exponential enrichment (SELEX). These aptamers bind to specific molecules with high affinity (Wu and Kwon, 2016). In this case, the target molecules are fluorophores in their inactive state. Upon binding to the aptamer, the fluorophore stabilizes, and the aptamer-target complex lights up and emits high-intensity fluorescence. Since the sequence of fluorescent RNA aptamers is determined during the SELEX process, they can be utilized by transfecting cells with plasmids encoding aptamer-tagged RNA, followed by the addition of the target small molecule to enable fluorescence (Shui et al., 2012). Fluorescent RNA aptamers are among the most commonly used biosensors in RNA research. They are named after vegetables and fruits (e.g. Spinach, Broccoli, Mango, and Pepper) because their fluorescence properties resemble those of naturally occurring colors (Zhou and Zhang, 2022). There are specific fluorophores used for each fluorescent RNA aptamers such as DFHBI for Spinach or TO1-Biotin for Mango, enabling real-time visualization of RNA in live cells (Neubacher and Hennig, 2019). Broccoli offers improved fluorescence stability and has less cytotoxicity compared to Spinach, while Mango and Pepper provide distinct spectral properties and enhanced fluorescence brightness. These aptamers are widely used for studying RNA localization, dynamics, and real-time expression patterns in living systems, making them essential tools in cell biology and synthetic biology (Zhou and Zhang, 2022). To enhance their utility in developmental biology studies, these fluorescent RNA aptamers can be engineered to specifically bind developmental RNAs. Furthermore, incorporating fluorophores with distinct spectral properties enables multiplex high-resolution imaging (Sunbul et al., 2021), allowing simultaneous tracking of multiple RNAs involved in different developmental pathways. A notable example is a recently developed genetically encoded sensor based on a fluorogenic allosteric aptamer (FaApt), which enables sensitive imaging of the localization and dynamics of RNA targets in live cells during zebrafish embryonic development (Peng et al., 2023).
2.2. Fluorescence-Quenching RNA Aptamers
Due to the limited number of fluorophore-binding RNA aptamers, fluorescence-quenching RNA aptamers were developed. These platforms consist of a quencher-binding aptamer that functions independently of the fluorophore structure and facilitates multiplex RNA monitoring in living systems. When an RNA aptamer specifically binds to a quencher molecule (e.g., dinitroaniline), the fluorescent response is either enhanced or suppressed by modulating the fluorophore-quencher interaction. For instance, if the quencher is directly conjugated to fluorophore molecules, it forms a non-fluorescent molecular complex. When the aptamer binds to the quencher, it dissociates from the fluorophore, leading to an increase in fluorescence. Recent advances in fluorescence-quenching RNA aptamer technologies have expanded their potential for developmental biology applications. For instance, the RhoBAST:SpyRho system offers enhanced brightness and stability for tracking RNA localization during embryogenesis (Englert et al., 2023), while the Squash:DFQL-1T complex enables in vivo RNA imaging in mammalian models and could be applied to monitor gene expression during tissue differentiation (Chen et al., 2025).
Alternatively, some designs rely on aptamer conformational changes induced by external environmental factors, such as pH or ionic concentrations, which alter binding affinity for the quencher and result in fluorescence modulation (Arora et al., 2015). These aptamers are not primarily used to probe RNA biology itself (such as splicing or localization), but rather to sense biochemical and physiological signals that may affect or accompany developmental processes. Their high sensitivity and reversibility make them suitable for monitoring intracellular and environmental changes (e.g., pH or ions), or tracking metabolite levels in living cells (X. Liu et al., 2022; Wang et al., 2013). They have been applied in studies of metabolism, cell stress responses, and environmental sensing, all of which can intersect with developmental outcomes. These aptamers can be modified to detect specific molecular flux, such as neurotransmitters (Moraldo et al., 2022), that influence developmental processes. When used alongside other RNA sensors, coupling them with multi-channel imaging systems enables simultaneous monitoring of extrinsic factors and genetic programs driving development in species with high environmental sensitivity (Kudłak and Wieczerzak, 2020).
2.3. Molecular Beacon Biosensors
Molecular beacons (MBs) are hairpin-shaped RNA or DNA probes designed to fluoresce when hybridized to a specific target RNA sequence (Bidar et al., 2021). Their hairpin structure keeps the fluorophore and quencher at close proximity, suppressing fluorescence until the target RNA binds, separating the two and restoring fluorescence. These biosensors are highly sensitive and specific, making them ideal for applications such as real-time PCR, in situ hybridization, and RNA quantification in diagnostics and molecular biology (Bidar et al., 2021; Kuang et al., 2017). Molecular beacons can be customized to detect lineage-specific RNAs (Wile et al., 2014) or co-expressed developmental regulators as established for tracking endogenous maternal mRNAs in live Drosophila melanogaster egg chambers (Catrina et al., 2019). Furthermore, MB-based sensors have been engineered for chronological visualization of cell proliferation (Murata et al., 2021) and differentiation in developing mammalian tissues (Kang et al., 2011). Addition of photo-stable fluorescent dyes can improve their performance during long-term developmental studies (Nesterova et al., 2009).
2.4. FRET-Based RNA Sensors
Förster resonance energy transfer (FRET)-based RNA sensors utilize energy transfer between two fluorescent molecules working as a donor-acceptor pair, which are attached to RNA to detect conformational changes or interactions (Tomescu et al., 2014). It is worth noting that the acceptor exhibits fluorescence activity only when the donor is in close proximity typically within the range of 1 – 10 nm (Verma et al., 2023). When the RNA undergoes conformational changes, the distance between the donor-acceptor fluorophore pair shifts, altering the FRET signal (Trachman et al., 2020). These sensors are powerful tools for studying biomolecular interactions at extremely short distances, typically below 100 nm, making them more advantageous than conventional methods (Verma et al., 2023). They are primarily used in structural biology to study RNA folding, ribozyme activity, and RNA-protein interactions, providing real-time insights into dynamic molecular processes (Pitchiaya et al., 2014). FRET-based sensors can be adapted to detect conformational changes in RNAs that mediate developmental processes or evolve structural roles across species. Probes can be fine-tuned to monitor the dynamics of RNA-protein interactions critical for developmental transitions. These applications fall within the category of intrinsic RNA biology. However, in a different context, FRET sensors can also be engineered to detect extrinsic biophysical cues, such as mechanical forces, by linking donor-acceptor pairs to mechanically responsive RNA structures or associated molecules. Such sensors can also be engineered with donor-acceptor pairs optimized for deep-tissue imaging (Trachman et al., 2020), as developed for mechanical measurements in embryonic tissues (Loffet et al., 2023). For example, FRET-based approaches have been repurposed to study external factors such as mechanical tension in embryonic patterning in frogs (Eroshkin et al., 2024) and zygotic genome activation in evolutionary diverged ascidians (Wei et al., 2024), using custom-engineered RNA-FRET hybrids. It is important to note that these represent conceptually distinct applications of FRET technology: one focused on RNA-level biology, and the other on sensing biophysical influences on developmental processes.
3. Enzymatic and Electrochemical Sensors
3.1. Catalytic RNA Biosensors
Catalytic RNA biosensors, or ribozymes, are natural or artificially engineered RNA molecules which are able to catalyze biochemical reactions, such as self-cleavage or substrate cleavage, in response to specific ligands (Li et al., 2023). These biosensors are used for detecting small molecules such as ATP, GTP, or environmental pollutants (Frommer et al., 2015). By coupling ribozyme activity to a detectable signal (e.g., fluorescence or color change), they provide a versatile biosensing platform for both fundamental research and practical applications in environmental studies. In developmental biology research, catalytic ribozymes can be modified not only to sense specific metabolites (Winkler et al., 2004) or track the distribution of target molecules in tissues (Osman et al., 2024) but also to efficiently manipulate gene expression in a spatiotemporal manner, as recently demonstrated in
C. elegans (Fang et al., 2023) and
D. melanogaster (Nyberg et al., 2024). For instance, enhanced hammerhead ribozymes (EhhRzs), originally optimized for therapeutic mRNA cleavage, represent a promising class of catalytic RNA biosensors that could be adapted for spatiotemporal gene regulation or transcript imaging in developmental biology models due to their high turnover efficiency under physiological conditions (Myers and Sullivan, 2025). Another interesting example is the massively parallel engineering of ribozyme-based RNA devices in mammalian cells, which produced highly responsive ligand-activated ribozyme switches; these could be adapted for developmental biology to enable precise, spatiotemporal control of gene expression or dynamic reporting of molecular cues in embryos and developing tissues (Xiang et al., 2019). In zebrafish, the RiboFlip system uses a recombinase-activated hammerhead ribozyme to conditionally knock down gene expression with spatiotemporal precision and fluorescent cell labeling (Juan et al., 2025). Furthermore, progresses in ribozyme-guided genetic manipulation techniques can promote their application in developmental studies, as shown in a ribozyme–guide RNA–ribozyme (RGR) method established in plant models (Gao et al., 2015). Other modifications, such as enhancing the speed of their catalytic reactions while maintaining specificity may also provide real-time insights into tissue patterning and regeneration. Moreover, ribozymes can be coupled with modular detection systems, such as split reporters, which is discussed in
Section 3.2 (see below).
3.2. Split RNA Aptamers and Ribozymes
Split RNA aptamers and ribozymes consist of two RNA fragments that become functional only when a target RNA bridges them (Debiais et al., 2020; Gambill et al., 2023). This bridging restores the structure of the aptamer or ribozyme, activating fluorescence or catalytic activity. These biosensors are highly specific and are used for detecting rare RNA sequences, monitoring alternative splicing events, and studying RNA-protein interactions (Debiais et al., 2020). A recent example of a split ribozyme is the platform developed in Arabidopsis and Nicotiana, which enables RNA-triggered fluorescent reporting of gene expression in plant tissues during development (Gambill et al., 2023). While fully ligand-responsive split aptamer–ribozyme systems have yet to be applied in vivo, such designs could build on these modular architectures to enable spatiotemporal biosensing in developmental models. Split aptamers and ribozymes can be engineered to detect alternative splicing events that generate developmental isoforms (Furukawa et al., 2016; Gambill et al., 2023). Incorporating switches that activate only in response to lineage-specific RNAs ensures high specificity (Schmidt and Smolke, 2021). Additionally, these biosensors can be integrated into synthetic gene circuits to dynamically monitor RNA regulation such as morphogenesis (Alam et al., 2017).
3.3. Electrochemical RNA Sensors
Typically, electrochemical RNA biosensors consist of electrodes functionalized with oligonucleotide probes, including either single-stranded DNA or RNA, that are complementary to the sequence of target RNA molecules. The hybridization of target RNAs with probe molecules generates an electrical signal, which can be measured using electrochemical techniques such as voltammetry, amperometry, and impedance spectroscopy. Regardless of the electrochemical approach used, the generated signal is highly dependent on several factors, including the electroactivity of the target and probe molecules, the ionic strength of the sample, and the presence of redox reporters such as methylene blue, and ferrocene (Islam et al., 2017; Kang et al., 2009). These sensors are portable and highly sensitive, making them ideal for point-of-care diagnostics (Hashem et al., 2022). Common applications include detecting miRNAs or RNA biomarkers associated with cancers and infectious diseases in clinical samples (Mikaeeli Kangarshahi et al., 2024). Electrochemical sensors can be adapted for developmental biology research by designing probes that detect specific RNA types for instance miRNA (Hamidi-Asl et al., 2013), or RNA modifications such as m6A or adenosine-to-inosine editing (Campuzano et al., 2019). RNA-based electrochemical assays provide a precise real-time and label-free alternative to fluorescence methods, enabling accurate intracellular sensing in living organisms (Ino et al., 2018). Integrating nano-biointerfaces can improve sensitivity, making these biosensors suitable for small sample sizes like single cells or early embryos. A recent study demonstrated the use of an electrochemical aptamer-based sensor to directly and continuously monitor ATP release from astrocytes in a three-dimensional culture system, highlighting the potential of such platforms for real-time molecular sensing in physiologically relevant environments (Santos-Cancel et al., 2019).
