Submitted:
24 June 2025
Posted:
24 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Historical Background of Bacteriophage Research
1.2. Modern Applications and the Need for Phage Engineering
2. Biological Foundations of Phage Engineering
2.1. Phage Structure and Morphology
2.2. Phage Life Cycles and Genome Packaging Mechanisms
2.3. Host Recognition and Engineering of Receptor-Binding Proteins
| Phage name | Phage type | Original RBPs/ Host | New RBPs/ Host | Introduced modification | Purpose and Application | References |
|---|---|---|---|---|---|---|
| Ur-l | Temperate | LamB, OmpC | OmpC and LPS | P2-STF | Genetic engineering of bacteria in the mouse gut /Transduction capsid/dCas9 base editor | [22] |
| Ur-l | Temperate | LamB, OmpC | OmpC and O-antigen (O157) | STF tail fiber that recognize O-antigen (O157) | Eliminate STEC/ Transduction capsid/ Cas12a | [23] |
| P2 | Temperate | LPS | Shigilla flexneri M90T, Escherichia coli O-antigen O157 | Hybrid long tail fiber (gpHG or gpH only) P1-S’ and P1-U’, PhiV10 tail spike protein, respectively | Eliminate STEC/ Transduction capsid/Cas9 | [35] |
| P2vir1 | Lytic | LPS | Salmonella (OmpC) | P2-gpH and S16- gp37 | Expand the Host range | [24] |
| T7 | Lytic | Rough LPS | - | Selective mutations in HRDRs of gp17 | Expand host range/ Transduction capsid | [61] |
| T7 | Lytic | Rough LPS | Kellebisella | Swapping gp11, gp12, and gp17 with those of phage K11 | Expand host range | [76] |
| T3 | Lytic | - | Escherichia coli (BW25113) | Swapping T3-gp17 with T7-gp17 | Expand host range | [78] |
| a15 | Lytic | LPS | Tsx | Knock-In gp38 of phage a17 | Expand host range/ Cas delivery | [25] |
| Others | R-type pyocin | Pseudomonas | Escherichia coli O-antigen (O157) | Utilize the C-terminal of the phiV10 tail spike protein | Expand host range/ Bacteria killing agent | [77] |
| Others | Nisin- nanoparticles | - | MRSA | RBPs of Staphylococcal phage Sb-1 | Expand host range/ Bacteria killing agent | [42] |
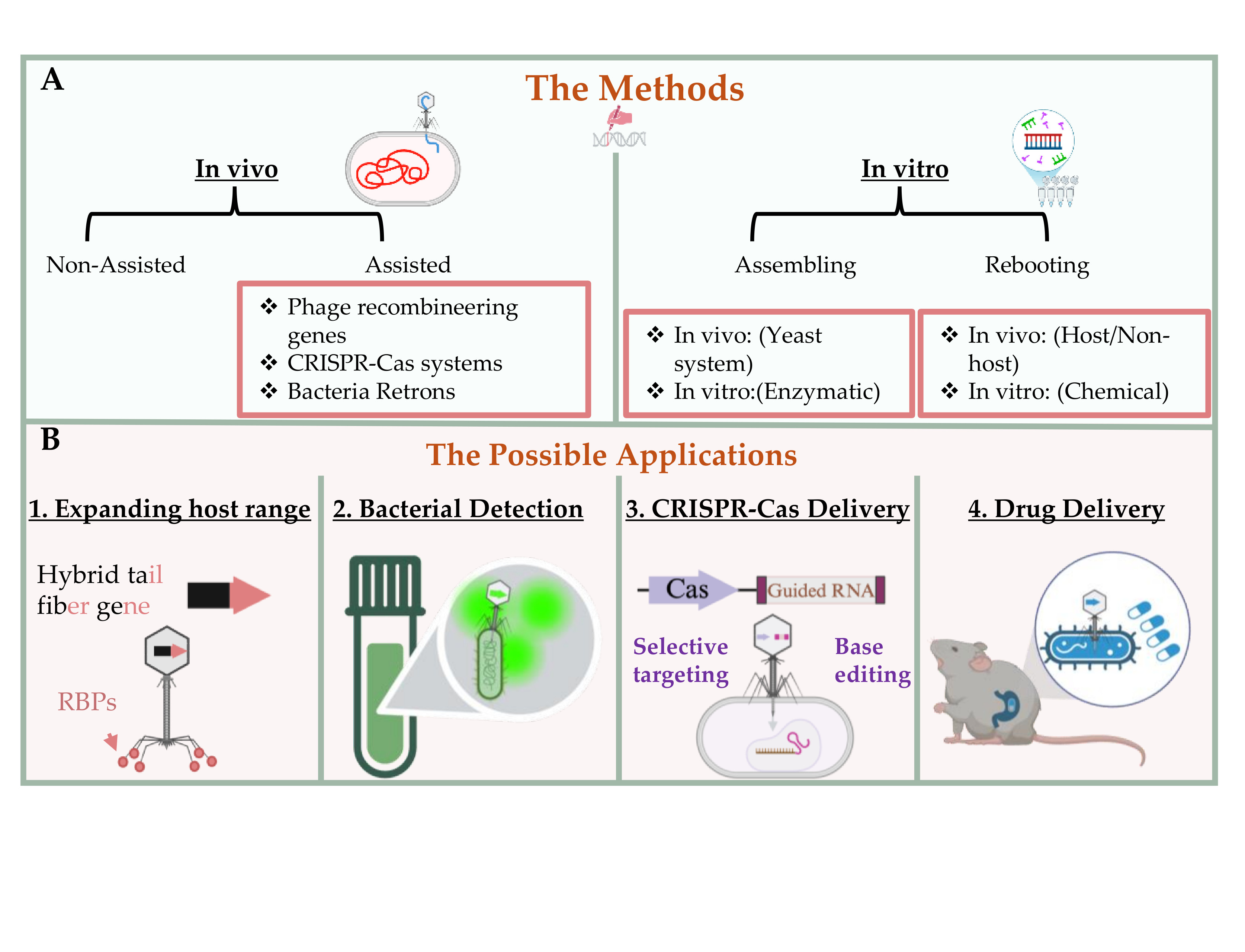
3. Engineering Strategies for Bacteriophages
3.1. Genetic Engineering Approaches
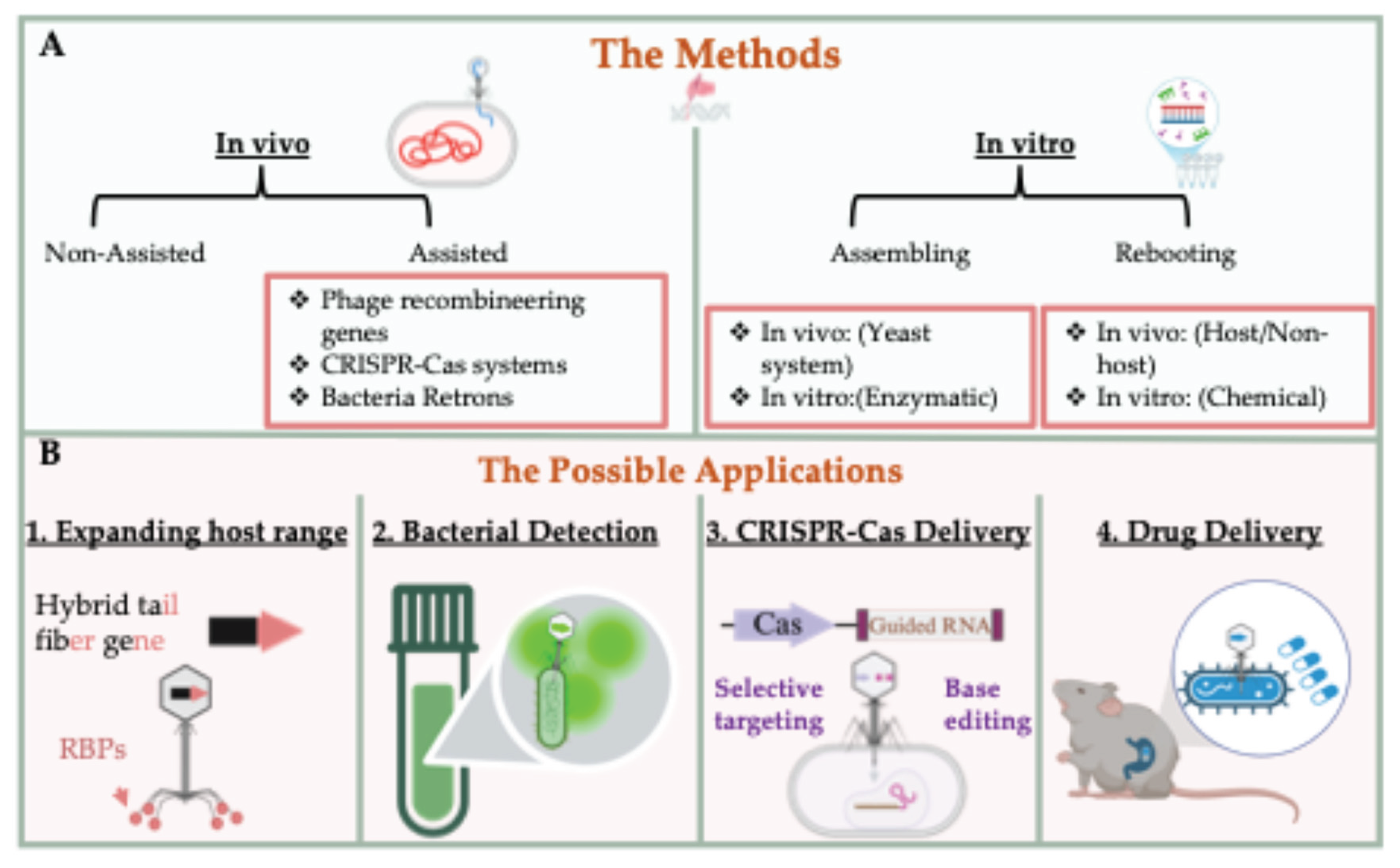
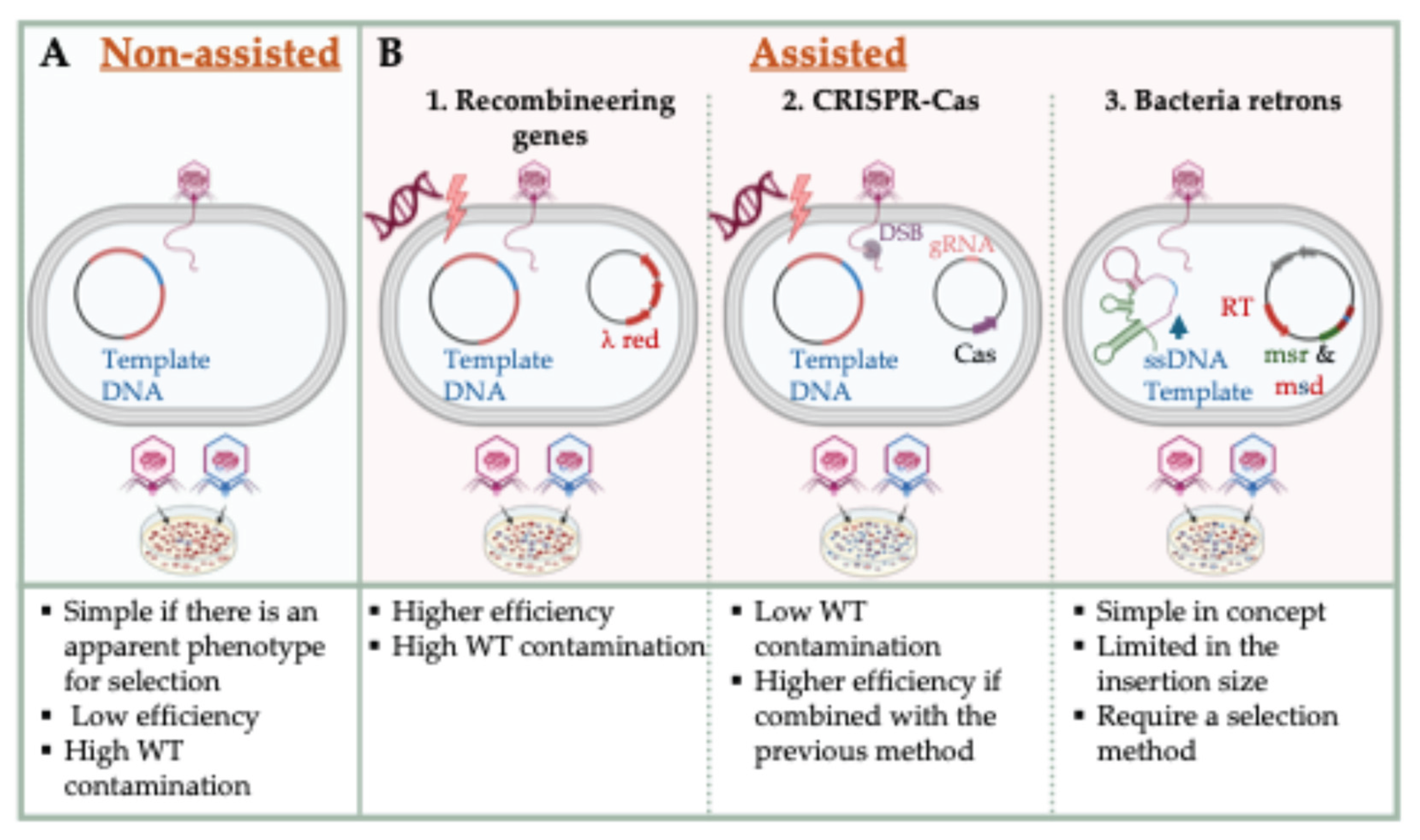
3.1.1. Phage Recombineering System
3.1.2. CRISPR-Cas-Assisted Phage Editing
3.1.3. Retron-Mediated Genome Modification
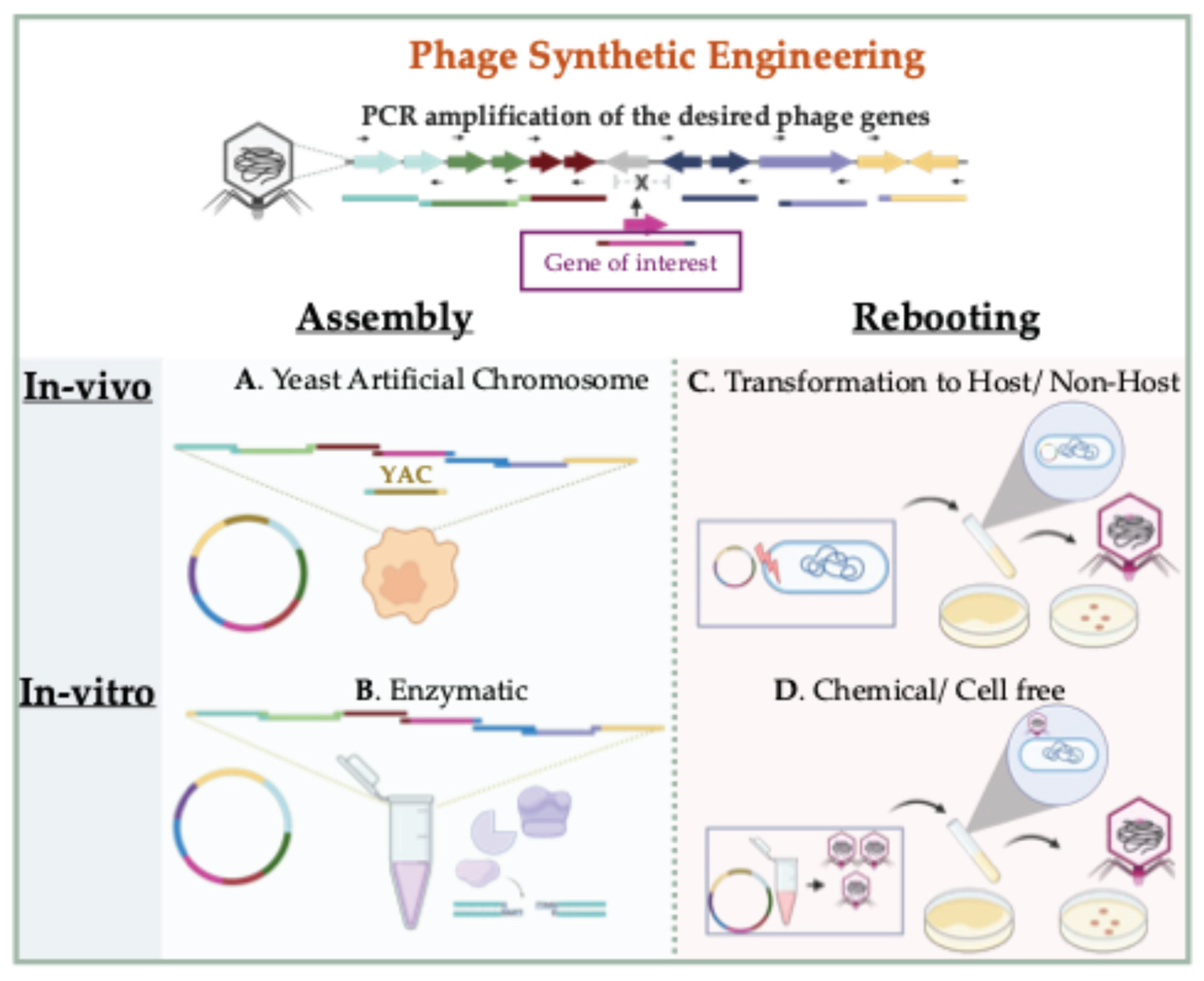
3.2. In Vitro Synthetic Engineering Platforms
3.2.1. Synthetic Assembly of Phage Genomes
3.2.2. Rebooting Engineered Phage
4. Applications and Innovations in Engineered Phage Platforms
4.1. Host Range Expansion and Targeting
4.2. CRISPR-Cas Delivery via Phage Capsids
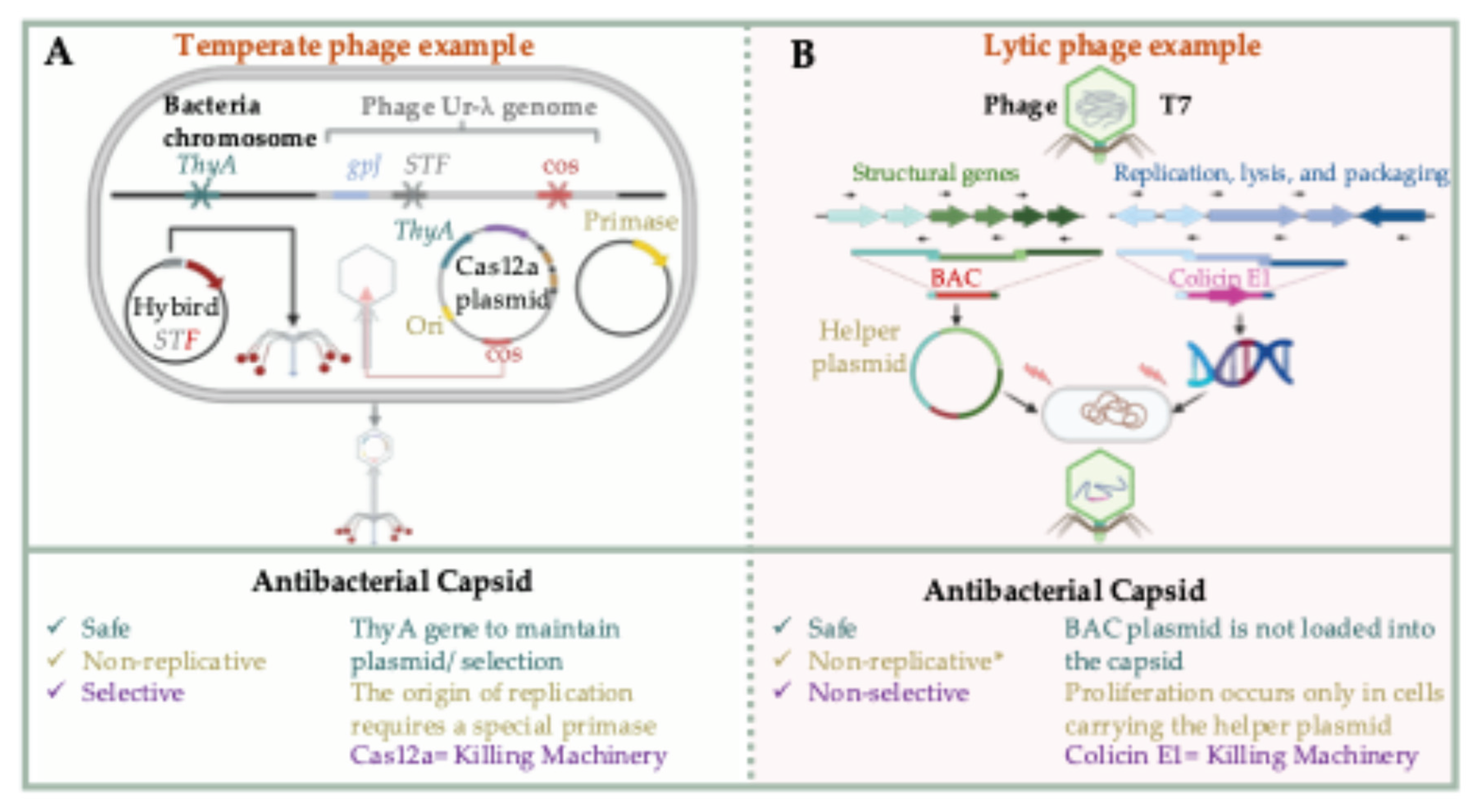
4.3. Biocontained Non-Replicative Phage Therapeutics
4.4. Diagnostic and Antimicrobial Payload Delivery
5. Conclusions and Future Perspectives
Author Contributions
Funding and Acknowledgment
Conflicts of Interest
References
- Hankin, ME. L’action Bactéricide Des Eaux de La et Du Gange Sur Le Vibrion Du Choléra. Ann. Inst. Pasteur 1896, 10. [Google Scholar]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J. Bacteriophage Therapy. Antimicrob Agents Chemother 2001, 45, 649–659. [Google Scholar] [CrossRef]
- Twort, F.W. An investigation on the nature of ultra-microscopic viruses. The Lancet 1915, 186, 1241–1243. [Google Scholar] [CrossRef]
- D’Herelle F. On an invisible microbe antagonistic toward dysenteric bacilli: brief note by Mr. F. D’Herelle, presented by Mr. Roux. 1917. Res Microbiol. 2007 Sep;158(7):553-4. [CrossRef]
- Wollman, E. The phenomenon of twort-d’hérelle and its significance. The Lancet 1935, 226, 1312–1314. [Google Scholar] [CrossRef]
- Chanishvili, N. Phage Therapy—History from Twort and d’Herelle Through Soviet Experience to Current Approaches. Adv Virus Res 2012, 83, 3–40. [Google Scholar] [CrossRef]
- Peitzman, S.J. Félix d’Hérelle and the Origins of Molecular Biology (Review). Bull Hist Med 2001, 75. [Google Scholar] [CrossRef]
- Summers, W.C. The Strange History of Phage Therapy. Bacteriophage 2012, 2, 130–133. [Google Scholar] [CrossRef]
- Hershey, A.D.; Chase, M.M. Independent Functions of Viral Protein and Nucleic Acid in Growth of Bacteriophage. J. Gen. Physiol. 1952, 36, 39–56. [Google Scholar] [CrossRef]
- Van Valen, D.; Wu, D.; Chen, Y.J.; Tuson, H.; Wiggins, P.; Phillips, R. A Single-Molecule Hershey-Chase Experiment. Current Biology 2012, 22, 1339–1343. [Google Scholar] [CrossRef]
- Hershey, A.D.; Rotman, R. Genetic Recombination between Host-Range and Plaque-Type Mutants of Bacteriophage in Single Bacterial Cells. Genetics 1949, 34, 44. [Google Scholar] [CrossRef]
- Harshey, R.M. The Mu Story: How a Maverick Phage Moved the Field Forward. Mob DNA 2012, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Pingoud, A.; Wilson, G.G.; Wende, W. Type II Restriction Endonucleases—A Historical Perspective and More. Nucleic Acids Res 2014, 42, 7489–7527. [Google Scholar] [CrossRef] [PubMed]
- Rossi, R.; Montecucco, A.; Ciarrocchi, G.; Biamonti, G. Functional Characterization of the T4 DNA Ligase: A New Insight into the Mechanism of Action; Oxford University Press, 1997; Vol. 25.
- Sanger, F.; Air, G.M.; Barrell, B.G.; Brown, N.L.; Coulson, A.R.; Fiddes, J.C.; Hutchison, C.A.; Slocombe, P.M.; Smith, M. Nucleotide Sequence of Bacteriophage Φx174 DNA. Nature 1977, 265, 687–695. [Google Scholar] [CrossRef]
- Navarro, F.; Muniesa, M. Phages in the Human Body. Front Microbiol 2017, 8. [Google Scholar] [CrossRef]
