1. Introduction
Technology and digitalization have dramatically altered research, moving it towards faster, automated, collaborative, and data-driven methodologies. However, scientists now face significant challenges in managing the vast volume of research data and associated files produced throughout scientific studies. These files include scripts, tests, descriptions of scientific workflows, variables from experimental data, execution environments (both software and hardware), manuscript files and other related materials.[
1]
Neuroimaging studies are no exception to this trend. These investigations are particularly notable for their intricate workflows, which include multiple stages such as data collection, data preparation, experimental setup, analysis execution and statistical thresholding, documentation, and publication of findings. The complexity of managing these resources is increased by the use of multiple data sources and a variety of data formats and contents. This complexity in combination with lack of effective standards and high degree of operations make it difficult to effectively track and manage the information while ensuring the reproducibility of results.[
2,
3,
32]
There are several tools available to support research workflows by ensuring traceability and version control. For example, Git is utilized for managing code and scripts, while platforms such as Google Docs, Dropbox, Microsoft office and Overleaf facilitate documentation. Additionally, data management solutions like XNAT[
4] and Flywheel[
5] are effective for data storage as well as analysis. However, these tools often function in isolation, and a comprehensive system that integrates all of these resources into a single platform is currently lacking.
Current scientific data management practices largely focus on the dissemination and archiving of final results, focusing on how these outcomes should be stored and indexed within existing repositories and strategies. This emphasis is crucial for tackling reproducibility issues in neuroscience, particularly neuroimaging. It is essential to develop a system that researchers can employ throughout the research process to maintain immutable traceability from the original data through each transformation and analysis, extending to the results prepared for publication.[
6]
A solution to this challenge is the implementation of Electronic Lab Notebook (ELN). Examples of widely used ELNs include LabArchives[
7], LabInform[
8], SciNote[
9], eLabFTW[
10], Benchling[
11], eLabNext[
12], and Genemod[
13]. These platforms are designed to support the management of the datasets, inventory, project documentation, and collaborative workflows.
Despite their potential advantages, the adoption of ELNs within research projects presents several significant challenges. Key issues include high subscription costs, limited integration with widely used research tools, non-standardized regulations, and a general lack of time or interest among researchers to adopt new applications.[
14] Additionally, technical limitations such as restricted memory and storage capacity pose further obstacles, particularly when managing large and complex datasets. Security concerns also emerge when dealing with sensitive or proprietary information. While ELNs provide streamlined data organization and documentation, they frequently lack support for critical features such as cross-platform traceability and verifiable authorship certification. These shortcomings impede the ability to ensure ownership and transparency in scientific research.
Therefore, this study proposed an application, IntegriLAB to incorporate widely used traditional repositories, specifically DataLad[
15], GitHub[
16], and Overleaf[
17] for managing project data, source code, and documentation. This integration enables researchers to continue using familiar platforms while enhancing project tracking capabilities. This ELN creates a centralized environment where all aspects of a research project—data collection, pre-processing, analysis, and reporting are systematically logged and monitored in real time.
A key feature of this system is its ability to track project status through comprehensive logs that document every modification, update, and milestone. This transparency allows researchers, collaborators, and reviewers to trace each step of the study, facilitating better reproducibility and accountability.
Furthermore, the proposed ELN integrates LabTrace - a blockchain-based platform built on Algorand[
18], to certify each stage of the research process by securely recording timestamps and cryptographic signatures, ensuring transparency, data integrity, and an immutable audit trail. This enhances trust and credibility in research outcomes by providing verifiable proof of authorship and data provenance.
The remainder of this manuscript is structured as follows. The next section presents a background review, outlining the key challenges in neuroimaging research, the shortcomings of existing solutions for tracking research workflow, and the potential of blockchain technology to address these gaps. This is followed by a detailed description of the proposed application’s architecture, including its core functionalities, data privacy features, and mechanisms to enhance research transparency, along with its development and deployment processes. A real-world case study illustrating the application of the platform in a neuroimaging research context is presented thereafter. The discussion section explores the implications of the findings, and last section concludes the paper by outlining directions for future research.