3.4. Field-Effect Transistor (FET)-Based RNA Biosensors:
FET-based RNA biosensors leverage the properties of semiconductor-based transistors to detect RNA molecules through changes in electrical conductivity upon RNA binding (Panahi and Ghafar-Zadeh, 2023). These FET biosensors are typically consist of two conductive terminals (a source and a drain electrode) that allows current flow through the transistor, a semiconducting channel (usually composed of nanomaterials such as graphene, and silicone) that serves as highly sensitive transducing surfaces, and a gate electrode that modulates the electrical response upon probe-target interactions (Sung and Koo, 2021; Tian et al., 2018). RNA detection occurs when complementary probes on the FET surface hybridize with target RNA, altering the charge distribution in the semiconductor channel. This results in a change in gate conductance or the change in source-drain current, which correlates with the concentration of the target RNA. FET biosensors are known for their label-free detection, rapid response, high sensitivity, and potential for miniaturization, making them ideal for diagnostics and real-time RNA quantification in small samples (Zhang et al., 2024). To adapt FET-based biosensors for developmental biology research, probes can be designed to target or activate developmentally relevant RNAs, such as transcripts involved in developmental morphogenesis like genes encoding bone morphogenetic proteins (BMPs) (Wang et al., 2017). Their integration with microfluidics would allow these sensors to analyze RNA from single cells or small tissue samples, which is particularly useful in early developmental stages. Such a method has recently been proposed for studies of neural tissue development (Buentello et al., 2024). Furthermore, increasing specificity through multiplexed arrays can enable simultaneous detection of multiple RNAs involved in organ-specific developmental pathways (Sadeghzade et al., 2024), which has potentials for investigations of gene regulatory networks across species.
4. Environmental and Metabolite-Responsive Sensors
4.1. Riboswitch biosensors
Riboswitch biosensors are naturally occurring, or engineered RNA elements that regulate gene expression in response to binding small molecules such as S-adenosylmethionine (SAM), flavin mononucleotide (FMN), or cyclic-di-GMP to their aptamer aptamer domain (Sherwood and Henkin, 2016). Upon ligand binding, the riboswitch undergoes a conformational change that either activates or represses a downstream reporter gene expression which results in the change of fluorescence, color, as well as enzymatic activity (Wu et al., 2023). Engineered riboswitches are increasingly used in synthetic biology to design cells that can sense and respond to specific metabolites (Hallberg et al., 2017). They are particularly useful in metabolic pathway studies, microbial engineering, and environmental monitoring, where they provide insights into the cellular regulation of key biochemical compounds. Riboswitches can be engineered (e.g., combined with CRISPR guided methods) to detect metabolites or ions critical for developmental checkpoints and function across all kingdoms of life, not just bacterial species (Galizi and Jaramillo, 2019). For instance, small molecule-inducible riboswitches have been engineered to enable reversible, dose-dependent control of gene expression in mammalian systems, including in vivo applications in mice, suggesting their potential utility for temporal regulation in developmental biology studies (Rovira et al., 2023). Interestingly, a recent study engineered splice-modulating riboswitches to control exon skipping in a developmentally relevant gene, enabling chemically inducible isoform switching in mammalian cells and offering a novel approach for regulating gene expression through alternative splicing (Bruckhoff et al., 2025). Furthermore, the multicolor riboswitch-based platforms for RNA imaging RNA in live cells have already been established in mammals (Braselmann et al., 2018). Coupling these biosensors with light-activated switches allows precise spatiotemporal control over biosensor activity (L. Zhang et al., 2023), enabling detailed studies of metabolic regulation during embryogenesis and organogenesis (R. Zheng et al., 2024).
4.2. RNA Thermometers
RNA thermometers are natural RNA molecules that undergo conformational changes in response to temperature fluctuations (Narberhaus et al., 2006). These structural changes can expose or hide ribosome binding sites, modulating translation in a temperature-dependent manner. As a result, they can regulate the expression of a fluorescent or luminescent reporter, as well as an enzyme molecule that produces a color change. RNA thermometers are commonly found in bacteria, where they regulate stress responses, such as heat shock protein expression, and contribute to in pathogenesis during infections (Sharma et al., 2022). They are used in microbiology to study how organisms adapt to thermal stress and environmental changes. It is important to note that RNA thermometers are designed to detect extrinsic physical signals, in this case, temperature, rather than intrinsic RNA processing events. RNA thermometers can be optimized to monitor temperature-sensitive developmental processes and underlying mechanisms of -adaptation and acclimatization to temperature in animals (Somero, 2018) and plants (Thomas et al., 2022). Engineering them to respond to small temperature fluctuations increases their utility for studying phenotypic plasticity in diverse environmental conditions, such as those affecting embryos of aquatic species (Salinas and Jayasundara, 2022).
4.3. RNA Sensors for Reactive Oxygen Species (ROS)
RNA sensors for reactive oxygen species (ROS) detect oxidative molecules, such as hydrogen peroxide or superoxide, which are indicators of cellular stress (Buchser et al., 2023). These biosensors are engineered RNA molecules, which interact with ROS and undergo either oxidation-induced conformational changes, or modifications that can alter their folding, stability, interactions with proteins, as well as base-pairing and subsequent translation disruption. These alterations can activate reporters (e.g. fluorescence, colorimetric, electrochemical signals) upon exposure to ROS, providing a quantitative measure of oxidative stress levels. They are applied in studies of diseases like cancer, neurodegeneration, and aging, where oxidative damage plays a significant role (Khodarev, 2019). These sensors are primarily used to detect extrinsic biochemical conditions, specifically oxidative stress, rather than intrinsic RNA functions such as splicing or localization. ROS sensors can be improved to detect specific types of oxidative stress linked to developmental events (Terzi et al., 2021), as recently shown in immune cells during inflammation (Aich et al., 2024). Incorporating self-calibrating mechanisms ensures precise quantification of ROS levels (Murphy et al., 2022), which can be instrumental during tissue repair or organ formation. These sensors can also be integrated with genetic circuits that activate stress response genes for real-time feedback (Huang et al., 2024).
4.4. tRNA-Based Biosensors
Transfer RNA (tRNA)-based biosensors utilize modified tRNA molecules or tRNA-derived fragments to detect specific biological targets including amino acids, metabolites, ions, and cellular stress markers (Gupta and Laxman, 2020; Sun et al., 2020). By coupling tRNA activity with a reporting system, such as fluorescent aptamers or riboswitches, to induce a fluorescent, colorimetric, or electrochemical signal, these biosensors provide insights into translation and metabolic regulation under various conditions, particularly in studies of nutrient availability and stress responses (Manna et al., 2021). tRNA-based biosensors can be adapted to detect amino acids, metabolites, or environmental changes that influence translation during developmental processes. These sensors can be engineered to include fluorescent or enzymatic reporters that activate upon interaction with specific metabolites, such as methionine or tryptophan, which are essential for protein synthesis during early development. Modifications to the tRNA-binding domain can enable the detection of species-specific codon usage or translational regulation mechanisms (Jiang et al., 2023). In additon, these biosensors can be fine-tuned to monitor dynamic translation rates in differentiating cells or tissues (Lateef et al., 2022), revealing how translation regulation affects gene expression patterns over developmental timelines.
5. Sequence-Specific Detection Sensors
5.1. CRISPR-Cas13/ and -Cas12a RNA Sensors
CRISPR-based RNA sensors are important molecular systems that function based on the clustered regularly interspaced short palindromic repeats (CRISPR) concept for specific RNA detection. Unlike the well-known CRISPR-Cas9 that works for DNA editing in genome, both Cas13 and Cas12a proteins have a programmable nature to detect specific RNA sequences (Aman et al., 2020). Cas13 is an RNA-guided endonuclease that recognizes target RNAs and activates collateral cleavage of nearby reporter molecules, generating fluorescent, colorimetric, or electrochemical signals. However, Cas12a, primarily a DNA-targeting CRISPR enzyme, can be adapted for RNA detection by designing RNA-specific guide RNAs (Wani et al., 2024). These biosensors are pivotal in diagnostics, especially for detecting viral RNAs (e.g., SARS-CoV-2, Zika virus) and other disease-specific biomarkers. Their high sensitivity, specificity, and rapid detection capabilities make them widely adoptable for point-of-care diagnostics and pathogen surveillance. CRISPR-Cas sensors can be tailored to detect transient RNAs involved in early developmental processes. Introducing switches that link target RNA detection to reporter activation (e.g., fluorescent and luminescent signals) ensures real-time monitoring (Yang et al., 2019). Furthermore, programmable guide RNAs can also target isoforms or transcripts that mark specific cell fates (Gupta et al., 2022). A recent example of their application is the development of a system that combines the programmable RNA-targeting CRISPR–Csm complex with multiplexed guide RNAs for direct and efficient visualization of single RNA molecules during neuronal development (Xia et al., 2025).
5.2. Toehold Switches
Toehold switches are synthetic RNA elements that regulate translation in response to the presence of a specific RNA trigger (Zhao et al., 2021). The RNA molecules form a hairpin structure that blocks ribosome access until a complementary RNA sequence binds to the “toehold” region, unwinding the hairpin and initiating translation. Toehold switches are highly modular and programmable, making them suitable for diagnostic applications, such as pathogen detection and biomarker sensing (Yarra et al., 2023). Their versatility also allows their use in constructing synthetic gene circuits for precise control of gene expression in synthetic biology. Toehold switches can be re-engineered to detect maternal RNAs or early zygotic transcripts unique to developmental pathways (McNerney et al., 2019), to design programmable in situ amplification for multiplexed imaging of mRNA expression during development (Choi et al., 2010), and to sense and regulate developmental genes (R. Liu et al., 2022). Another interesting example is a study demonstrating a synthetic toehold switch engineered for microRNA detection in different mammalian cell types at various cellular states (S. Wang et al., 2019). Adding feedback loops that regulate protein synthesis upon RNA detection can provide dynamic insights into post-transcriptional regulation during development (Weldemichael et al., 2022). Furthermore, these biosensors can be integrated into synthetic gene networks for in vivo validation (Falgenhauer et al., 2022).
5.3. RNA aptamer-Based Sensors for Post-Transcriptional Modifications
RNA aptamer-based sensors can detect post-transcriptional modifications such as methylation (e.g. N6-methyladenosine m6A, 5-methylcytosine M5C), pseudouridylation, and inosine editing , which can alter RNA functionality (Han et al., 2024; Liu et al., 2024; Q. Zhang et al., 2023b). These sensors use engineered RNA aptamers that selectively bind to modified RNA bases, often coupled with a fluorescent, colorimetric, or electrochemical reporter to signal the presence of modifications (Q. Zhang et al., 2023b). They are commonly used to study epitranscriptomic regulation in real-time, providing insights into how RNA modifications influence translation, stability, and cellular signaling. These sensors are particularly valuable in exploring disease-related RNA modifications, such as those linked to cancer or neurological disorders (Liu et al., 2024). In developmental biology, RNA aptamer-based sensors can be adapted to investigate how RNA modifications regulate developmental processes. For example, sensors specific to m6A could be used to study its role in controlling RNA decay or translation during embryogenesis (Vong et al., 2021). Another recent example involves a novel fluorescent light-up aptamer (FLAP) system designed to detect the process of m6A RNA demethylation in living cells during tissue development (Zhou et al., 2025). Enhancements such as integrating dual reporters for live imaging or combining with CRISPR systems for RNA modification mapping can expand their utility (Han et al., 2024). In addition, delivery methods like microinjection for embryos or tissue-specific promoters for in vivo expression can make these biosensors suitable for developmental models. Tracking modification patterns across species can also provide insights into evolutionary adaptations of regulatory mechanisms.