- Breitbart, M.; Rohwer, F. Here a Virus, There a Virus, Everywhere the Same Virus? Trends Microbiol 2005, 13, 278–284. [Google Scholar] [CrossRef]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage Diversity, Genomics and Phylogeny. Nature Reviews Microbiology 2020 18:3 2020, 18, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Uyttebroek, S.; Chen, B.; Onsea, J.; Ruythooren, F.; Debaveye, Y.; Devolder, D.; Spriet, I.; Depypere, M.; Wagemans, J.; Lavigne, R.; et al. Safety and Efficacy of Phage Therapy in Difficult-to-Treat Infections: A Systematic Review. Lancet Infect Dis 2022, 22, e208–e220. [Google Scholar] [CrossRef]
- Azam, A.H.; Tan, X.E.; Veeranarayanan, S.; Kiga, K.; Cui, L. Bacteriophage Technology and Modern Medicine. Antibiotics 2021, Vol. 10, Page 999 2021, 10, 999. [Google Scholar] [CrossRef]
- Pirnay, J.P.; Djebara, S.; Steurs, G.; Griselain, J.; Cochez, C.; De Soir, S.; Glonti, T.; Spiessens, A.; Vanden Berghe, E.; Green, S.; et al. Personalized Bacteriophage Therapy Outcomes for 100 Consecutive Cases: A Multicentre, Multinational, Retrospective Observational Study. Nature Microbiology 2024 9:6 2024, 9, 1434–1453. [Google Scholar] [CrossRef]
- Brödel, A.K.; Charpenay, L.H.; Galtier, M.; Fuche, F.J.; Terrasse, R.; Poquet, C.; Havránek, J.; Pignotti, S.; Krawczyk, A.; Arraou, M.; et al. In Situ Targeted Base Editing of Bacteria in the Mouse Gut. Nature 2024 632:8026 2024, 632, 877–884. [Google Scholar] [CrossRef]
- Galtier, M.; Krawczyk, A.; Fuche, F.J.; Charpenay, L.H.; Stzepourginski, I.; Pignotti, S.; Arraou, M.; Terrasse, R.; Brödel, A.K.; Poquet, C.; et al. Treatment of STEC Infection via CRISPR-Cas Targeted Cleavage of the Shiga Toxin Gene in Animal Models. bioRxiv 2025, 2025.02.28.640725. [CrossRef]
- Cunliffe, T.G.; Parker, A.L.; Jaramillo, A. Pseudotyping Bacteriophage P2 Tail Fibers to Extend the Host Range for Biomedical Applications. ACS Synth Biol 2022, 11, 3207–3215. [Google Scholar] [CrossRef] [PubMed]
- Gencay, Y.E.; Jasinskytė, D.; Robert, C.; Semsey, S.; Martínez, V.; Petersen, A.Ø.; Brunner, K.; de Santiago Torio, A.; Salazar, A.; Turcu, I.C.; et al. Engineered Phage with Antibacterial CRISPR–Cas Selectively Reduce E. Coli Burden in Mice. Nature Biotechnology 2023 42:2 2023, 42, 265–274. [Google Scholar] [CrossRef]
- Lu, T.K.; Collins, J.J. Dispersing Biofilms with Engineered Enzymatic Bacteriophage; Proc. Natl. Acad. Sci. U.S.A. (2007). 104 (27) 11197-11202. [CrossRef]
- Cheng, L.; Deng, Z.; Tao, H.; Song, W.; Xing, B.; Liu, W.; Kong, L.; Yuan, S.; Ma, Y.; Wu, Y.; et al. Harnessing Stepping-Stone Hosts to Engineer, Select, and Reboot Synthetic Bacteriophages in One Pot. Cell Reports Methods 2022, 2, 100217. [Google Scholar] [CrossRef]
- Eghbalpoor, F.; Gorji, M.; Alavigeh, M.Z.; Moghadam, M.T. Genetically Engineered Phages and Engineered Phage-Derived Enzymes to Destroy Biofilms of Antibiotics Resistance Bacteria. Heliyon 2024, 10, e35666. [Google Scholar] [CrossRef] [PubMed]
- Meile, S.; Du, J.; Staubli, S.; Grossmann, S.; Koliwer-Brandl, H.; Piffaretti, P.; Leitner, L.; Matter, C.I.; Baggenstos, J.; Hunold, L.; et al. Engineered Reporter Phages for Detection of Escherichia Coli, Enterococcus, and Klebsiella in Urine. Nature Communications 2023 14:1 2023, 14, 1–15. [Google Scholar] [CrossRef]
- Kim, J.; Kim, M.; Kim, S.; Ryu, S. Sensitive Detection of Viable Escherichia Coli O157:H7 from Foods Using a Luciferase-Reporter Phage PhiV10lux. Int J Food Microbiol 2017, 254, 11–17. [Google Scholar] [CrossRef]
- Tamura, A.; Azam, A.H.; Nakamura, T.; Lee, K.; Iyoda, S.; Kondo, K.; Ojima, S.; Chihara, K.; Yamashita, W.; Cui, L.; et al. Synthetic Phage-Based Approach for Sensitive and Specific Detection of Escherichia Coli O157. Communications Biology 2024 7:1 2024, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pulkkinen, E.M.; Hinkley, T.C.; Nugen, S.R. Utilizing in Vitro DNA Assembly to Engineer a Synthetic T7 Nanoluc Reporter Phage for Escherichia Coli Detection. Integr Biol (Camb) 2019, 11, 63–68. [Google Scholar] [CrossRef]
- Baker, Z.R.; Zhang, Y.; Zhang, H.; Franklin, H.C.; Serpa, P.B.S.; Southard, T.; Li, L.; Hsu, B.B. Sustained in Situ Protein Production and Release in the Mammalian Gut by an Engineered Bacteriophage. Nat Biotechnol 2025, 1–10. [Google Scholar] [CrossRef]
- Dhasmana, N.; Ram, G.; McAllister, K.N.; Chupalova, Y.; Lopez, P.; Ross, H.F.; Novick, R.P. Dynamics of Antibacterial Drone Establishment in Staphylococcus Aureus: Unexpected Effects of Antibiotic Resistance Genes. mBio 2021, 12. [Google Scholar] [CrossRef]
- Fa-Arun, J.; Huan, Y.W.; Darmon, E.; Wang, B. Tail-Engineered Phage P2 Enables Delivery of Antimicrobials into Multiple Gut Pathogens. ACS Synth Biol 2023, 12, 596–607. [Google Scholar] [CrossRef] [PubMed]
- Kiga, K.; Tan, X.E.; Ibarra-Chávez, R.; Watanabe, S.; Aiba, Y.; Sato’o, Y.; Li, F.Y.; Sasahara, T.; Cui, B.; Kawauchi, M.; et al. Development of CRISPR-Cas13a-Based Antimicrobials Capable of Sequence-Specific Killing of Target Bacteria. Nat Commun 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Shimamori, Y.; Tan, X.E.; Li, F.Y.; Nishikawa, Y.; Watanabe, S.; Sasahara, T.; Miyanaga, K.; Aiba, Y.; Veeranarayanan, S.; Thitiananpakorn, K.; et al. Efficient Synthesis of CRISPR-Cas13a-Antimicrobial Capsids against MRSA Facilitated by Silent Mutation Incorporation. Scientific Reports 2024 14:1 2024, 14, 1–10. [Google Scholar] [CrossRef]