2. Background Study
In today’s evolving research landscape, there is an increasing emphasis on openness, collaboration, and the seamless integration of tools, technologies, and administrative workflows. The rapid expansion of digital solutions has given rise to a wide array of applications designed to streamline various aspects of the research lifecycle. This section explores the key challenges researchers face and reviews existing tools, laying the groundwork for the introduction of IntegriLAB - the motivation behind its development and the needs it aims to address.
2.1. Challenges in Reproducibility of Neuroimaging Research
Reproducibility, defined as the ability to obtain consistent results using the same data and code[
19], remains a cornerstone of scientific integrity. In neuroimaging research, however, reproducibility challenges persist, undermining the credibility and reliability of findings.[
20,
21,
22,
31] As illustrated in
Figure 1, these issues span multiple stages of the neuroimaging workflow - including data acquisition, preprocessing, statistical analysis, visualization, and dissemination. Each stage is characterized by high data complexity, multiple dependencies, and domain-specific tools, contributing to variability across laboratories and studies.
A significant barrier to reproducibility is the absence of standardized protocols for conducting and analyzing neuroimaging experiments.[
20] Differences in scanner hardware, acquisition parameters, preprocessing pipelines, and statistical methodologies often result in divergent outcomes, even when analyzing similar datasets.[
21,
22] Moreover, many studies fail to comprehensively share scripts, metadata, or computational environments - making it difficult to independently verify or replicate results.[
22]
The complexity of neuroimaging workflows also leads to fragmented data management practices. Research files are commonly dispersed across cloud services, local servers, and external repositories, often stored with inconsistent naming conventions, directory structures, and metadata standards. These inefficiencies hinder version control, compromise data provenance, and introduce avoidable redundancy.[
23] In collaborative settings, particularly in multi-institutional or interdisciplinary projects, ambiguities in authorship and data ownership can further complicate accountability and raise concerns about intellectual property rights.[
24]
2.2. Electronic Lab Notebooks (ELNs)
In response to these challenges, ELNs have emerged as digital replacements for traditional paper records, offering structured, searchable, and collaborative platforms for documenting research activities.[
14,
25] Popular ELNs, such as LabArchives, SciNote, Benchling, Genemod, and open-source alternatives like eLabFTW and LabInform, provide features including project documentation, version control, collaborative access, API integrations, and support for structured data entry. In terms of deployment, some platforms are exclusively cloud-based, while others support local installation, enabling institutions to store data on their own infrastructure for greater control and privacy.
Despite these advantages, most ELNs are not tailored to the specific demands of high-throughput, computationally intensive fields like neuroimaging. Key limitations include a lack of native support for domain-specific software (e.g., FreeSurfer, FSL, SPM) and minimal integration with programming environments such as MATLAB, Python, or R. Consequently, researchers are often forced to manage workflows and documentation across separate platforms, leading to potential misalignments in data, code, and results. Moreover, commercial ELNs may impose restrictive data storage quotas—for example, LabArchives’ professional edition limits users to 100 GB, which is often inadequate for neuroimaging datasets.[
26] Critically, while ELNs may track changes to documents or files, they rarely offer cryptographically verifiable audit trails or immutable certifications of authorship or data provenance - features increasingly essential for promoting transparency and trust in collaborative research.
2.3. Green Blockchain Technology
By leveraging blockchain technology, we can effectively address these existing limitations in current systems. Blockchain offers decentralized and tamper-proof ledgers that use cryptographic algorithms to record transactions. This innovation ensures secure, transparent, and verifiable documentation of research activities.[
27] However, traditional blockchain architectures, particularly those based on Proof-of-Work (PoW) consensus mechanisms, are highly energy-intensive, raising environmental and scalability concerns. PoW relies on miners solving complex cryptographic puzzles, a process that demands significant computational resources and electricity. This energy-intensive design, while critical for maintaining the security and integrity of the network, poses environmental challenges.[
28]
To mitigate these concerns, the concept of green blockchain has emerged. Green blockchains adopt energy-efficient consensus protocols that significantly reduce greenhouse gas emissions, ensuring that environmental sustainability is maintained alongside data integrity.[
29] One of the leading platforms in this space is Algorand, which employs a Pure Proof-of-Stake (PPoS) consensus mechanism. This approach enables high-speed, low-energy operations while maintaining security, decentralization, and verifiability. Notably, Algorand operates as a carbon-negative blockchain, making it well-suited for integration into environmentally conscious research infrastructures.[
28]
Building on these capabilities, we developed IntegriLAB - a blockchain-based integrated digital lab platform designed to promote reproducible, transparent, and trustworthy research in neuroimaging and other data-intensive scientific domains. The following sections present the system architecture, implementation strategy, and deployment configurations of IntegriLAB, along with a real-world case study demonstrating its application in a neuroimaging research context.