6. Structural and Inter-Cellular Communication Sensors
6.1. Circular RNA Sensors
Circular RNA (CircRNA) sensors utilize unique engineered non-coding RNAs that form into a closed-loop structure, making them resistant to degradation and enabling them to function as long-lasting biosensing platforms for detecting, and analyzing circRNA expression in real-time (Ji et al., 2021; Zhao et al., 2023). These sensors often contain aptamer sequences that bind specific targets, generating a fluorescent or electrochemical signal. Moreover, they can be integrated with colorimetric and CRISPR-based detection systems (Litke and Jaffrey, 2019; Zhao et al., 2023). They are particularly valuable for long-term monitoring of RNA expression in live cells, especially in challenging environments where linear RNA sensors may degrade. CircRNA sensors can be customized with binding motifs that detect conserved RNAs critical for lineage specification (Dodbele et al., 2021; Zhao et al., 2023). During mammalian tissue development, for instance, circRNA-based constructs have been engineered to both monitor and manipulate cellular functions (Litke and Jaffrey, 2023, 2019; Zhao et al., 2023). Their inherent stability makes them ideal for tracking developmental transcripts over prolonged timeframes. Furthermore, adding modular reporters tailored to specific model organisms enhances their versatility (Zhou et al., 2024).
6.2. MS2 System
The MS2 system is an RNA-based biosensor platform uses MS2 bacteriophage coat proteins that specifically bind to RNA stem-loop structures, tagging RNA molecules for visualization (Tutucci et al., 2017). These coat proteins can be fused with fluorescent, colorimetric, and electrochemical reporter tags enabling researchers to track RNA localization, movement, and interactions in live cells (Cawte et al., 2020; Chen et al., 2019). The MS2 system is a staple tool in RNA biology, particularly for studying RNA transport, stability, and translation dynamics in eukaryotic cells. It can also be combined with synthetic promoters that activate exclusively in specific developmental stages (Lefebvre and Lécuyer, 2018). This system enables precise monitoring of RNA transport and localization, addressing key developmental biology questions such as the wavelike spatial and temporal regulation of gene expression during embryogenesis(Mau et al., 2023). Another potential application is the creation of live MS2 reporters to enable imaging-based modeling of gene regulatory networks that drive developmental patterning (Keenan et al., 2022). Recent examples of MS2-based systems for tracking RNA targets during embryonic development have been established in invertebrate models such as C. elegans (Hu et al., 2023) and Drosophila (Beadle et al., 2024), as well as in a vertebrate model, zebrafish (Eck et al., 2024). Similar approaches have been implemented to study tissue development in mammalian cells (Pichon et al., 2020) and in the plant model Arabidopsis thaliana (Hani et al., 2021).
6.3. Exosomal RNA Sensors
Exosomal RNA sensors are designed to detect RNA molecules encapsulated within exosomes, which are small extracellular vesicles involved in cell-to-cell communication and carry various biomolecules such as proteins, lipids, and nucleic acids (e.g., mRNA, microRNA, lncRNA, circRNA) (L. Zheng et al., 2024). These sensors typically use hybridization techniques to bind target RNA molecules, utilizing fluorescent electrochemical, colorimetric, and CRISPR-based transducing platforms (Yin et al., 2023). They are widely used in cancer diagnostics to study how tumor cells communicate and transfer genetic material through exosomes, as well as to understand the role of exosomal RNAs in disease progression (Yin et al., 2023). Exosomal RNA sensors can be tailored to detect RNAs that are selectively packaged into extracellular vesicles during developmental or stress-related processes (Wong et al., 2024). These sensors can include RNA aptamers that hybridize with conserved developmental RNAs, such as miRNAs involved in intercellular signaling (Pavani et al., 2022). Fluorescent or electrochemical reporters can be integrated for sensitive detection of RNA content in exosomes derived from developing tissues. Furthermore, engineering sensors to detect lineage-specific RNA markers in exosomes could provide real-time insights into cell-to-cell communication during embryogenesis (Moros et al., 2021). It is important to note that exosomal RNA sensors are primarily used to monitor extracellular RNA involved in intercellular communication, rather than intracellular RNA processing or regulation. These biosensors are particularly impactful in developmental biology research by enabling the study of how exosomal RNAs mediate developmental signaling across species and environmental contexts.
6.4. miRNA Sensors
MicroRNAs (miRNAs) are short non-coding RNAs (approximately 18–25 nucleotides) that play an important role in post-transcriptional gene regulation by targeting mRNAs for degradation or translational repression (Huang et al., 2017). MicroRNA (miRNA) sensors are designed to detect specific miRNAs using complementary RNA or single-stranded DNA sequences, which hybridize with target miRNA molecules. These biosensors can be integrated with fluorescence, electrochemical, colorimetric, and CRISPR-based transducing platforms, providing highly sensitive detection of miRNA levels in real-time (Wang et al., 2022; Zhang et al., 2019; Zhong and Sczepanski, 2019). miRNA sensors are extensively used in cancer diagnostics to study miRNA dysregulation, which is often linked to tumor development, progression, and metastasis. They also provide a non-invasive tools to monitor miRNAs as biomarkers in liquid biopsies (Yu et al., 2024). Furthermore, miRNA sensors can be engineered to detect miRNAs that regulate cell-type specific developmental transitions (X. W. Wang et al., 2019). Enhancements in their sensitivity, such as tandem binding sites, ensures accurate detection of low-abundance miRNAs (Huang et al., 2017). Moreover, multiplexed biosensing platforms allow for the simultaneous tracking of miRNA networks, which indicates their roles in coordinating complex developmental processes. Recent studies have applied miRNA sensors with quantifiable readouts in developmental models (Song et al., 2020). In Ciona embryos, a CRISPR-based miRNA sensor (MICR-ON) was developed in which miRNA-dependent activation of a guide RNA triggers RFP expression, enabling real-time visualization of endogenous miRNA activity during embryogenesis (Wang et al., 2021). In Arabidopsis, transgenic fluorescent miRNA sensor lines have enabled spatiotemporal visualization of miRNA activity during zygotic and somatic embryogenesis (Wójcik, 2020). In zebrafish, fluorescent and luminescent miRNA sensors have been used to track miRNAs during development and tissue regeneration (Fan et al., 2024; Liu et al., 2020; Moro et al., 2019; Sheng et al., 2022). In human iPSCs, genetically encoded endoribonuclease-mediated miRNA sensors provided real-time detection of cell-state–specific miRNAs and guided sequential differentiation toward the hematopoietic lineage (Wang et al., 2024).
6.5. LncRNA Sensors
LncRNA sensors are biosensors designed to detect long non-coding RNAs (lncRNAs), a class of RNA molecules over 200 nucleotides in length that play critical regulatory roles in gene expression, chromatin organization, and developmental processes. These sensors typically rely on sequence-specific hybridization or aptamer-based detection systems that recognize unique lncRNA sequences (Fasciano et al., 2023; Yao et al., 2025; Q. Zhang et al., 2023a). They can be coupled with fluorescent, electrochemical, colorimetric, and CRISPR-based signaling platforms to non-invasively quantify and visualize lncRNAs in cells and tissues in real time. In developmental biology research, lncRNA sensors can be modified to target lineage-specific or developmentally regulated lncRNAs (Srinivas et al., 2023). For instance, incorporating sequence motifs specific to lncRNAs involved in embryogenesis, such as those influencing Hox gene clusters, can enhance their relevance. To improve their functionality in developmental systems, these sensors can be coupled with multiplex platforms to detect multiple lncRNAs simultaneously (Srinivas et al., 2023). Furthermore, using advanced delivery systems such as nanoparticle carriers can facilitate efficient introduction into embryos or specific tissues in model organisms like zebrafish (Sarfraz et al., 2023). Enhanced sensitivity and spatial resolution would allow researchers to track lncRNA dynamics during critical developmental transitions.
7. Limitations and Challenges
While RNA-based biosensors offer immense potential for advancing developmental biology, their practical implementation in living, developing organisms presents several challenges. These limitations arise from factors such as sensor biocompatibility, delivery efficiency, temporal stability, spatial resolution, and target specificity within the dynamic and complex embryonic environment (see
Table 2).
Sensor delivery and expression: One of the primary challenges is the efficient delivery of RNA biosensors into embryonic tissues. Unlike cultured cells, developing embryos are often protected by extra-embryonic structures or membranes, making microinjection or electroporation technically demanding. Genetically encoded sensors, such as fluorescent aptamers, toehold switches, or riboswitches, require integration into the genome or stable mRNA delivery, which can trigger innate immune responses or induce developmental perturbations if not tightly controlled (Galizi and Jaramillo, 2019; Hallberg et al., 2017; Micura and Höbartner, 2020; Yoo et al., 2020). For non-genetically encoded sensors (e.g., molecular beacons, electrochemical sensors), ensuring stable localization and uptake without toxicity remains a hurdle.
Intracellular stability and degradation: Many RNA-based biosensors are susceptible to nuclease-mediated degradation (Su and Hammond, 2020; Zhang et al., 2021), especially in early embryos where RNA turnover is high (Barckmann and Simonelig, 2013). Engineering chemically modified RNAs or incorporating protective structural elements (e.g., pseudoknots, locked nucleic acids) can mitigate this (Kornienko et al., 2024; Larkey et al., 2020; Napoletano et al., 2024), but these modifications may alter sensor kinetics or affinity which requires further validation investigations.
Signal fidelity and background noise: In vivo environments are biochemically complex, and background fluorescence, redox fluctuations, and ion concentration changes can interfere with sensor readout. This is especially problematic for FRET sensors, aptamer-based fluorescent reporters, and quenching-based sensors, where high signal-to-noise ratios are essential for real-time imaging (Alkhamis et al., 2023; Chu et al., 2024; Du et al., 2022). Photobleaching and autofluorescence in live embryos may further complicate long-term imaging with these sensors.
Tissue-specific accessibility and resolution: Different embryonic tissues exhibit variable permeability, autofluorescence, and metabolic activity, influencing sensor performance. Deep-tissue detection remains a particular challenge for optical sensors like fluorescent aptamers or MS2 systems (Chen et al., 2023; Lu et al., 2023; Markey et al., 2021; Saw and Song, 2025). Engineering sensors compatible with infrared or near-infrared fluorophores or using biosensors with electrochemical or bioluminescent readouts may help circumvent these limitations (Braselmann et al., 2020; Markey et al., 2021).
Target specificity and modularity: Some biosensors (e.g., riboswitches, ribozymes, CRISPR-based systems) offer programmable specificity, but their in vivo activation may be context-dependent (Boussebayle et al., 2019; Chau et al., 2020; Dykstra et al., 2022; Qian et al., 2022). For example, CRISPR-Cas13-based sensors may suffer from off-target cleavage or require precise timing to avoid unintended developmental effects (Abudayyeh et al., 2017). Fine-tuning specificity while minimizing background activity is an ongoing engineering challenge.