- Li, F.Y.; Tan, X.E.; Shimamori, Y.; Kiga, K.; Veeranarayanan, S.; Watanabe, S.; Nishikawa, Y.; Aiba, Y.; Sato’o, Y.; Miyanaga, K.; et al. Phagemid-Based Capsid System for CRISPR-Cas13a Antimicrobials Targeting Methicillin-Resistant Staphylococcus Aureus. Commun Biol 2024, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mcgillin, M.; Tokman, J.I.; Hsu, E.; Alcaine, S.D.; Denes, T.G. Assessment of Resistance to Colicinogenic Synthetic Phage Antimicrobial System. Microbiol Spectr 2024, 12. [Google Scholar] [CrossRef]
- Kiga, K.; Sato’o, Y.; Tan, X.-E.; Miyanaga, K.; Nguyen, H.M.; Li, F.-Y.; Azam, A.H.; Veeranarayanan, S.; Watanabe, S.; Aiba, Y.; et al. Development of a Non-Replicative Phage-Based DNA Delivery System and Its Application to Antimicrobial Therapies. PNAS Nexus 2025. [CrossRef]
- Zhu, J.; Batra, H.; Ananthaswamy, N.; Mahalingam, M.; Tao, P.; Wu, X.; Guo, W.; Fokine, A.; Rao, V.B. Design of Bacteriophage T4-Based Artificial Viral Vectors for Human Genome Remodeling. Nature Communications 2023 14:1 2023, 14, 1–19. [Google Scholar] [CrossRef]
- Wan, H.; Zhong, X.; Yang, S.; Deng, J.; Song, X.; Liu, Y.; Li, Y.; Yin, Z.; Zhao, X. Enhancing the Therapeutic Potential of Peptide Antibiotics Using Bacteriophage Mimicry Strategies. Advanced Science 2024, 12. [Google Scholar] [CrossRef]
- He, Y.; Chen, J. CRISPR/Cas9-Mediated Genome Editing of T4 Bacteriophage for High-Throughput Antimicrobial Susceptibility Testing. Anal Chem 2024, 96. [Google Scholar] [CrossRef]
- Huan, Y.W.; Torraca, V.; Brown, R.; Fa-Arun, J.; Miles, S.L.; Oyarzún, D.A.; Mostowy, S.; Wang, B. P1 Bacteriophage-Enabled Delivery of CRISPR-Cas9 Antimicrobial Activity Against Shigella Flexneri. ACS Synth Biol 2023, 12, 709–721. [Google Scholar] [CrossRef]
- Valencia-Toxqui, G.; Ramsey, J. How to Introduce a New Bacteriophage on the Block: A Short Guide to Phage Classification. J Virol 2024, 98. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. 5500 Phages Examined in the Electron Microscope. Arch Virol 2007, 152, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.M.; Watanabe, S.; Sharmin, S.; Kawaguchi, T.; Tan, X.E.; Wannigama, D.L.; Cui, L. RNA and Single-Stranded DNA Phages: Unveiling the Promise from the Underexplored World of Viruses. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Albrycht, K.; Rynkiewicz, A.A.; Harasymczuk, M.; Barylski, J.; Zielezinski, A. Daily Reports on Phage-Host Interactions. Front Microbiol 2022, 13, 946070. [Google Scholar] [CrossRef]
- Yuan, Y.; Gao, M. Jumbo Bacteriophages: An Overview. Front Microbiol 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Michniewski, S.; Rihtman, B.; Cook, R.; Jones, M.A.; Wilson, W.H.; Scanlan, D.J.; Millard, A. A New Family of “Megaphages” Abundant in the Marine Environment. ISME Communications 2021, 1. [Google Scholar] [CrossRef]
- Zrelovs, N.; Dislers, A.; Kazaks, A. Motley Crew: Overview of the Currently Available Phage Diversity. Front Microbiol 2020, 11. [Google Scholar] [CrossRef]
- Olszak, T.; Latka, A.; Roszniowski, B.; Valvano, M.A.; Drulis-Kawa, Z. Phage Life Cycles Behind Bacterial Biodiversity. Curr Med Chem 2017, 24, 3987–4001. [Google Scholar] [CrossRef]
- Shkoporov, A.N.; Khokhlova, E. V.; Stephens, N.; Hueston, C.; Seymour, S.; Hryckowian, A.J.; Scholz, D.; Ross, R.P.; Hill, C. Long-Term Persistence of CrAss-like Phage CrAss001 Is Associated with Phase Variation in Bacteroides Intestinalis. BMC Biol 2021, 19, 1–16. [Google Scholar] [CrossRef]
- Dougherty, P.E.; Bernard, C.; Carstens, A.B.; Stanford, K.; McAllister, T.A.; Rocha, E.P.C.; Hansen, L.H. Persistent Virulent Phages Exist in Bacterial Isolates. bioRxiv 2025, 2024.12.31.630880. [CrossRef]
- Łobocka, M.B.; Rose, D.J.; Plunkett, G.; Rusin, M.; Samojedny, A.; Lehnherr, H.; Yarmolinsky, M.B.; Blattner, F.R. Genome of Bacteriophage P1. J Bacteriol 2004, 186, 7032. [Google Scholar] [CrossRef]
- Marvin, D.A.; Symmons, M.F.; Straus, S.K. Structure and Assembly of Filamentous Bacteriophages. Prog Biophys Mol Biol 2014, 114, 80–122. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, H.; Morita, M. Phage DNA Packaging. Genes to Cells 1997, 2, 537–545. [Google Scholar] [CrossRef]
- Casjens, S.R.; Gilcrease, E.B. Determining DNA Packaging Strategy by Analysis of the Termini of the Chromosomes in Tailed-Bacteriophage Virions. Methods Mol Biol 2009, 502, 91–111. [Google Scholar] [CrossRef] [PubMed]
- Merrill, B.D.; Ward, A.T.; Grose, J.H.; Hope, S. Software-Based Analysis of Bacteriophage Genomes, Physical Ends, and Packaging Strategies. BMC Genomics 2016, 17, 1–16. [Google Scholar] [CrossRef]
- Garneau, J.R.; Depardieu, F.; Fortier, L.C.; Bikard, D.; Monot, M. PhageTerm: A Tool for Fast and Accurate Determination of Phage Termini and Packaging Mechanism Using next-Generation Sequencing Data. Sci Rep 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Apjok, G.; Számel, M.; Christodoulou, C.; Seregi, V.; Vásárhelyi, B.M.; Stirling, T.; Eszenyi, B.; Sári, T.; Vidovics, F.; Nagrand, E.; et al. Characterization of Antibiotic Resistomes by Reprogrammed Bacteriophage-Enabled Functional Metagenomics in Clinical Strains. Nature Microbiology 2023 8:3 2023, 8, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Dunne, M.; Prokhorov, N.S.; Loessner, M.J.; Leiman, P.G. Reprogramming Bacteriophage Host Range: Design Principles and Strategies for Engineering Receptor Binding Proteins. Curr Opin Biotechnol 2021, 68, 272–281. [Google Scholar] [CrossRef]
- Taslem Mourosi, J.; Awe, A.; Guo, W.; Batra, H.; Ganesh, H.; Wu, X.; Zhu, J. Understanding Bacteriophage Tail Fiber Interaction with Host Surface Receptor: The Key “Blueprint” for Reprogramming Phage Host Range. International Journal of Molecular Sciences 2022, Vol. 23, Page 12146 2022, 23, 12146. [Google Scholar] [CrossRef]
- Leprince, A.; Mahillon, J. Phage Adsorption to Gram-Positive Bacteria. Viruses 2023, 15, 196. [Google Scholar] [CrossRef]
- Bennett, N.J.; Gagic, D.; Sutherland-Smith, A.J.; Rakonjac, J. Characterization of a Dual-Function Domain That Mediates Membrane Insertion and Excision of Ff Filamentous Bacteriophage. J Mol Biol 2011, 411, 972–985. [Google Scholar] [CrossRef]
- Kleinbeck, F.; Kuhn, A. Membrane Insertion of the M13 Minor Coat Protein G3p Is Dependent on Yidc and the Secayeg Translocase. Viruses 2021, 13, 1414. [Google Scholar] [CrossRef]
- Chang, C.; Guo, W.; Yu, X.; Guo, C.; Zhou, N.; Guo, X.; Huang, R.L.; Li, Q.; Zhu, Y. Engineered M13 Phage as a Novel Therapeutic Bionanomaterial for Clinical Applications: From Tissue Regeneration to Cancer Therapy. Mater Today Bio 2023, 20, 100612. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide Endotoxins. Annu Rev Biochem 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Washizaki, A.; Yonesaki, T.; Otsuka, Y. Characterization of the Interactions between Escherichia Coli Receptors, LPS and OmpC, and Bacteriophage T4 Long Tail Fibers. Microbiologyopen 2016, 5, 1003–1015. [Google Scholar] [CrossRef]
- Montag, D.; Schwarz, H.; Henning, U. A Component of the Side Tail Fiber of Escherichia Coli Bacteriophage Lambda Can Functionally Replace the Receptor-Recognizing Part of a Long Tail Fiber Protein of the Unrelated Bacteriophage T4. J Bacteriol 1989, 171, 4378–4384. [Google Scholar] [CrossRef] [PubMed]
- González-García, V.A.; Pulido-Cid, M.; Garcia-Doval, C.; Bocanegra, R.; Van Raaij, M.J.; Martín-Benito, J.; Cuervo, A.; Carrascosa, J.L. Conformational Changes Leading to T7 DNA Delivery upon Interaction with the Bacterial Receptor. Journal of Biological Chemistry 2015, 290, 10038–10044. [Google Scholar] [CrossRef]
- Witte, S.; Zinsli, L. V.; Gonzalez-Serrano, R.; Matter, C.I.; Loessner, M.J.; van Mierlo, J.T.; Dunne, M. Structural and Functional Characterization of the Receptor Binding Proteins of Escherichia Coli O157 Phages EP75 and EP335. Comput Struct Biotechnol J 2021, 19, 3416–3426. [Google Scholar] [CrossRef] [PubMed]
- Oats, M.F.; Coronel-Aguilera, C.P.; Applegate, B.M.; Csonka, L.N.; Bhunia, A.K.; Gehring, A.G.; Paoli, G.C. Determination of the Infection Dynamics of Escherichia Coli O157:H7 by Bacteriophage ΦV10. Foods 2025, Vol. 14, Page 617 2025, 14, 617. [Google Scholar] [CrossRef]
- Marti, R.; Zurfluh, K.; Hagens, S.; Pianezzi, J.; Klumpp, J.; Loessner, M.J. Long Tail Fibres of the Novel Broad-Host-Range T-Even Bacteriophage S16 Specifically Recognize Salmonella OmpC. Mol Microbiol 2013, 87, 818–834. [Google Scholar] [CrossRef]
- Baxa, U.; Steinbacher, S.; Miller, S.; Weintraub, A.; Huber, R.; Seckler, R. Interactions of Phage P22 Tails with Their Cellular Receptor, Salmonella O-Antigen Polysaccharide. Biophys J 1996, 71, 2040–2048. [Google Scholar] [CrossRef]
- Ando, H.; Lemire, S.; Pires, D.P.; Lu, T.K. Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst 2015, 1, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Scholl, D.; Cooley, M.; Williams, S.R.; Gebhart, D.; Martin, D.; Bates, A.; Mandrell, R. An Engineered R-Type Pyocin Is a Highly Specific and Sensitive Bactericidal Agent for the Food-Borne Pathogen Escherichia Coli O157:H7. Antimicrob Agents Chemother 2009, 53, 3074–3080. [Google Scholar] [CrossRef] [PubMed]
- Mitsunaka, S.; Yamazaki, K.; Pramono, A.K.; Ikeuchi, M.; Kitao, T.; Ohara, N.; Kubori, T.; Nagai, H.; Ando, H. Synthetic Engineering and Biological Containment of Bacteriophages. Proc Natl Acad Sci U S A 2022, 119. [Google Scholar] [CrossRef] [PubMed]
- Luria, S.E.; Dulbecco, R. Genetic Recombinations Leading to Production of Active Bacteriophage from Ultraviolet Inactivated Bacteriophage Particles. Genetics 1949, 34, 93. [Google Scholar] [CrossRef]
- Duong, M.M.; Carmody, C.M.; Ma, Q.; Peters, J.E.; Nugen, S.R. Optimization of T4 Phage Engineering via CRISPR/Cas9. Sci Rep 2020, 10. [Google Scholar] [CrossRef]
- Yosef, I.; Goren, M.G.; Globus, R.; Molshanski-Mor, S.; Qimron, U. Extending the Host Range of Bacteriophage Particles for DNA Transduction. Mol Cell 2017, 66, 721–728. [Google Scholar] [CrossRef]
- Isaev, A.; Andriianov, A.; Znobishcheva, E.; Zorin, E.; Morozova, N.; Severinov, K. Editing of Phage Genomes—Recombineering-Assisted SpCas9 Modification of Model Coliphages T7, T5, and T3. Molecular Biology 2022 56:6 2022, 56, 801–815. [Google Scholar] [CrossRef]
- Yuan, S.; Li, Y.; Kou, C.; Sun, Y.C.; Ma, Y. CRISPR/Cas12a-Based Genome Editing for Cyanophage of Anabeana Sp. Synth Syst Biotechnol 2025, 10, 140–147. Synth Syst Biotechnol 2025, 10, 140–147. [Google Scholar] [CrossRef]
- Lin, Z.; Li, H.; He, L.; Jing, Y.; Pistolozzi, M.; Wang, T.; Ye, Y. Efficient Genome Editing for Pseudomonas Aeruginosa Using CRISPR-Cas12a. Gene 2021, 790, 145693. [Google Scholar] [CrossRef]
- Guan, J.; Oromí-Bosch, A.; Mendoza, S.D.; Karambelkar, S.; Berry, J.D.; Bondy-Denomy, J. Bacteriophage Genome Engineering with CRISPR–Cas13a. Nat Microbiol 2022, 7, 1956–1966. [Google Scholar] [CrossRef]