8. Conclusion and Future Directions
The integration of RNA biosensors into developmental biology holds great promise, and ongoing efforts continue to explore and expand their potential in this field (see
Figure 4). To bridge this gap, a multidisciplinary effort is required to refine these tools for in vivo applications and adapt them to the unique challenges of studying dynamic developmental processes. One of the most critical steps forward is the development of biosensors with enhanced sensitivity, specificity, and stability in living systems. Current RNA biosensors must be optimized to function within complex embryonic environments, where fluctuating molecular conditions can interfere with signal fidelity. Advances in nanotechnology, molecular engineering, and bioinformatics will be essential for improving sensor design and ensuring their robustness in long-term studies. Moreover, scaling up the use of RNA biosensors in model organisms requires tailored genetic engineering strategies. This includes integrating biosensor expression directly into transgenic animal models and refining gene-editing techniques such as CRISPR-based activation or inhibition systems. These approaches will allow for precise spatiotemporal tracking of RNA species without disrupting normal developmental processes. Another important future direction is the expansion of RNA biosensor applications beyond simple detection. While existing sensors effectively track RNA localization and modifications, they should be evolved into functional tools that manipulate RNA activity in real-time, providing a means to dissect causative relationships between RNA regulation and developmental outcomes. The development of dual-function biosensors, capable of both sensing and modulating RNA pathways, could revolutionize how researchers investigate RNA’s role in cell fate determination, tissue organization, and morphogenesis. Importantly, both biosensors that monitor intrinsic RNA biology (such as expression, splicing, or localization) and those that detect extrinsic factors (such as pH, ROS, or mechanical stress) offer complementary insights into developmental processes and should be advanced in parallel. For these innovations to become mainstream in developmental biology, collaboration between bioengineers, computational biologists, and developmental researchers is essential. Efforts should be made to standardize protocols, create open-source databases for RNA biosensor applications, and develop computational models to predict RNA behavior in vivo. Furthermore, incorporating artificial intelligence and machine learning into RNA biosensor analysis could improve real-time data interpretation, allowing for more accurate and automated insights into developmental processes. Ultimately, the widespread adoption of RNA biosensors in developmental biology will require not only technological advancements but also a shift in research paradigms. As these tools become more refined and accessible, they have the potential to unlock new dimensions of RNA regulation, leading to groundbreaking discoveries in embryology and regenerative medicine. By taking these strategic steps, the field can harness RNA biosensors as powerful instruments for deciphering the molecular blueprint of life itself.
References
- Abudayyeh, O.O., Gootenberg, J.S., Essletzbichler, P., Han, S., Joung, J., Belanto, J.J., Verdine, V., Cox, D.B.T., Kellner, M.J., Regev, A., Lander, E.S., Voytas, D.F., Ting, A.Y., Zhang, F., 2017. RNA targeting with CRISPR–Cas13. Nat. 2017 5507675 550, 280–284. [CrossRef]
- Aich, A., Boshnakovska, A., Witte, S., Gall, T., Unthan-Fechner, K., Yousefi, R., Chowdhury, A., Dahal, D., Methi, A., Kaufmann, S., Silbern, I., Prochazka, J., Nichtova, Z., Palkova, M., Raishbrook, M., Koubkova, G., Sedlacek, R., Tröder, S.E., Zevnik, B., Riedel, D., Michanski, S., Möbius, W., Ströbel, P., Lüchtenborg, C., Giavalisco, P., Urlaub, H., Fischer, A., Brügger, B., Jakobs, S., Rehling, P., 2024. Defective mitochondrial COX1 translation due to loss of COX14 function triggers ROS-induced inflammation in mouse liver. Nat. Commun. 2024 151 15, 1–20. [CrossRef]
- Alam, K.K., Tawiah, K.D., Lichte, M.F., Porciani, D., Burke, D.H., 2017. A Fluorescent Split Aptamer for Visualizing RNA-RNA Assembly in Vivo. ACS Synth. Biol. 6, 1710–1721. [CrossRef]
- Alkhamis, O., Canoura, J., Willis, C., Wang, L., Perry, J., Xiao, Y., 2023. Comparison of Aptamer Signaling Mechanisms Reveals Disparities in Sensor Response and Strategies to Eliminate False Signals. J. Am. Chem. Soc. 145, 12407–12422. [CrossRef]
- Aman, R., Mahas, A., Mahfouz, M., 2020. Nucleic Acid Detection Using CRISPR/Cas Biosensing Technologies. ACS Synth. Biol. 9, 1226–1233. [CrossRef]
- Arora, A., Sunbul, M., Jäschke, A., 2015. Dual-colour imaging of RNAs using quencher- and fluorophore-binding aptamers. Nucleic Acids Res. 43, e144–e144. [CrossRef]
- Barckmann, B., Simonelig, M., 2013. Control of maternal mRNA stability in germ cells and early embryos. Biochim. Biophys. Acta - Gene Regul. Mech. 1829, 714–724. [CrossRef]
- Barta, A., Jantsch, M.F., 2017. RNA in Disease and development. RNA Biol. 14, 457–459. [CrossRef]
- Beadle, L.F., Sutcliffe, C., Ashe, H.L., 2024. A simple MiMIC-based approach for tagging endogenous genes to visualise live transcription in Drosophila. Development 151. [CrossRef]
- Bidar, N., Amini, M., Oroojalian, F., Baradaran, B., Hosseini, S.S., Shahbazi, M.A., Hashemzaei, M., Mokhtarzadeh, A., Hamblin, M.R., de la Guardia, M., 2021. Molecular beacon strategies for sensing purpose. TrAC Trends Anal. Chem. 134, 116143. [CrossRef]
- Boussebayle, A., Torka, D., Ollivaud, S., Braun, J., Bofill-Bosch, C., Dombrowski, M., Groher, F., Hamacher, K., Suess, B., 2019. Next-level riboswitch development—implementation of Capture-SELEX facilitates identification of a new synthetic riboswitch. Nucleic Acids Res. 47, 4883–4895. [CrossRef]
- Braselmann, E., Rathbun, C., Richards, E.M., Palmer, A.E., 2020. Illuminating RNA Biology: Tools for Imaging RNA in Live Mammalian Cells. Cell Chem. Biol. 27, 891–903. [CrossRef]
- Braselmann, E., Wierzba, A.J., Polaski, J.T., Chromiński, M., Holmes, Z.E., Hung, S.T., Batan, D., Wheeler, J.R., Parker, R., Jimenez, R., Gryko, D., Batey, R.T., Palmer, A.E., 2018. A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells. Nat. Chem. Biol. 2018 1410 14, 964–971. [CrossRef]
- Bruckhoff, R.W., Becker, O., Steinhilber, D., Suess, B., 2025. Potential and Optimization of Mammalian Splice Riboswitches for the Regulation of Exon Skipping-Dependent Gene Expression and Isoform Switching within the ALOX5 Gene. ACS Synth. Biol. 14. [CrossRef]
- Buchser, R., Sweet, P., Anantharaman, A., Contreras, L., 2023. RNAs as Sensors of Oxidative Stress in Bacteria. Annu. Rev. Chem. Biomol. Eng. 14, 265–281. [CrossRef]
- Buentello, D.C., Garcia-Corral, M., Trujillo-De Santiago, G., Alvarez, M.M., 2024. Neuron(s)-on-a-Chip: A Review of the Design and Use of Microfluidic Systems for Neural Tissue Culture. IEEE Rev. Biomed. Eng. 17, 243–263. [CrossRef]
- Campuzano, S., Pedrero, M., Yánez-Sedeño, P., Pingarrón, J.M., 2019. Advances in Electrochemical (Bio)Sensing Targeting Epigenetic Modifications of Nucleic Acids. Electroanalysis 31, 1816–1832. [CrossRef]
- Catrina, I.E., Bayer, L. V., Omar, O.S., Bratu, D.P., 2019. Visualizing and Tracking Endogenous mRNAs in Live <em>Drosophila melanogaster</em> Egg Chambers. JoVE (Journal Vis. Exp. 2019, e58545. [CrossRef]
- Cawte, A.D., Unrau, P.J., Rueda, D.S., 2020. Live cell imaging of single RNA molecules with fluorogenic Mango II arrays. Nat. Commun. 2020 111 11, 1–11. [CrossRef]
- Chau, T.H.T., Mai, D.H.A., Pham, D.N., Le, H.T.Q., Lee, E.Y., 2020. Developments of Riboswitches and Toehold Switches for Molecular Detection-Biosensing and Molecular Diagnostics. Int. J. Mol. Sci. 21. [CrossRef]
- Chen, W., Zhao, X., Yang, N., Li, X., 2023. Single mRNA Imaging with Fluorogenic RNA Aptamers and Small-molecule Fluorophores. Angew. Chemie 135, e202209813. [CrossRef]
- Chen, X., Zhang, D., Su, N., Bao, B., Xie, X., Zuo, F., Yang, L., Wang, H., Jiang, L., Lin, Q., Fang, M., Li, N., Hua, X., Chen, Z., Bao, C., Xu, J., Du, W., Zhang, L., Zhao, Y., Zhu, L., Loscalzo, J., Yang, Y., 2019. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat. Biotechnol. 2019 3711 37, 1287–1293. [CrossRef]
- Chen, Z., Chen, W., Xu, C., Song, H., Ji, X., Jiang, H., Duan, H., Li, Z., Gao, W., Yao, T., Zhang, Z., He, L., Yin, Y., Yang, N., Tian, W., Wu, J., Li, X., 2025. Near-infrared fluorogenic RNA for in vivo imaging and sensing. Nat. Commun. 2025 161 16, 1–15. [CrossRef]
- Choi, H.M.T., Chang, J.Y., Trinh, L.A., Padilla, J.E., Fraser, S.E., Pierce, N.A., 2010. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010 2811 28, 1208–1212. [CrossRef]
- Chu, J., Ejaz, A., Lin, K.M., Joseph, M.R., Coraor, A.E., Drummond, D.A., Squires, A.H., 2024. Single-molecule fluorescence multiplexing by multi-parameter spectroscopic detection of nanostructured FRET labels. Nat. Nanotechnol. 2024 198 19, 1150–1157. [CrossRef]
- Debiais, M., Lelievre, A., Smietana, M., Müller, S., 2020. Splitting aptamers and nucleic acid enzymes for the development of advanced biosensors. Nucleic Acids Res. 48, 3400–3422. [CrossRef]
- Dodbele, S., Mutlu, N., Wilusz, J.E., 2021. Best practices to ensure robust investigation of circular RNAs: pitfalls and tips. EMBO Rep. 22. [CrossRef]
- Du, J., Dartawan, R., Rice, W., Gao, F., Zhou, J.H., Sheng, J., 2022. Fluorescent Platforms for RNA Chemical Biology Research. Genes 2022, Vol. 13, Page 1348 13, 1348. [CrossRef]