- Adler, B.A.; Hessler, T.; Cress, B.F.; Lahiri, A.; Mutalik, V.K.; Barrangou, R.; Banfield, J.; Doudna, J.A. Broad-Spectrum CRISPR-Cas13a Enables Efficient Phage Genome Editing. Nature Microbiology 2022 7:12 2022, 7, 1967–1979. [Google Scholar] [CrossRef] [PubMed]
- Fishman, C.B.; Crawford, K.D.; Bhattarai-Kline, S.; Poola, D.; Zhang, K.; González-Delgado, A.; Rojas-Montero, M.; Shipman, S.L. Continuous Multiplexed Phage Genome Editing Using Recombitrons. Nat Biotechnol 2024. [CrossRef]
- Levrier, A.; Karpathakis, I.; Nash, B.; Bowden, S.D.; Lindner, A.B.; Noireaux, V. PHEIGES: All-Cell-Free Phage Synthesis and Selection from Engineered Genomes. Nat Commun 2024, 15. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.J.; Ripley, L.S. The Spectrum of Acridine Resistant Mutants of Bacteriophage T4 Reveals Cryptic Effects of the TsL141 DNA Polymerase Allele on Spontaneous Mutagenesis. Genetics 1998, 148, 1655. [Google Scholar] [CrossRef]
- Filsinger, G.T.; Wannier, T.M.; Pedersen, F.B.; Lutz, I.D.; Zhang, J.; Stork, D.A.; Debnath, A.; Gozzi, K.; Kuchwara, H.; Volf, V.; et al. Characterizing the Portability of Phage-Encoded Homologous Recombination Proteins. Nature Chemical Biology 2021 17:4 2021, 17, 394–402. [Google Scholar] [CrossRef]
- Marinelli, L.J.; Piuri, M.; Swigoňová, Z.; Balachandran, A.; Oldfield, L.M.; van Kessel, J.C.; Hatfull, G.F. BRED: A Simple and Powerful Tool for Constructing Mutant and Recombinant Bacteriophage Genomes. PLoS One 2008, 3, e3957. [Google Scholar] [CrossRef] [PubMed]
- Poteete, A.R. What Makes the Bacteriophage λ Red System Useful for Genetic Engineering: Molecular Mechanism and Biological Function. FEMS Microbiol Lett 2001, 201, 9–14. [Google Scholar] [CrossRef]
- Clark, A.J.; Margulies, A.D. Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc Natl Acad Sci U S A 1965, 53, 451–459. [Google Scholar] [CrossRef]
- Maresca, M.; Erler, A.; Fu, J.; Friedrich, A.; Zhang, Y.; Stewart, A.F. Single-Stranded Heteroduplex Intermediates in λ Red Homologous Recombination. BMC Mol Biol 2010, 11, 1–15. [Google Scholar] [CrossRef]
- Marinelli, L.J.; Hatfull, G.F.; Piuri, M. Recombineering. Bacteriophage 2012, 2, 5–14. [Google Scholar] [CrossRef]
- Subramanian, K.; Rutvisuttinunt, W.; Scott, W.; Myers, R.S. The Enzymatic Basis of Processivity in Lambda Exonuclease. Nucleic Acids Res 2003, 31, 1585–1596. [Google Scholar] [CrossRef] [PubMed]
- Erler, A.; Wegmann, S.; Elie-Caille, C.; Bradshaw, C.R.; Maresca, M.; Seidel, R.; Habermann, B.; Muller, D.J.; Stewart, A.F. Conformational Adaptability of Redβ during DNA Annealing and Implications for Its Structural Relationship with Rad52. J Mol Biol 2009, 391, 586–598. [Google Scholar] [CrossRef] [PubMed]
- Court, R.; Cook, N.; Saikrishnan, K.; Wigley, D. The Crystal Structure of Lambda-Gam Protein Suggests a Model for RecBCD Inhibition. J Mol Biol 2007, 371, 25–33. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-Step Inactivation of Chromosomal Genes in Escherichia Coli K-12 Using PCR Products. Proc Natl Acad Sci U S A 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Karlinsey, J.E. Λ-Red Genetic Engineering in Salmonella Enterica Serovar Typhimurium. Methods Enzymol 2007, 421, 199–209. [Google Scholar] [CrossRef]
- Zhang, Y.; Buchholz, F.; Muyrers, J.P.P.; Francis Stewart, A. A New Logic for DNA Engineering Using Recombination in Escherichia Coli. Nat Genet 1998, 20, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, A.B.; Rattray, A.J.; Bubunenko, M.; Thomason, L.C.; Court, D.L. In Vivo Recombineering of Bacteriophage λ by PCR Fragments and Single-Strand Oligonucleotides. Virology 2004, 319, 185–189. [Google Scholar] [CrossRef]
- Serra-Moreno, R.; Acosta, S.; Hernalsteens, J.P.; Jofre, J.; Muniesa, M. Use of the Lambda Red Recombinase System to Produce Recombinant Prophages Carrying Antibiotic Resistance Genes. BMC Mol Biol 2006, 7, 1–12. [Google Scholar] [CrossRef]
- Fehér, T.; Karcagi, I.; Blattner, F.R.; Pósfai, G. Bacteriophage Recombineering in the Lytic State Using the Lambda Red Recombinases. Microb Biotechnol 2012, 5, 466–476. [Google Scholar] [CrossRef]
- Thomason, L.C.; Sawitzke, J.A.; Li, X.; Costantino, N.; Court, D.L. Recombineering: Genetic Engineering in Bacteria Using Homologous Recombination. Curr Protoc Mol Biol 2014, 106, 1. [Google Scholar] [CrossRef]
- Derbise, A.; Lesic, B.; Dacheux, D.; Ghigo, J.M.; Carniel, E. A Rapid and Simple Method for Inactivating Chromosomal Genes in Yersinia. FEMS Immunol Med Microbiol 2003, 38, 113–116. [Google Scholar] [CrossRef]
- Lesic, B.; Rahme, L.G. Use of the Lambda Red Recombinase System to Rapidly Generate Mutants in Pseudomonas Aeruginosa. BMC Mol Biol 2008, 9, 1–9. [Google Scholar] [CrossRef]
- Muyrers, J.P.P.; Zhang, Y.; Buchholz, F.; Stewart, A.F. RecE/RecT and Redα/Redβ Initiate Double-Stranded Break Repair by Specifically Interacting with Their Respective Partners. Genes Dev 2000, 14, 1971. [Google Scholar] [CrossRef] [PubMed]
- Muyrers, J.P.P.; Zhang, Y.; Stewart, A.F. Techniques: Recombinogenic Engineering–New Options for Cloning and Manipulating DNA. Trends Biochem Sci 2001, 26, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Van Kessel, J.C.; Hatfull, G.F. Recombineering in Mycobacterium Tuberculosis. Nature Methods 2006 4:2 2006, 4, 147–152. [Google Scholar] [CrossRef]