- Dykstra, P.B., Kaplan, M., Smolke, C.D., 2022. Engineering synthetic RNA devices for cell control. Nat. Rev. Genet. 2022 234 23, 215–228. [CrossRef]
- Eck, E., Moretti, B., Schlomann, B.H., Bragantini, J., Lange, M., Zhao, X., VijayKumar, S., Valentin, G., Loureiro, C., Soroldoni, D., Royer, L.A., Oates, A.C., Garcia, H.G., 2024. Single-cell transcriptional dynamics in a living vertebrate. bioRxiv 2024.01.03.574108. [CrossRef]
- Englert, D., Burger, E.M., Grün, F., Verma, M.S., Lackner, J., Lampe, M., Bühler, B., Schokolowski, J., Nienhaus, G.U., Jäschke, A., Sunbul, M., 2023. Fast-exchanging spirocyclic rhodamine probes for aptamer-based super-resolution RNA imaging. Nat. Commun. 2023 141 14, 1–13. [CrossRef]
- Eroshkin, F.M., Fefelova, E.A., Bredov, D. V., Orlov, E.E., Kolyupanova, N.M., Mazur, A.M., Sokolov, A.S., Zhigalova, N.A., Prokhortchouk, E.B., Nesterenko, A.M., Zaraisky, A.G., 2024. Mechanical Tensions Regulate Gene Expression in the Xenopus laevis Axial Tissues. Int. J. Mol. Sci. 25, 870. [CrossRef]
- Falgenhauer, E., Mückl, A., Schwarz-Schilling, M., Simmel, F.C., 2022. Transcriptional Interference in Toehold Switch-Based RNA Circuits. ACS Synth. Biol. 11, 1735–1745. [CrossRef]
- Fan, J., Liu, X., Duan, Z., Zhao, H., Chang, Z., Li, L., 2024. The Regulatory Role of miRNAs in Zebrafish Fin Regeneration. Int. J. Mol. Sci. 25, 10542. [CrossRef]
- Fang, J., Wang, J., Wang, Y., Liu, X., Chen, B., Zou, W., 2023. Ribo-On and Ribo-Off tools using a self-cleaving ribozyme allow manipulation of endogenous gene expression in C. elegans. Commun. Biol. 2023 61 6, 1–13. [CrossRef]
- Fasciano, S., Luo, S., Wang, S., 2023. Long non-coding RNA (lncRNA) MALAT1 in regulating osteogenic and adipogenic differentiation using a double-stranded gapmer locked nucleic acid nanobiosensor. Analyst 148, 6261–6273. [CrossRef]
- Frommer, J., Appel, B., Müller, S., 2015. Ribozymes that can be regulated by external stimuli. Curr. Opin. Biotechnol. 31, 35–41. [CrossRef]
- Furukawa, A., Tanaka, T., Furuta, H., Matsumura, S., Ikawa, Y., 2016. Use of a Fluorescent Aptamer RNA as an Exonic Sequence to Analyze Self-Splicing Ability of a Group I Intron from Structured RNAs. Biol. 2016, Vol. 5, Page 43 5, 43. [CrossRef]
- Galizi, R., Jaramillo, A., 2019. Engineering CRISPR guide RNA riboswitches for in vivo applications. Curr. Opin. Biotechnol. 55, 103–113. [CrossRef]
- Gambill, L., Staubus, A., Mo, K.W., Ameruoso, A., Chappell, J., 2023. A split ribozyme that links detection of a native RNA to orthogonal protein outputs. Nat. Commun. 2023 141 14, 1–12. [CrossRef]
- Gao, Y., Zhang, Y., Zhang, D., Dai, X., Estelle, M., Zhao, Y., 2015. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc. Natl. Acad. Sci. U. S. A. 112, 2275–2280. [CrossRef]
- Gupta, R., Ghosh, A., Chakravarti, R., Singh, R., Ravichandiran, V., Swarnakar, S., Ghosh, D., 2022. Cas13d: A New Molecular Scissor for Transcriptome Engineering. Front. Cell Dev. Biol. 10, 866800. [CrossRef]
- Gupta, R., Laxman, S., 2020. tRNA wobble-uridine modifications as amino acid sensors and regulators of cellular metabolic state. Curr. Genet. 66, 475–480. [CrossRef]
- Hallberg, Z.F., Su, Y., Kitto, R.Z., Hammond, M.C., 2017. Engineering and in vivo applications of riboswitches. Annu. Rev. Biochem. 86, 515–539. [CrossRef]
- Halstead, J.M., Lionnet, T., Wilbertz, J.H., Wippich, F., Ephrussi, A., Singer, R.H., Chao, J.A., 2015. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science (80-. ). 347, 1367–1370. [CrossRef]
- Hamidi-Asl, E., Palchetti, I., Hasheminejad, E., Mascini, M., 2013. A review on the electrochemical biosensors for determination of microRNAs. Talanta 115, 74–83. [CrossRef]
- Han, Y., Yang, D., Jiang, S., Zhao, S., Ma, F., Zhang, C. yang, 2024. Recent advance in optical single-molecule detection of methylation modification and methyl-modifying enzymes. TrAC Trends Anal. Chem. 172, 117553. [CrossRef]
- Hani, S., Cuyas, L., David, P., Secco, D., Whelan, J., Thibaud, M.C., Merret, R., Mueller, F., Pochon, N., Javot, H., Faklaris, O., Maréchal, E., Bertrand, E., Nussaume, L., 2021. Live single-cell transcriptional dynamics via RNA labelling during the phosphate response in plants. Nat. plants 7, 1050–1064. [CrossRef]
- Hashem, A., Hossain, M.A.M., Marlinda, A.R., Mamun, M. Al, Sagadevan, S., Shahnavaz, Z., Simarani, K., Johan, M.R., 2022. Nucleic acid-based electrochemical biosensors for rapid clinical diagnosis: advances, challenges, and opportunities. Crit. Rev. Clin. Lab. Sci. 59, 156–177. [CrossRef]
- Hu, Y., Xu, J., Gao, E., Fan, X., Wei, J., Ye, B., Xu, S., Ma, W., 2023. Enhanced single RNA imaging reveals dynamic gene expression in live animals. Elife 12. [CrossRef]
- Huang, J., Xue, S., Teixeira, A.P., Fussenegger, M., 2024. A Gene-Switch Platform Interfacing with Reactive Oxygen Species Enables Transcription Fine-Tuning by Soluble and Volatile Pharmacologics and Food Additives. Adv. Sci. 11, 2306333. [CrossRef]
- Huang, K., Doyle, F., Wurz, Z.E., Tenenbaum, S.A., Hammond, R.K., Caplan, J.L., Meyers, B.C., 2017. FASTmiR: an RNA-based sensor for in vitro quantification and live-cell localization of small RNAs. Nucleic Acids Res. 45, e130–e130. [CrossRef]
- Ino, K., Nashimoto, Y., Taira, N., Azcon, J.R., Shiku, H., 2018. Intracellular Electrochemical Sensing. Electroanalysis 30, 2195–2209. [CrossRef]
- Islam, M.N., Masud, M.K., Haque, M.H., Hossain, M.S. Al, Yamauchi, Y., Nguyen, N.T., Shiddiky, M.J.A., 2017. RNA Biomarkers: Diagnostic and Prognostic Potentials and Recent Developments of Electrochemical Biosensors. Small Methods 1, 1700131. [CrossRef]
- Ji, D., Lyu, K., Zhao, H., Kwok, C.K., 2021. Circular L-RNA aptamer promotes target recognition and controls gene activity. Nucleic Acids Res. 49, 7280–7291. [CrossRef]
- Jiang, H.K., Ambrose, N.L., Chung, C.Z., Wang, Y.S., Söll, D., Tharp, J.M., 2023. Split aminoacyl-tRNA synthetases for proximity-induced stop codon suppression. Proc. Natl. Acad. Sci. U. S. A. 120, e2219758120. [CrossRef]
- Jones, K.A., Zinkus-Boltz, J., Dickinson, B.C., 2019. Recent advances in developing and applying biosensors for synthetic biology. Nano Futur. 3, 042002. [CrossRef]
- Juan, T., Molina, T., Xie, L., Papadopoulou, S., Cardoso, B., Jha, S.G., Stainier, D.Y.R., 2025. A recombinase-activated ribozyme to knock down endogenous gene expression in zebrafish. PLOS Genet. 21, e1011594. [CrossRef]
- Kang, D., Zuo, X., Yang, R., Xia, F., Plaxco, K.W., White, R.J., 2009. Comparing the properties of electrochemical-based DNA sensors employing different redox tags. Anal. Chem. 81, 9109–9113. [CrossRef]
- Kang, W.J., Cho, Y.L., Chae, J.R., Lee, J.D., Choi, K.J., Kim, S., 2011. Molecular beacon-based bioimaging of multiple microRNAs during myogenesis. Biomaterials 32, 1915–1922. [CrossRef]
- Keenan, S.E., Avdeeva, M., Yang, L., Alber, D.S., Wieschaus, E.F., Shvartsman, S.Y., 2022. Dynamics of Drosophila endoderm specification. Proc. Natl. Acad. Sci. U. S. A. 119, e2112892119. [CrossRef]
- Khodarev, N.N., 2019. Intracellular RNA Sensing in Mammalian Cells: Role in Stress Response and Cancer Therapies. Int. Rev. Cell Mol. Biol. 344, 31–89. [CrossRef]
- Kornienko, I. V., Aramova, O.Y., Tishchenko, A.A., Rudoy, D. V., Chikindas, M.L., 2024. RNA Stability: A Review of the Role of Structural Features and Environmental Conditions. Molecules 29, 5978. [CrossRef]
- Kuang, T., Chang, L., Peng, X., Hu, X., Gallego-Perez, D., 2017. Molecular Beacon Nano-Sensors for Probing Living Cancer Cells. Trends Biotechnol. 35, 347–359. [CrossRef]
- Kudłak, B., Wieczerzak, M., 2020. Aptamer based tools for environmental and therapeutic monitoring: A review of developments, applications, future perspectives. Crit. Rev. Environ. Sci. Technol. 50, 816–867. [CrossRef]
- Larkey, N.E., Phillips, J.L., Jang, H.S., Kolluri, S.K., Burrows, S.M., 2020. Small RNA Biosensor Design Strategy to Mitigate Off-Analyte Response. ACS Sensors 5, 377–384. [CrossRef]
- Lateef, O.M., Akintubosun, M.O., Olaoba, O.T., Samson, S.O., Adamczyk, M., 2022. Making Sense of “Nonsense” and More: Challenges and Opportunities in the Genetic Code Expansion, in the World of tRNA Modifications. Int. J. Mol. Sci. 2022, Vol. 23, Page 938 23, 938. [CrossRef]
- Lefebvre, F.A., Lécuyer, É., 2018. Flying the RNA Nest: Drosophila Reveals Novel Insights into the Transcriptome Dynamics of Early Development. J. Dev. Biol. 2018, Vol. 6, Page 5 6, 5. [CrossRef]
- Li, W., Xu, Y., Zhang, Y., Li, P., Zhu, X., Feng, C., 2023. Cell-Free Biosensing Genetic Circuit Coupled with Ribozyme Cleavage Reaction for Rapid and Sensitive Detection of Small Molecules. ACS Synth. Biol. 12, 1657–1666. [CrossRef]
- Litke, J.L., Jaffrey, S.R., 2023. Designing and Expressing Circular RNA Aptamers to Regulate Mammalian Cell Biology. Methods Mol. Biol. 2570, 223–234. [CrossRef]
- Litke, J.L., Jaffrey, S.R., 2019. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 2019 376 37, 667–675. [CrossRef]
- Liu, M. hao, Zhao, N. ning, Yu, W. tong, Qiu, J.G., Jiang, B.H., Zhang, Y., Zhang, C.Y., 2024. Construction of a label-free fluorescent biosensor for homogeneous detection of m6A eraser FTO in breast cancer tissues. Talanta 272, 125784. [CrossRef]
- Liu, R., Zhong, Y., Chen, R., Chu, C., Liu, G., Zhou, Y., Huang, Y., Fang, Z., Liu, H., 2022. m6A reader hnRNPA2B1 drives multiple myeloma osteolytic bone disease. Theranostics 12, 7760–7774. [CrossRef]
- Liu, X., Wang, T., Wu, Y., Tan, Y., Jiang, T., Li, K., Lou, B., Chen, L., Liu, Y., Liu, Z., 2022. Aptamer based probes for living cell intracellular molecules detection. Biosens. Bioelectron. 208, 114231. [CrossRef]
- Liu, Y., Zhu, Z., Ho, I.H.T., Shi, Y., Li, J., Wang, X., Chan, M.T.V., Cheng, C.H.K., 2020. Genetic Deletion of miR-430 Disrupts Maternal-Zygotic Transition and Embryonic Body Plan. Front. Genet. 11, 568845. [CrossRef]