- Van Kessel, J.C.; Hatfull, G.F. Efficient Point Mutagenesis in Mycobacteria Using Single-Stranded DNA Recombineering: Characterization of Antimycobacterial Drug Targets. Mol Microbiol 2008, 67, 1094–1107. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.; Amarir-Bouhram, J.; Faure, G.; Petit, M.A.; Guerois, R. Detection of Novel Recombinases in Bacteriophage Genomes Unveils Rad52, Rad51 and Gp2.5 Remote Homologs. Nucleic Acids Res 2010, 38, 3952–3962. [Google Scholar] [CrossRef]
- Swingle, B.; Bao, Z.; Markel, E.; Chambers, A.; Cartinhour, S. Recombineering Using RecTE from Pseudomonas Syringae. Appl Environ Microbiol 2010, 76, 4960–4968. [Google Scholar] [CrossRef]
- Jacobs, W.R.; Snapper, S.B.; Tuckinan, M.; Bloom, B.R. Mycobacteriophage Vector Systems. Rev Infect Dis 1989, 11, S404–S410. [Google Scholar] [CrossRef]
- Pearson, R.E.; Jurgensen, S.; Sarkis, G.J.; Hatfull, G.F.; Jacobs, W.R. Construction of D29 Shuttle Phasmids and Luciferase Reporter Phages for Detection of Mycobacteria. Gene 1996, 183, 129–136. [Google Scholar] [CrossRef]
- Sarkis, G.J.; Jacobs, W.R.; Hatfulll, G.F. L5 Luciferase Reporter Mycobacteriophages: A Sensitive Tool for the Detection and Assay of Live Mycobacteria. Mol Microbiol 1995, 15, 1055–1067. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, K.S.; Guerrero-Bustamante, C.A.; Dedrick, R.M.; Ko, C.C.; Freeman, K.G.; Aull, H.G.; Divens, A.M.; Rock, J.M.; Zack, K.M.; Hatfull, G.F. CRISPY-BRED and CRISPY-BRIP: Efficient Bacteriophage Engineering. Scientific Reports 2021 11:1 2021, 11, 1–6. [Google Scholar] [CrossRef]
- Yin, J.; Zheng, W.; Gao, Y.; Jiang, C.; Shi, H.; Diao, X.; Li, S.; Chen, H.; Wang, H.; Li, R.; et al. Single-Stranded DNA-Binding Protein and Exogenous RecBCD Inhibitors Enhance Phage-Derived Homologous Recombination in Pseudomonas. iScience 2019, 14, 1. [Google Scholar] [CrossRef]
- Bao, Z.; Cartinhour, S.; Swingle, B. Substrate and Target Sequence Length Influence RecTEPsy Recombineering Efficiency in Pseudomonas Syringae. PLoS One 2012, 7, e50617. [Google Scholar] [CrossRef]
- Zheng, W.; Xia, Y.; Wang, X.; Gao, S.; Zhou, D.; Fu, J.; Li, R.; Yin, J. Cascade-Cas3 Facilitates High-Accuracy Genome Engineering in Pseudomonas Using Phage-Encoded Homologous Recombination. Engineering Microbiology 2022, 2, 100046. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Liu, J. Scarless and Sequential Gene Modification in Pseudomonas Using PCR Product Flanked by Short Homology Regions. BMC Microbiol 2010, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yan, B.; Chen, W.; Zhang, X.; Liu, Z.; Zhang, Q.; Li, L.; Hu, M.; Zhao, X.; Xu, X.; et al. Development of the CRISPR-Cas12a System for Editing of Pseudomonas Aeruginosa Phages. iScience 2024, 27, 110210. [Google Scholar] [CrossRef]
- Hatoum-Aslan, A. Phage Genetic Engineering Using CRISPR–Cas Systems. Viruses 2018, 10. [Google Scholar] [CrossRef]
- Mahler, M.; Costa, A.R.; van Beljouw, S.P.B.; Fineran, P.C.; Brouns, S.J.J. Approaches for Bacteriophage Genome Engineering. Trends Biotechnol 2023, 41, 669–685. [Google Scholar] [CrossRef]
- Hoshiga, F.; Yoshizaki, K.; Takao, N.; Miyanaga, K.; Tanji, Y. Modification of T2 Phage Infectivity toward Escherichia Coli O157:H7 via Using CRISPR/Cas9. FEMS Microbiol Lett 2019, 366, 41. [Google Scholar] [CrossRef]
- Dionne, E.N.; Cornely, K. An Application of Mycobacteriophage Genome Engineering Using Bacteriophage Recombineering with Electroporated DNA (BRED) and CRISPR Cas-9 Systems. The FASEB Journal 2022, 36. [Google Scholar] [CrossRef]
- Choi, S.Y.; Romero-Calle, D.X.; Cho, H.G.; Bae, H.W.; Cho, Y.H. Use of Cas9 Targeting and Red Recombination for Designer Phage Engineering. Journal of Microbiology 2024, 62, 1–10. [Google Scholar] [CrossRef]
- Heler, R.; Samai, P.; Modell, J.W.; Weiner, C.; Goldberg, G.W.; Bikard, D.; Marraffini, L.A. Cas9 Specifies Functional Viral Targets during CRISPR-Cas Adaptation. Nature 2015, 519, 199. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Jakimo, N.; Lee, J.; Amrani, N.; Rodríguez, T.; Koseki, S.R.T.; Tysinger, E.; Qing, R.; Hao, S.; Sontheimer, E.J.; et al. An Engineered ScCas9 with Broad PAM Range and High Specificity and Activity. Nat Biotechnol 2020, 38, 1154–1158. [Google Scholar] [CrossRef]
- Zhao, L.; Koseki, S.R.T.; Silverstein, R.A.; Amrani, N.; Peng, C.; Kramme, C.; Savic, N.; Pacesa, M.; Rodríguez, T.C.; Stan, T.; et al. PAM-Flexible Genome Editing with an Engineered Chimeric Cas9. Nat Commun 2023, 14, 1–8. [Google Scholar] [CrossRef]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained Genome Targeting with Near-PAMless Engineered CRISPR-Cas9 Variants. Science (1979) 2020, 368, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Hibshman, G.N.; Bravo, J.P.K.; Hooper, M.M.; Dangerfield, T.L.; Zhang, H.; Finkelstein, I.J.; Johnson, K.A.; Taylor, D.W. Unraveling the Mechanisms of PAMless DNA Interrogation by SpRY-Cas9. Nat Commun 2024, 15. [Google Scholar] [CrossRef]
- Tao, P.; Wu, X.; Tang, W.C.; Zhu, J.; Rao, V. Engineering of Bacteriophage T4 Genome Using CRISPR-Cas9. ACS Synth Biol 2017, 6, 1952. [Google Scholar] [CrossRef]
- Fernbach, J.; Baggenstos, J.; Riedo, J.; McCallin, S.; Loessner, M.J.; Kilcher, S. CRISPR-Cas9 Enables Efficient Genome Engineering of the Strictly Lytic, Broad Host-Range Staphylococcal Bacteriophage K. bioRxiv 2024, 2024.03.19.585701. [CrossRef]
- Shen, J.; Zhou, J.; Chen, G.-Q.; Xiu, Z.-L. Efficient Genome Engineering of a Virulent Klebsiella Bacteriophage Using CRISPR-Cas9. 2018.