- Loffet, E.A., Durel, J.F., Nerurkar, N.L., 2023. Evo-Devo Mechanobiology: The Missing Link. Integr. Comp. Biol. 63, 1455–1473. [CrossRef]
- Lu, X., Kong, K.Y.S., Unrau, P.J., 2023. Harmonizing the growing fluorogenic RNA aptamer toolbox for RNA detection and imaging. Chem. Soc. Rev. 52, 4071–4098. [CrossRef]
- Manna, S., Kellenberger, C.A., Hallberg, Z.F., Hammond, M.C., 2021. Live Cell Imaging Using Riboswitch–Spinach tRNA Fusions as Metabolite-Sensing Fluorescent Biosensors. Methods Mol. Biol. 2323, 121–140. [CrossRef]
- Markey, F.B., Parashar, V., Batish, M., 2021. Methods for spatial and temporal imaging of the different steps involved in RNA processing at single-molecule resolution. Wiley Interdiscip. Rev. RNA 12. [CrossRef]
- Mau, C., Rudolf, H., Strobl, F., Schmid, B., Regensburger, T., Palmisano, R., Stelzer, E.H.K., Taher, L., El-Sherif, E., 2023. How enhancers regulate wavelike gene expression patterns. Elife 12. [CrossRef]
- McNerney, M.P., Zhang, Y., Steppe, P., Silverman, A.D., Jewett, M.C., Styczynski, M.P., 2019. Point-of-care biomarker quantification enabled by sample-specific calibration. Sci. Adv. 5. [CrossRef]
- Micura, R., Höbartner, C., 2020. Fundamental studies of functional nucleic acids: aptamers, riboswitches, ribozymes and DNAzymes. Chem. Soc. Rev. 49, 7331–7353. [CrossRef]
- Mikaeeli Kangarshahi, B., Naghib, S.M., Rabiee, N., 2024. DNA/RNA-based electrochemical nanobiosensors for early detection of cancers. Crit. Rev. Clin. Lab. Sci. 61, 473–495. [CrossRef]
- Moraldo, C., Vuille-dit-Bille, E., Shkodra, B., Kloter, T., Nakatsuka, N., 2022. Aptamer-modified biosensors to visualize neurotransmitter flux. J. Neurosci. Methods 365, 109386. [CrossRef]
- Moro, A., Driscoll, T.P., Boraas, L.C., Armero, W., Kasper, D.M., Baeyens, N., Jouy, C., Mallikarjun, V., Swift, J., Ahn, S.J., Lee, D., Zhang, J., Gu, M., Gerstein, M., Schwartz, M., Nicoli, S., 2019. MicroRNA-dependent regulation of biomechanical genes establishes tissue stiffness homeostasis. Nat. Cell Biol. 2019 213 21, 348–358. [CrossRef]
- Moros, M., Fergola, E., Marchesano, V., Mutarelli, M., Tommasini, G., Miedziak, B., Palumbo, G., Ambrosone, A., Tino, A., Tortiglione, C., 2021. The Aquatic Invertebrate Hydra vulgaris Releases Molecular Messages Through Extracellular Vesicles. Front. Cell Dev. Biol. 9, 788117. [CrossRef]
- Morris, K. V., Mattick, J.S., 2014. The rise of regulatory RNA. Nat. Rev. Genet. 2014 156 15, 423–437. [CrossRef]
- Murata, Y., Jo, J.I., Tabata, Y., 2021. Molecular Beacon Imaging to Visualize Ki67 mRNA for Cell Proliferation Ability. Tissue Eng. Part A 27, 526–535. [CrossRef]
- Murphy, M.P., Bayir, H., Belousov, V., Chang, C.J., Davies, K.J.A., Davies, M.J., Dick, T.P., Finkel, T., Forman, H.J., Janssen-Heininger, Y., Gems, D., Kagan, V.E., Kalyanaraman, B., Larsson, N.G., Milne, G.L., Nyström, T., Poulsen, H.E., Radi, R., Van Remmen, H., Schumacker, P.T., Thornalley, P.J., Toyokuni, S., Winterbourn, C.C., Yin, H., Halliwell, B., 2022. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 2022 46 4, 651–662. [CrossRef]
- Myers, J.M., Sullivan, J.M., 2025. Enhanced hammerhead ribozyme turnover rates: Reevaluating therapeutic space for small catalytic RNAs. Mol. Ther. Nucleic Acids 36, 102431. [CrossRef]
- Napoletano, S., Battista, E., Netti, P.A., Causa, F., 2024. MicroLOCK: Highly stable microgel biosensor using locked nucleic acids as bioreceptors for sensitive and selective detection of let-7a. Biosens. Bioelectron. 260, 116406. [CrossRef]
- Narberhaus, F., Waldminghaus, T., Chowdhury, S., 2006. RNA thermometers. FEMS Microbiol. Rev. 30, 3–16. [CrossRef]
- Nesterova, I. V., Erdem, S.S., Pakhomov, S., Hammer, R.P., Soper, S.A., 2009. Phthalocyanine dimerization-based molecular beacons using near-IR fluorescence. J. Am. Chem. Soc. 131, 2432–2433. [CrossRef]
- Neubacher, S., Hennig, S., 2019. RNA Structure and Cellular Applications of Fluorescent Light-Up Aptamers. Angew. Chemie Int. Ed. 58, 1266–1279. [CrossRef]
- Nyberg, K.G., Navales, F.G., Keles, E., Nguyen, J.Q., Hertz, L.M., Carthew, R.W., 2024. Robust and heritable knockdown of gene expression using a self-cleaving ribozyme in Drosophila. Genetics 227. [CrossRef]
- Osman, E.A., Rynes, T.P., Wang, Y.L., Mruk, K., McKeague, M., McKeague, M., 2024. Non-invasive single cell aptasensing in live cells and animals. Chem. Sci. 15. [CrossRef]
- Palchetti, I., Mascini, M., 2008. Nucleic acid biosensors for environmental pollution monitoring. Analyst 133, 846–854. [CrossRef]
- Panahi, A., Ghafar-Zadeh, E., 2023. Emerging Field-Effect Transistor Biosensors for Life Science Applications. Bioeng. 2023, Vol. 10, Page 793 10, 793. [CrossRef]
- Pavani, K.C., Meese, T., Pascottini, O.B., Guan, X.F., Lin, X., Peelman, L., Hamacher, J., van Nieuwerburgh, F., Deforce, D., Boel, A., Heindryckx, B., Tilleman, K., van Soom, A., Gadella, B.M., Hendrix, A., Smits, K., 2022. Hatching is modulated by microRNA-378a-3p derived from extracellular vesicles secreted by blastocysts. Proc. Natl. Acad. Sci. U. S. A. 119, e2122708119. [CrossRef]
- Peng, Y., Shu, L., Deng, X., Huang, X., Mo, X., Du, F., Tang, Z., 2023. Live-Cell Imaging of Endogenous RNA with a Genetically Encoded Fluorogenic Allosteric Aptamer. Anal. Chem. 95, 13762–13768. [CrossRef]
- Pichon, X., Robert, M.C., Bertrand, E., Singer, R.H., Tutucci, E., 2020. New Generations of MS2 Variants and MCP Fusions to Detect Single mRNAs in Living Eukaryotic Cells. Methods Mol. Biol. 2166, 121–144. [CrossRef]
- Pitchiaya, S., Heinicke, L.A., Custer, T.C., Walter, N.G., 2014. Single molecule fluorescence approaches shed light on intracellular RNAs. Chem. Rev. 114, 3224–3265. [CrossRef]
- Qian, Y., Li, J., Zhao, S., Matthews, E.A., Adoff, M., Zhong, W., An, X., Yeo, M., Park, C., Yang, X., Wang, B.S., Southwell, D.G., Huang, Z.J., 2022. Programmable RNA sensing for cell monitoring and manipulation. Nat. 2022 6107933 610, 713–721. [CrossRef]
- Rosa, A., Ciaudo, C., Sumazin, P., Fazi, F., 2021. Editorial: The RNA Revolution in Embryonic Development and Cell Differentiation in Health and Disease. Front. Cell Dev. Biol. 9, 715341. [CrossRef]
- Rovira, E., Moreno, B., Razquin, N., Blázquez, L., Hernández-Alcoceba, R., Fortes, P., Pastor, F., 2023. Engineering U1-Based Tetracycline-Inducible Riboswitches to Control Gene Expression in Mammals. ACS Nano 17, 23331–23346. [CrossRef]
- Sadeghzade, S., Hooshiar, M.H., Akbari, H., Tajer, M.H.M., Sahneh, K.K., Ziaei, S.Y., Jalali, F., Akouchakian, E., 2024. Recent advances in Organ-on-a-Chip models: How precision engineering integrates cutting edge technologies in fabrication and characterization. Appl. Mater. Today 38, 102231. [CrossRef]
- Salinas, S., Jayasundara, N., 2022. Thermal plasticity in marine organisms. Mar. Biol. A Funct. Approach to Ocean. their Org. 327–348. [CrossRef]
- Santos-Cancel, M., Simpson, L.W., Leach, J.B., White, R.J., 2019. Direct, Real-Time Detection of Adenosine Triphosphate Release from Astrocytes in Three-Dimensional Culture Using an Integrated Electrochemical Aptamer-Based Sensor. ACS Chem. Neurosci. 10, 2070–2079. [CrossRef]
- Sarfraz, N., Lee, H.J., Rice, M.K., Moscoso, E., Shafik, L.K., Glasgow, E., Ranjit, S., Lambeck, B.J., Braselmann, E., 2023. Establishing Riboglow-FLIM to visualize noncoding RNAs inside live zebrafish embryos. Biophys. Reports 3. [CrossRef]
- Saw, P.E., Song, E., 2025. RNA-Based Imaging System. RNA Ther. Hum. Dis. 459–488. [CrossRef]
- Schmidt, C.M., Smolke, C.D., 2021. A convolutional neural network for the prediction and forward design of ribozyme-based gene-control elements. Elife 10. [CrossRef]
- Sharma, P., Mondal, K., Kumar, S., Tamang, S., Najar, I.N., Das, S., Thakur, N., 2022. RNA thermometers in bacteria: Role in thermoregulation. Biochim. Biophys. Acta - Gene Regul. Mech. 1865, 194871. [CrossRef]
- Sheng, J., Gong, J., Shi, Y., Wang, X., Liu, D., 2022. MicroRNA-22 coordinates vascular and motor neuronal pathfinding via sema4 during zebrafish development. Open Biol. 12. [CrossRef]
- Sherwood, A. V., Henkin, T.M., 2016. Riboswitch-Mediated Gene Regulation: Novel RNA Architectures Dictate Gene Expression Responses. Annu. Rev. Microbiol. 70, 361–374. [CrossRef]
- Shui, B., Ozer, A., Zipfel, W., Sahu, N., Singh, A., Lis, J.T., Shi, H., Kotlikoff, M.I., 2012. RNA aptamers that functionally interact with green fluorescent protein and its derivatives. Nucleic Acids Res. 40. [CrossRef]
- Somero, G.N., 2018. RNA thermosensors: How might animals exploit their regulatory potential? J. Exp. Biol. 221. [CrossRef]
- Song, Y., Xu, Z., Wang, F., 2020. Genetically Encoded Reporter Genes for MicroRNA Imaging in Living Cells and Animals. Mol. Ther. Nucleic Acids 21, 555–567. [CrossRef]
- Srinivas, T., Siqueira, E., Guil, S., 2023. Techniques for investigating lncRNA transcript functions in neurodevelopment. Mol. Psychiatry 2023 294 29, 874–890. [CrossRef]
- Su, Y., Hammond, M.C., 2020. RNA-based fluorescent biosensors for live cell imaging of small molecules and RNAs. Curr. Opin. Biotechnol. 63, 157–166. [CrossRef]
- Sun, X., Li, Q., Wang, Y., Zhou, W., Guo, Y., Chen, J., Zheng, P., Sun, J., Ma, Y., 2020. Isoleucyl-tRNA synthetase mutant based whole-cell biosensor for high-throughput selection of isoleucine overproducers. Biosens. Bioelectron. 172, 112783–112783. [CrossRef]
- Sunbul, M., Lackner, J., Martin, A., Englert, D., Hacene, B., Grün, F., Nienhaus, K., Nienhaus, G.U., Jäschke, A., 2021. Super-resolution RNA imaging using a rhodamine-binding aptamer with fast exchange kinetics. Nat. Biotechnol. 2021 396 39, 686–690. [CrossRef]