- Tao, P.; Wu, X.; Tang, W.C.; Zhu, J.; Rao, V. Engineering of Bacteriophage T4 Genome Using CRISPR-Cas9. ACS Synth Biol 2017, 6, 1952–1961. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Liu, Y.; Tao, P. Genetic Engineering of Bacteriophage Using CRISPR-Cas12a. 2025, 43–53. [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 Is a Single-Component Programmable RNA-Guided RNA-Targeting CRISPR Effector. Science (1979) 2016, 353. [Google Scholar] [CrossRef] [PubMed]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell 2015, 60, 385–397. [Google Scholar] [CrossRef] [PubMed]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.D.; Tjian, R.; Doudna, J.A. Two Distinct RNase Activities of CRISPR-C2c2 Enable Guide-RNA Processing and RNA Detection. Nature 2016 538:7624 2016, 538, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Yee, T.; Furuichi, T.; Inouye, S.; Inouye, M. Multicopy Single-Stranded DNA Isolated from a Gram-Negative Bacterium, Myxococcus Xanthus. Cell 1984, 38, 203–209. [Google Scholar] [CrossRef]
- Dhundale, A.; Lampson, B.; Furuichi, T.; Inouye, M.; Inouye, S. Structure of MsDNA from Myxococcus Xanthus: Evidence for a Long, Self-Annealing RNA Precursor for the Covalently Linked, Branched RNA. Cell 1987, 51, 1105–1112. [Google Scholar] [CrossRef]
- Hsu, M.Y.; Inouye, S.; Inouye, M. Structural Requirements of the RNA Precursor for the Biosynthesis of the Branched RNA-Linked Multicopy Single-Stranded DNA of Myxococcus Xanthus. Journal of Biological Chemistry 1989, 264, 6214–6219. [Google Scholar] [CrossRef]
- Lampson, B.C.; Inouye, M.; Inouye, S. Reverse Transcriptase with Concomitant Ribonuclease H Activity in the Cell-Free Synthesis of Branched RNA-Linked MsDNA of Myxococcus Xanthus. Cell 1989, 56, 701–707. [Google Scholar] [CrossRef]
- Lampson, B.; Inouye, M.; Inouye, S. The MsDNAs of Bacteria. Prog Nucleic Acid Res Mol Biol 2001, 67, 65–91. [Google Scholar] [CrossRef]
- Simon, A.J.; Ellington, A.D.; Finkelstein, I.J. Retrons and Their Applications in Genome Engineering. Nucleic Acids Res 2019, 47, 11007–11019. [Google Scholar] [CrossRef]
- Millman, A.; Bernheim, A.; Stokar-Avihail, A.; Fedorenko, T.; Voichek, M.; Leavitt, A.; Oppenheimer-Shaanan, Y.; Sorek, R. Bacterial Retrons Function In Anti-Phage Defense. Cell 2020, 183, 1551–1561.e12. [Google Scholar] [CrossRef]
- Lopez, S.C.; Crawford, K.D.; Lear, S.K.; Bhattarai-Kline, S.; Shipman, S.L. Precise Genome Editing across Kingdoms of Life Using Retron-Derived DNA. Nat Chem Biol 2022, 18, 199–206. [Google Scholar] [CrossRef] [PubMed]
- González-Delgado, A.; Lopez, S.C.; Rojas-Montero, M.; Fishman, C.B.; Shipman, S.L. Simultaneous Multi-Site Editing of Individual Genomes Using Retron Arrays. Nat Chem Biol 2024, 20, 1482–1492. [Google Scholar] [CrossRef] [PubMed]
- Jaschke, P.R.; Lieberman, E.K.; Rodriguez, J.; Sierra, A.; Endy, D. A Fully Decompressed Synthetic Bacteriophage ØX174 Genome Assembled and Archived in Yeast. Virology 2012, 434, 278–284. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Engler, C.; Kandzia, R.; Marillonnet, S. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLoS One 2008, 3. [Google Scholar] [CrossRef]
- Nozaki, S. Rapid and Accurate Assembly of Large DNA Assisted by In Vitro Packaging of Bacteriophage. ACS Synth Biol 2022, 11, 4113–4122. [Google Scholar] [CrossRef] [PubMed]
- Garenne, D.; Haines, M.C.; Romantseva, E.F.; Freemont, P.; Strychalski, E.A.; Noireaux, V. Cell-Free Gene Expression. Nature Reviews Methods Primers 2021, 1, 1–18. [Google Scholar] [CrossRef]
- Rustad, M.; Eastlund, A.; Jardine, P.; Noireaux, V. Cell-Free TXTL Synthesis of Infectious Bacteriophage T4 in a Single Test Tube Reaction. Synth Biol 2018, 3. [Google Scholar] [CrossRef]
- Burke, D.T.; Carle, G.F.; Olson, M. V. Cloning of Large Segments of Exogenous DNA into Yeast by Means of Artificial Chromosome Vectors. Science (1979) 1987, 236, 806–812. [Google Scholar] [CrossRef]
- Frazer, L.A.N.; O’Keefe, R.T. A New Series of Yeast Shuttle Vectors for the Recovery and Identification of Multiple Plasmids from Saccharomyces Cerevisiae. Yeast 2007, 24, 777–789. [Google Scholar] [CrossRef]
- Gibson, D.G.; Benders, G.A.; Axelrod, K.C.; Zaveri, J.; Algire, M.A.; Moodie, M.; Montague, M.G.; Venter, J.C.; Smith, H.O.; Hutchison, C.A. One-Step Assembly in Yeast of 25 Overlapping DNA Fragments to Form a Complete Synthetic Mycoplasma Genitalium Genome. Proceedings of the National Academy of Sciences 2008, 105, 20404–20409. [Google Scholar] [CrossRef] [PubMed]
- Brownstein, B.H.; Silverman, G.A.; Little, R.D.; Burke, D.T.; Korsmeyer, S.J.; Schlessinger, D.; Olson, M. V. Isolation of Single-Copy Human Genes from a Library of Yeast Artificial Chromosome Clones. Science (1979) 1989, 244, 1348–1351. [Google Scholar] [CrossRef]
- Ipoutcha, T.; Racharaks, R.; Huttelmaier, S.; Wilson, C.J.; Ozer, E.A.; Hartmann, E.M. A Synthetic Biology Approach to Assemble and Reboot Clinically Relevant Pseudomonas Aeruginosa Tailed Phages. Microbiol Spectr 2024, 12. [Google Scholar] [CrossRef]
- Kristensen, C.S.; Petersen, A.Ø.; Kilstrup, M.; van der Helm, E.; Takos, A. Cell-Free Synthesis of Infective Phages from in Vitro Assembled Phage Genomes for Efficient Phage Engineering and Production of Large Phage Libraries. Synth Biol 2024, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; He, L.; Guo, Y.; Wang, T.; Ye, Y.; Lin, Z. Synthesis of Headful Packaging Phages Through Yeast Transformation-Associated Recombination. Viruses 2025, 17, 45. [Google Scholar] [CrossRef] [PubMed]
- Assad-Garcia, N.; D’Souza, R.; Buzzeo, R.; Tripathi, A.; Oldfield, L.M.; Vashee, S.; Fouts, D.E. Cross-Genus “Boot-Up” of Synthetic Bacteriophage in Staphylococcus Aureus by Using a New and Efficient DNA Transformation Method. Appl Environ Microbiol 2022, 88. [Google Scholar] [CrossRef]
- Gibson, D.G. Enzymatic Assembly of Overlapping DNA Fragments. Methods Enzymol 2011, 498, 349–361. [Google Scholar] [CrossRef]
- Pryor, J.M.; Potapov, V.; Bilotti, K.; Pokhrel, N.; Lohman, G.J.S. Rapid 40 Kb Genome Construction from 52 Parts through Data-Optimized Assembly Design. ACS Synth Biol 2022, 11, 2036–2042. [Google Scholar] [CrossRef]
- Kilcher, S.; Studer, P.; Muessner, C.; Klumpp, J.; Loessner, M.J.; Adhya, S. Cross-Genus Rebooting of Custom-Made, Synthetic Bacteriophage Genomes in L-Form Bacteria. Proc Natl Acad Sci U S A 2018, 115, 567–572. [Google Scholar] [CrossRef]
- Nirenberg, M.W.; Matthaei, J.H. The Dependence of Cell-Free Protein Synthesis in E. Coli upon Naturally Occurring or Synthetic Polyribonucleotides. Proc Natl Acad Sci U S A 1961, 47, 1588–1602. [Google Scholar] [CrossRef]
- Carlson, E.D.; Gan, R.; Hodgman, C.E.; Jewett, M.C. Cell-Free Protein Synthesis: Applications Come of Age. Biotechnol Adv 2012, 30, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Gregorio, N.E.; Levine, M.Z.; Oza, J.P. A User’s Guide to Cell-Free Protein Synthesis. Methods and Protocols 2019, Vol. 2, Page 24 2019, 2, 24. [Google Scholar] [CrossRef] [PubMed]
- Pizarro-Bauerle, J.; Ando, H. Engineered Bacteriophages for Practical Applications. Biol Pharm Bull 2020, 43, 240–249. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef]
- Borin, J.M.; Avrani, S.; Barrick, J.E.; Petrie, K.L.; Meyer, J.R. Coevolutionary Phage Training Leads to Greater Bacterial Suppression and Delays the Evolution of Phage Resistance. Proc Natl Acad Sci U S A 2021, 118, e2104592118. [Google Scholar] [CrossRef]
- Hodyra-Stefaniak, K.; Lahutta, K.; Majewska, J.; Kaźmierczak, Z.; Lecion, D.; Harhala, M.; Kęska, W.; Owczarek, B.; Jończyk-Matysiak, E.; Kłopot, A.; et al. Bacteriophages Engineered to Display Foreign Peptides May Become Short-Circulating Phages. Microb Biotechnol 2019, 12, 730–741. [Google Scholar] [CrossRef]