- Sung, D., Koo, J., 2021. A review of BioFET’s basic principles and materials for biomedical applications. Biomed. Eng. Lett. 11, 85–96. [CrossRef]
- Terzi, A., Alam, S.M.S., Suter, D.M., 2021. ROS Live Cell Imaging During Neuronal Development. J. Vis. Exp. 2021, 10.3791/62165. [CrossRef]
- Thomas, S.E., Balcerowicz, M., Chung, B.Y.W., 2022. RNA structure mediated thermoregulation: What can we learn from plants? Front. Plant Sci. 13, 938570. [CrossRef]
- Tian, M., Xu, S., Zhang, J., Wang, X., Li, Z., Liu, H., Song, R., Yu, Z., Wang, J., 2018. RNA Detection Based on Graphene Field-Effect Transistor Biosensor. Adv. Condens. Matter Phys. 2018, 8146765. [CrossRef]
- Tomescu, A.I., Robb, N.C., Hengrung, N., Fodor, E., Kapanidis, A.N., 2014. Single-molecule FRET reveals a corkscrew RNA structure for the polymerase-bound influenza virus promoter. Proc. Natl. Acad. Sci. U. S. A. 111, E3335–E3342. [CrossRef]
- Trachman, R.J., Cojocaru, R., Wu, D., Piszczek, G., Ryckelynck, M., Unrau, P.J., Ferré-D’Amaré, A.R., 2020. Structure-Guided Engineering of the Homodimeric Mango-IV Fluorescence Turn-on Aptamer Yields an RNA FRET Pair. Structure 28, 776-785.e3. [CrossRef]
- Tutucci, E., Vera, M., Biswas, J., Garcia, J., Parker, R., Singer, R.H., 2017. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat. Methods 2017 151 15, 81–89. [CrossRef]
- Verma, A.K., Noumani, A., Yadav, A.K., Solanki, P.R., 2023. FRET Based Biosensor: Principle Applications Recent Advances and Challenges. Diagnostics (Basel, Switzerland) 13. [CrossRef]
- Vong, Y.H., Sivashanmugam, L., Leech, R., Zaucker, A., Jones, A., Sampath, K., 2021. The RNA-binding protein Igf2bp3 is critical for embryonic and germline development in zebrafish. PLOS Genet. 17, e1009667. [CrossRef]
- Wang, J., Cui, C., Nan, H., Yu, Y., Xiao, Y., Poon, E., Yang, G., Wang, X., Wang, C., Li, L., Boheler, K.R., Ma, X., Cheng, X., Ni, Z., Chen, M., 2017. Graphene Sheet-Induced Global Maturation of Cardiomyocytes Derived from Human Induced Pluripotent Stem Cells. ACS Appl. Mater. Interfaces 9, 25929–25940. [CrossRef]
- Wang, L., Hast, K., Aggarwal, T., Baci, M., Hong, J., Izgu, E.C., 2022. MicroRNA detection in biologically relevant media using a split aptamer platform. Bioorg. Med. Chem. 69, 116909. [CrossRef]
- Wang, L., Xu, W., Zhang, S., Gundberg, G.C., Zheng, C.R., Wan, Z., Mustafina, K., Caliendo, F., Sandt, H., Kamm, R., Weiss, R., 2024. Sensing and guiding cell-state transitions by using genetically encoded endoribonuclease-mediated microRNA sensors. Nat. Biomed. Eng. 2024 812 8, 1730–1743. [CrossRef]
- Wang, S., Emery, N.J., Liu, A.P., 2019. A Novel Synthetic Toehold Switch for MicroRNA Detection in Mammalian Cells. ACS Synth. Biol. 8, 1079–1088. [CrossRef]
- Wang, X.W., Hu, L.F., Hao, J., Liao, L.Q., Chiu, Y.T., Shi, M., Wang, Y., 2019. A microRNA-inducible CRISPR–Cas9 platform serves as a microRNA sensor and cell-type-specific genome regulation tool. Nat. Cell Biol. 2019 214 21, 522–530. [CrossRef]
- Wang, Y., Li, Z., Weber, T.J., Hu, D., Lin, C.T., Li, J., Lin, Y., 2013. In situ live cell sensing of multiple nucleotides exploiting DNA/RNA aptamers and graphene oxide nanosheets. Anal. Chem. 85, 6775–6782. [CrossRef]
- Wang, Z., Sun, X., Zhang, X., Dong, B., Yu, H., 2021. Development of a miRNA Sensor by an Inducible CRISPR-Cas9 Construct in Ciona Embryogenesis. Mol. Biotechnol. 63, 613–620. [CrossRef]
- Wani, A.K., Akhtar, N., Mir, T. ul G., Chopra, C., Singh, R., Hong, J.C., Kadam, U.S., 2024. CRISPR/Cas12a-based biosensors for environmental monitoring and diagnostics. Environ. Technol. Innov. 34, 103625. [CrossRef]
- Wei, J., Zhang, W., Jiang, A., Peng, H., Zhang, Q., Li, Y., Bi, J., Wang, L., Liu, P., Wang, J., Ge, Y., Zhang, L., Yu, H., Li, L., Wang, S., Leng, L., Chen, K., Dong, B., 2024. Temporospatial hierarchy and allele-specific expression of zygotic genome activation revealed by distant interspecific urochordate hybrids. Nat. Commun. 2024 151 15, 1–11. [CrossRef]
- Weldemichael, T., Asemoloye, M.D., Marchisio, M.A., 2022. Feedforward Loops: Evolutionary Conserved Network Motifs Redesigned for Synthetic Biology Applications. Appl. Sci. 2022, Vol. 12, Page 8292 12, 8292. [CrossRef]
- Wile, B.M., Ban, K., Yoon, Y.S., Bao, G., 2014. Molecular beacon–enabled purification of living cells by targeting cell type–specific mRNAs. Nat. Protoc. 2014 910 9, 2411–2424. [CrossRef]
- Winkler, W.C., Nahvi, A., Roth, A., Collins, J.A., Breaker, R.R., 2004. Control of gene expression by a natural metabolite-responsive ribozyme. Nat. 2004 4286980 428, 281–286. [CrossRef]
- Wójcik, A.M., 2020. Research Tools for the Functional Genomics of Plant miRNAs During Zygotic and Somatic Embryogenesis. Int. J. Mol. Sci. 2020, Vol. 21, Page 4969 21, 4969. [CrossRef]
- Wong, C., Jurczak, E.M., Correspondence, R.R., Roy, R., 2024. Neuronal exosomes transport an miRISC cargo to preserve stem cell integrity during energy stress. Cell Rep. 43, 114851. [CrossRef]
- Wu, Y., Zhu, L., Li, S., Chu, H., Wang, X., Xu, W., 2023. High content design of riboswitch biosensors: All-around rational module-by-module design. Biosens. Bioelectron. 220, 114887. [CrossRef]
- Wu, Y.X., Kwon, Y.J., 2016. Aptamers: The “evolution” of SELEX. Methods 106, 21–28. [CrossRef]
- Xia, C., Colognori, D., Jiang, X.S., Xu, K., Doudna, J.A., 2025. Single-molecule live-cell RNA imaging with CRISPR–Csm. Nat. Biotechnol. 2025 1–8. [CrossRef]
- Xiang, J.S., Kaplan, M., Dykstra, P., Hinks, M., McKeague, M., Smolke, C.D., 2019. Massively parallel RNA device engineering in mammalian cells with RNA-Seq. Nat. Commun. 2019 101 10, 1–16. [CrossRef]
- Yang, L.Z., Wang, Y., Li, S.Q., Yao, R.W., Luan, P.F., Wu, H., Carmichael, G.G., Chen, L.L., 2019. Dynamic Imaging of RNA in Living Cells by CRISPR-Cas13 Systems. Mol. Cell 76, 981-997.e7. [CrossRef]
- Yao, Y., Yin, F., He, M., Cheng, W., Wang, Z., Xiang, Y., 2025. Multi-target recognition and DNAzyme signal amplification integrated functional nucleic acid framework for lncRNA in situ live cell imaging. Sensors Actuators B Chem. 422, 136599. [CrossRef]
- Yarra, S.S., Ashok, G., Mohan, U., 2023. “Toehold Switches; a foothold for Synthetic Biology.” Biotechnol. Bioeng. 120, 932–952. [CrossRef]
- Yin, S., Chen, A., Ding, Y., Song, J., Chen, R., Zhang, P., Yang, C., 2023. Recent advances in exosomal RNAs analysis towards diagnostic and therapeutic applications. TrAC Trends Anal. Chem. 158, 116840. [CrossRef]
- Yoo, H., Jo, H., Oh, S.S., 2020. Detection and beyond: challenges and advances in aptamer-based biosensors. Mater. Adv. 1, 2663–2687. [CrossRef]
- Yu, S., Lei, X., Qu, C., 2024. MicroRNA Sensors Based on CRISPR/Cas12a Technologies: Evolution From Indirect to Direct Detection. Crit. Rev. Anal. Chem. [CrossRef]
- Zhang, C., Miao, P., Sun, M., Yan, M., Liu, H., 2019. Progress in miRNA Detection Using Graphene Material–Based Biosensors. Small 15, 1901867. [CrossRef]
- Zhang, L., Xie, X., Djokovic, N., Nikolic, K., Kosenkov, D., Abendroth, F., Vázquez, O., 2023. Reversible Control of RNA Splicing by Photoswitchable Small Molecules. J. Am. Chem. Soc. 145, 12783–12792. [CrossRef]
- Zhang, Q., Ma, D., Wu, F., Standage-Beier, K., Chen, X., Wu, K., Green, A.A., Wang, X., 2021. Predictable control of RNA lifetime using engineered degradation-tuning RNAs. Nat. Chem. Biol. 2021 177 17, 828–836. [CrossRef]
- Zhang, Q., Su, C., Tian, X., Zhang, C.Y., 2023a. Corn-Based Fluorescent Light-Up Biosensors with Improved Signal-to-Background Ratio for Label-Free Detection of Long Noncoding RNAs. Anal. Chem. 95, 8097–8104. [CrossRef]
- Zhang, Q., Zhao, S., Su, C., Han, Q., Han, Y., Tian, X., Li, Y., Zhang, C.Y., 2023b. Construction of a Quantum-Dot-Based FRET Nanosensor through Direct Encoding of Streptavidin-Binding RNA Aptamers for N6-Methyladenosine Demethylase Detection. Anal. Chem. 95, 13201–13210. [CrossRef]
- Zhang, Z., Hu, J.J., Lin, S., Wu, J., Xia, F., Lou, X., 2024. Field effect transistor biosensors for healthcare monitoring. Interdiscip. Med. 2, e20240032. [CrossRef]
- Zhao, E.M., Mao, A.S., de Puig, H., Zhang, K., Tippens, N.D., Tan, X., Ran, F.A., Han, I., Nguyen, P.Q., Chory, E.J., Hua, T.Y., Ramesh, P., Thompson, D.B., Oh, C.Y., Zigon, E.S., English, M.A., Collins, J.J., 2021. RNA-responsive elements for eukaryotic translational control. Nat. Biotechnol. 2021 404 40, 539–545. [CrossRef]
- Zhao, N.N., Li, F.Z., Zhang, X., Liu, M., Cao, H., Zhang, C.Y., 2023. Construction of Genetically Encoded Light-Up RNA Aptamers for Label-free and Ultrasensitive Detection of CircRNAs in Cancer Cells and Tissues. Anal. Chem. 95, 8728–8734. [CrossRef]
- Zheng, L., Li, J., Li, Y., Sun, W., Ma, L. Le, Qu, F., Tan, W., 2024. Empowering Exosomes with Aptamers for Precision Theranostics. Small Methods 2400551. [CrossRef]
- Zheng, R., Xue, Z., You, M., 2024. Optogenetic Tools for Regulating RNA Metabolism and Functions. ChemBioChem 25, e202400615. [CrossRef]
- Zhong, W., Sczepanski, J.T., 2019. A Mirror Image Fluorogenic Aptamer Sensor for Live-Cell Imaging of MicroRNAs. ACS Sensors 4, 566–570. [CrossRef]
- Zhou, H., Zhang, S., 2022. Recent Development of Fluorescent Light-Up RNA Aptamers. Crit. Rev. Anal. Chem. 52, 1644–1661. [CrossRef]
- Zhou, Xiaolu, Gao, H., Xiang, L., Liu, H., Zhou, Xiang, 2025. Developments of the mDZ-RhoBAST Probe for Super-Resolution Imaging of FTO Demethylation in Live Cells. J. Am. Chem. Soc. [CrossRef]
- Zhou, Zhixin, Han, B., Wang, Y., Lin, N., Zhou, Zhongqiu, Zhang, Yuan, Bai, Y., Shen, L., Shen, Y., Zhang, Yuanjian, Yao, H., 2024. Fast and sensitive multivalent spatial pattern-recognition for circular RNA detection. Nat. Commun. 2024 151 15, 1–15. [CrossRef]
Figure 1.
Overview of RNA biosensor targets during development. Venn diagram categorizing RNA biosensors based on whether they detect intrinsic RNA biology processes (e.g., expression, localization, splicing) or extrinsic developmental cues (e.g., pH, temperature, ROS). Sensors in the overlapping region can be adapted to detect either type of signal, depending on design and application context.
Figure 1.
Overview of RNA biosensor targets during development. Venn diagram categorizing RNA biosensors based on whether they detect intrinsic RNA biology processes (e.g., expression, localization, splicing) or extrinsic developmental cues (e.g., pH, temperature, ROS). Sensors in the overlapping region can be adapted to detect either type of signal, depending on design and application context.
Figure 2.
Classification of RNA biosensors by function, mechanism, delivery, and cellular context. Radial diagrams illustrating four complementary classifications of RNA biosensors: (1) functional input class, reflecting the type of biological signal each sensor detects; (2) detection mechanism, representing the molecular basis by which sensors operate; (3) delivery and accessibility, categorizing how sensors are introduced and applied in experimental systems; and (4) cellular compatibility, describing the biological environments in which each sensor is functional. Some RNA biosensors, such as CRISPR-based sensors, have diverse capabilities and therefore appear in multiple classifications and sections to reflect their multifunctionality.
Figure 2.
Classification of RNA biosensors by function, mechanism, delivery, and cellular context. Radial diagrams illustrating four complementary classifications of RNA biosensors: (1) functional input class, reflecting the type of biological signal each sensor detects; (2) detection mechanism, representing the molecular basis by which sensors operate; (3) delivery and accessibility, categorizing how sensors are introduced and applied in experimental systems; and (4) cellular compatibility, describing the biological environments in which each sensor is functional. Some RNA biosensors, such as CRISPR-based sensors, have diverse capabilities and therefore appear in multiple classifications and sections to reflect their multifunctionality.
Figure 3.
Linking RNA biosensors to fundamental challenges in developmental biology. Radial diagram linking RNA biosensors to seven fundamental questions/challenges in developmental biology. Each colored sector represents a core question, with biosensors placed according to their relevance. Some biosensors appear in multiple sectors, reflecting their broad applicability in the field.
Figure 3.
Linking RNA biosensors to fundamental challenges in developmental biology. Radial diagram linking RNA biosensors to seven fundamental questions/challenges in developmental biology. Each colored sector represents a core question, with biosensors placed according to their relevance. Some biosensors appear in multiple sectors, reflecting their broad applicability in the field.
Figure 4.
Anticipated suitability of RNA-based biosensors in developmental biology research based on literature trends and practical considerations. The X-axis (“ease of use”) reflects the relative technical complexity of implementing each sensor in developmental biology contexts, based on a qualitative assessment of existing literature and practical considerations. The Y-axis (“stage of research”) represents the extent of exploration of each sensor in the existing developmental biology literature. These placements are not based on direct comparative analysis, as no such study has been conducted in any field. Rather, they represent a qualitative synthesis of the available literature. This figure is intended as a conceptual framework to guide interpretation, rather than a quantitative evaluation.
Figure 4.
Anticipated suitability of RNA-based biosensors in developmental biology research based on literature trends and practical considerations. The X-axis (“ease of use”) reflects the relative technical complexity of implementing each sensor in developmental biology contexts, based on a qualitative assessment of existing literature and practical considerations. The Y-axis (“stage of research”) represents the extent of exploration of each sensor in the existing developmental biology literature. These placements are not based on direct comparative analysis, as no such study has been conducted in any field. Rather, they represent a qualitative synthesis of the available literature. This figure is intended as a conceptual framework to guide interpretation, rather than a quantitative evaluation.
Table 1.
RNA-based biosensors, their mechanisms, unique attributes, and potential applications in developmental biology research.
Table 1.
RNA-based biosensors, their mechanisms, unique attributes, and potential applications in developmental biology research.
| RNA-Based Biosensors |
Function |
Most Common Usage |
Potential Developmental Applications |
| Spinach, Broccoli, Mango, and Pepper aptamers |
Fluoresce upon binding small molecules for RNA visualization |
Live-cell RNA imaging |
Real-time tracking of developmental RNAs |
| CRISPR-Cas13/Cas12a RNA sensors |
Detect RNA sequences via guide RNAs and produce signals |
Diagnostics for RNA biomarkers |
Detecting transient RNAs in gene networks (e.g., mapping m6A during development) |
| Riboswitch biosensors |
Bind small molecules and regulate gene expression structurally |
Metabolite sensing and gene regulation |
Studying conserved metabolic pathways during development |
| Toehold switches |
Regulate translation by hairpin unwinding upon RNA triggers |
Pathogen detection and synthetic biology |
Monitoring developmental stage-specific RNA triggers |
| Molecular Beacon biosensors |
Fluoresce when hybridizing with specific RNA sequences |
RNA quantification in diagnostics |
Visualizing developmental RNA patterns |
| MS2 system |
Tag RNA for visualization and tracking with phage coat proteins |
Studying RNA localization and stability |
Tracking RNA transport in cell differentiation |
| miRNA sensors |
Hybridize with microRNAs to produce measurable signals |
Cancer diagnostics and monitoring biomarkers |
Identifying roles of miRNAs in development |
| RNA thermometers |
Change conformation with temperature to regulate translation |
Studying bacterial thermal stress responses |
Exploring species-specific thermal effects on development |
| Fluorescence-quenching RNA aptamers |
Quench or enhance fluorescence based on environmental stimuli |
Environmental monitoring and metabolite tracking |
Examining metabolite-driven developmental changes |
| FRET-based RNA sensors |
Detect RNA conformational changes using FRET |
Visualizing RNA folding and interactions (e.g. real-time detection of m6A effects on RNA structure) |
Tracking RNA structural changes during development (e.g., m6A modifications effects on RNA folding) |
| Catalytic RNA biosensors (ribozymes) |
Catalyze reactions upon ligand binding for signal generation |
Detecting small molecules in RNA studies |
Detecting conserved signaling molecules in development |
| Exosomal RNA sensors |
Hybridize with RNA in exosomes for fluorescence or signals |
Cancer diagnostics and cell communication studies |
Studying RNA communication in development |
| Split RNA aptamers and ribozymes |
Activate fluorescence or catalysis when RNA fragments join |
Studying splicing and RNA-protein interactions |
Detecting splicing events at developmental level |
| RNA sensors for reactive oxygen species (ROS) |
Detect reactive oxygen species and fluoresce in response |
Researching oxidative stress in diseases |
Monitoring stress in regeneration or embryogenesis |
| Electrochemical RNA sensors |
Produce electrical signals upon RNA binding at electrodes |
Point-of-care diagnostics for RNA targets |
Detecting RNA biomarkers in development |
| Circular RNA sensors |
Resist degradation while binding targets to emit signals |
Long-term RNA expression monitoring |
Tracking stable RNAs in developmental contexts |
| tRNA-based biosensors |
Detect metabolites or amino acids via modified tRNA |
Exploring translation and metabolic regulation |
Studying amino acid-driven translation shifts |
Table 2.
Key limitations and challenges in the application of RNA biosensors in developmental biology research.
Table 2.
Key limitations and challenges in the application of RNA biosensors in developmental biology research.
| RNA-Based Biosensors |
Delivery Challenge |
Stability Issue |
Signal Limitations |
Tissue/Imaging Constraints |
Target Specificity Concern |
| Fluorescence-quenching RNA aptamers |
Synthetic RNA delivery required |
Moderate; prone to degradation in vivo |
Environmental sensitivity can cause false signals |
Auto-fluorescence interference in deep tissues |
Limited tuning for new targets |
| Ribozymes & Catalytic RNAs |
Often require stable expression or microinjection |
Variable, depends on structure and sequence |
Can produce background activity |
Requires amplification or reporter coupling |
Context-dependent activation |
| Molecular Beacon |
Challenging to deliver into intact embryos |
Sensitive to nucleases |
High background if improperly designed |
Difficult deep-tissue detection |
High for known targets, limited for novel RNAs |
| Electrochemical sensors |
Difficult in vivo; mostly external use |
Good extracellularly; intracellular stability is low |
Requires external instrumentation |
Limited spatial resolution |
High when using validated probes |
| ROS-RNA sensors |
Synthetic delivery or transfection |
Rapid degradation under stress conditions |
ROS fluctuation may confound signal |
Unspecific spatial resolution |
Low; generally detects ROS rather than RNA |
| RNA thermosensors |
Genetically encoded or synthetic |
Good if encoded; moderate if synthetic |
Temperature range may limit precision |
Affected by tissue heat gradients |
Low; responds to temperature not sequence |
| circRNA sensors |
Requires probe design or expression vectors |
Stable once formed |
Indirect; depends on backsplicing detection |
Tissue-specific expression complicates interpretation |
High if designed properly |
| miRNA sensors |
Requires careful delivery (e.g., nanoparticles) |
miRNA-probe interactions stable |
Signal influenced by miRNA abundance |
Tissue heterogeneity affects readout |
High with optimized probes |
| tRNA sensors |
Requires intracellular delivery |
Moderate; depends on structural mimicry |
Signal can be influenced by amino acid levels |
Requires normalization in complex tissues |
Moderate; depends on reporter design |
| Riboswitch |
Needs expression in host cells |
Good when chromosomally integrated |
Ligand availability may limit output |
May not work uniformly across tissues |
High if ligand is specific |
| Exosomal RNA sensors |
Requires vesicle isolation or extracellular sampling |
Stable in extracellular fluids |
Dependent on sample prep and purity |
No spatial resolution in tissue |
High for known exosomal RNAs |
| MS2 System |
Genetic tagging required |
Stable in vivo when fused with coat protein |
Signal dilution during division |
Limited in deep tissues unless optimized |
High for tagged transcripts |
| FRET sensors |
Genetic fusion or co-expression required |
Susceptible to photobleaching |
Sensitive to microenvironment changes |
Shallow imaging unless NIR optimized |
High for structured RNAs |
| CRISPR sensors |
Requires Cas protein and guide delivery |
Good if properly designed |
Collateral activity may cause noise |
Depends on delivery method and tissue |
High; programmable specificity |
| Fluorescent Aptamers |
Requires fluorophore and RNA co-expression |
Can be stabilized via aptamer design |
Background fluorescence possible |
Autofluorescence in thick tissues |
High when sequence-targeted |
| Toehold switches |
Needs transcription in cells |
Stable when encoded |
Minimal background with rational design |
Hard to tune expression across tissues |
High; modular and programmable |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).