- Kim, K.P.; Cha, J.D.; Jang, E.H.; Klumpp, J.; Hagens, S.; Hardt, W.D.; Lee, K.Y.; Loessner, M.J. PEGylation of Bacteriophages Increases Blood Circulation Time and Reduces T-Helper Type 1 Immune Response. Microb Biotechnol 2008, 1, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, K.; Abedon, S.T. Pharmacologically Aware Phage Therapy: Pharmacodynamic and Pharmacokinetic Obstacles to Phage Antibacterial Action in Animal and Human Bodies. Microbiology and Molecular Biology Reviews 2019, 83. [Google Scholar] [CrossRef]
- Cui, L.; Watanabe, S.; Miyanaga, K.; Kiga, K.; Sasahara, T.; Aiba, Y.; Tan, X.-E.; Veeranarayanan, S.; Thitiananpakorn, K.; Nguyen, H.M.; et al. A Comprehensive Review on Phage Therapy and Phage-Based Drug Development. Antibiotics 2024, Vol. 13, Page 870 2024, 13, 870. [Google Scholar] [CrossRef]
- Cui, L.; Veeranarayanan, S.; Thitiananpakorn, K.; Wannigama, D.L. Bacteriophage Bioengineering: A Transformative Approach for Targeted Drug Discovery and Beyond. Pathogens 2023, Vol. 12, Page 1179 2023, 12, 1179. [Google Scholar] [CrossRef]
- Veeranarayanan, S.; Azam, A.H.; Kiga, K.; Watanabe, S.; Cui, L. Bacteriophages as Solid Tumor Theragnostic Agents. International Journal of Molecular Sciences 2022, Vol. 23, Page 402 2021, 23, 402. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.P.; Kutter, E. Bacteriophages: It’s a Medicine, Jim, but Not as We Know It. Lancet Infect Dis 2021, 21, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.P.; Blasdel, B.G.; Bretaudeau, L.; Buckling, A.; Chanishvili, N.; Clark, J.R.; Corte-Real, S.; Debarbieux, L.; Dublanchet, A.; De Vos, D.; et al. Quality and Safety Requirements for Sustainable Phage Therapy Products. Pharm Res 2015, 32, 2173–2179. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Kiga, K.; Kondabagil, K.; Węgrzyn, A. Current and Future Directions in Bacteriophage Research for Developing Therapeutic Innovations. Sci Rep 2024, 14, 1–4. [Google Scholar] [CrossRef]
| Function | Phage name | Cargo | Targeted Host | Phage feature | References |
|---|---|---|---|---|---|
| Antibiofilm | CPB0329 | Dispersin B | Klebsiella pneumoniae | Replicative phage particles | [27] |
| Antimicrobial agent | T7 | Colicin E1 and Colicin M | Escherichia coli | Replicative phage particles | [39] |
| Antimicrobial agent | T7 | Colicin E1 | Escherichia coli | Non-Replicative particles * | [40] |
| Antimicrobial agent | T-even like phages | Cas (type I-E) | Pathogenic Escherichia coli | Replicative phage particles | [25] |
| Bacteria detection | T4 | LacZa | Escherichia coli | Replicative phage particles | [43] |
| Bacteria detection | T7 | NanoLuc luciferse | Escherichia coli | Replicative phage particles | [32] |
| Bacteria detection | PhiV10 | luxCDABE | Escherichia coli (Food samples]) | Replicative phage particles | [30] |
| Bacteria detection | E2, E4, EfS3, EfS7, K1and K4 | NanoLuc luciferse | Escherichia coli, Enterococcus spp., and Klebsiella spp. (Urine samples) | Replicative phage particles | [29] |
| Bacteria detection | vB_Eco4M-7 | HiBiT | STEC | Replicative phage particles | [31] |
| Bacteria genetic engineering | Ur-l | Base editor dCas9 | Escherichia coli (b-lactamase) | Non-replicative particles | [22] |
| Delivery of Antimicrobial peptide | Sb-1 | Nisin | MRSA | Phage structural components (Tail) | [42] |
| Drug delivery system for mammalian gut | T4 | Serpine B1a, Chaperone protein clpB | Nonpathogenic Escherichia coli | Replicative phage particles | [33] |
| Gene delivery system for the human cells | T4 | Gene editing, in situ protein expression and others | - | Phage structural components (Head) | [41] |
| Selective antimicrobial agent | Ur-l | Cas12a | STEC | Non-replicative particles | [23] |
| Selective antimicrobial agent | P1 | Cas9 | Escherichia coli | Non-replicative particles | [44] |
| Selective antimicrobial agent | Phi80, M13, 80a and Tan2 | Cas13a | Escherichia coli & Staphylococcus aureus | Non-replicative particles | [36,37,38] |
| Selective antimicrobial agent | P2 | Cas9 | STEC and Shigella flexneri | Non-replicative particles | [35] |
| Phage general information | Phage engineering method | In vivo Engineering conditions | In vitro (Synthetic) engineering conditions | Engineering purpose | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phage Host | Phage Name | Recombineering genes | Counter selection method | Assembly method | Rebooting method | Rebooting host | Intermediate host | Phage genome size Kbp | Modification | Purpose | ||
| Escherichia coli | Phage a15 | In vivo | - | - | - | - | - | - | - | Gene replacement of ~7 kp | Load Cas (type I-E) and gene for Tsx-binding adhesin | [25] |
| Escherichia coli | T7 | In vivo | Flp recombinase | Induced Phenotype *1 | BW25113ΔtrxA | Gene replacement | Tail fiber modification | [81] | ||||
| Escherichia coli | T4 | In vivo | - | Induced Phenotype *2 | - | - | - | - | Gene replacement | In situ protein expression within mammalian cells | [33] | |
| Escherichia coli | T4 | In vivo | - | CRISPR/ Cas9 | - | - | - | - | NanoLuc luciferase | Reporter gene | [80] | |
| Escherichia coli | T4 | In vivo*3 | - | CRISPR/ Cas9 or Cas12 | - | - | - | - | - | Eliminate phage DNA packaging to create an empty head | Load various cargoes to human cells | [41] |
| Escherichia coli | T3, T7, and T5 | In vivo | l-red | CRISPR/ Cas9 | - | - | - | - | Point substitutions, insertions, or deletions | Tail fiber modification | [82] | |
| Escherichia coli | P1 | In vivo | l-red | Selection Marker | - | - | - | Deletion of packaging region Dpac of plasmid phage P1 | Phage capsid construction | [44] | ||
| Escherichia coli | Ur-l | In vivo | l-red | CRISPR/ Cas9 | - | - | - | Deletion | Tail fiber modification | [22] | ||
| Klebsiella pneumoniae | T7 family and non-family Klebsiella pneumoniae phages | In vivo | l-red | CRISPR/ Cas9 | - | In-vivo | Escherichia coli DH10B | Yes | 41 to 46 | Either gene replacement of a non-essential ligase gene with dispersin B DspB or just gene insertion of the mentioned gene DspB | Distribute Biofilm | [27] |
| Anabaena | Cyanophage A-1(L) and A-4(L) | In vivo | - | CRISPR/ Cas12a | - | - | - | Deletion | Minimize genome reduction of 2400 bp | [83] | ||
| Pseudomonas aeruginosa | - | In vivo | l-red | CRISPR/ Cas12a | - | - | - | Deletion | 15kbp deletion | [84] | ||
| Pseudomonas aeruginosa | KZ | In vivo | - | CRISPR/ Cas13a and acrVIA1 | - | - | - | Insertion, deletion and fluorescent tagging | - | [85] | ||
| Escherichia coli | T4, T7 and EdH4 | In vivo | - | CRISPR/ Cas13a | - | - | - | - | - | Multi gene deletion and single base modification | - | [86] |
| Escherichia coli | T7 | In vivo | - | Recombitrons | - | - | - | - | Amino acid substitutions in gp17 | Expand host range | [87] | |
| Escherichia coli | T7 | Synthetic | - | - | NEBuilder HiFi DNA | In-vivo | Escherichia coli 10G | No | 39.937+ 0.977 | Insertion of NanoLuc luciferase | Reporter gene | [32] |
| Escherichia coli, Klebsiella and Yersinia | T7 family | Synthetic | - | - | YAC | In-vivo | Escherichia cloni 10G | Yes | 37 to 45 | Modify Tail fiber | Expand host range | [76] |
| Escherichia coli | T7 | Synthetic | - | - | Exonuclease only | TXTL | - | - | 39 | Modify Tail fiber | Expand host range | [88] |
| Salmonella | P22 | Synthetic | - | - | Gibson | In-vivo | Salmonella Typhimurium strain LT2 | - | - | Deletion of lytic cycle repressor c2 | Modify lifestyle | [78] |
| Mycobacterium | D29 | Synthetic | - | - | Gibson | In-vivo | M. smegmatis mc2155 | - | - | Gene replacement and insertion of NanoLuc luciferase gene | Reporter | |
| Escherichia coli | T7 | Synthetic | - | - | Gibson | TXTL | - | - | - | Gene replacement and insertion of LacZ operon | Reporter | |
| Salmonella | SP6 | Synthetic | - | - | Gibson | In vivo | Salmonella Typhimurium strain LT2 | - | - | Deletion of phage head | Biocontained Phages | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