3. IntegriLAB Overview
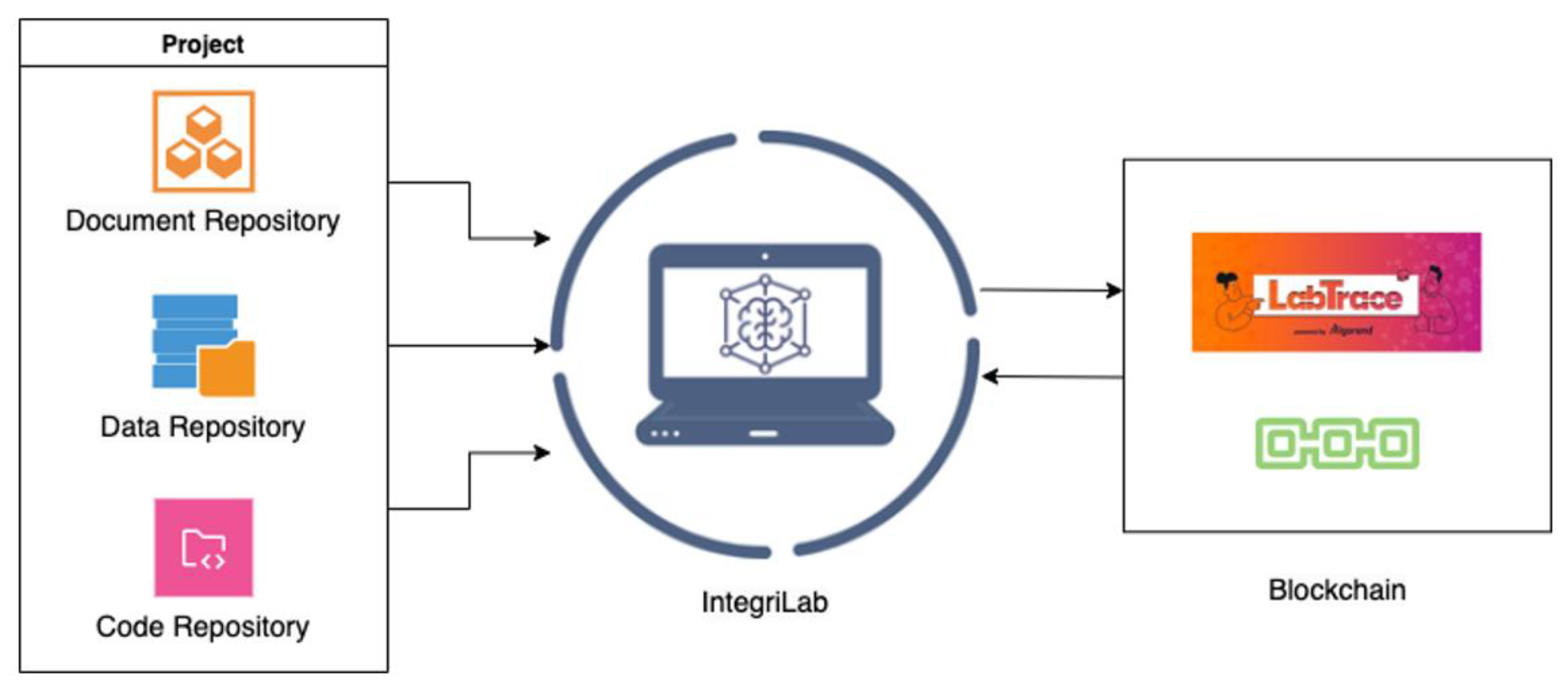
IntegriLAB is a web-based application designed to support and streamline the management of research projects. The platform offers a consolidated framework by integrating several widely used open-source tools (
Figure 2): Overleaf for collaborative manuscript writing, DataLad for data provenance and version control, and GitHub for source code management. This integration ensures that documentation, dataset histories, and analytical codes are consistently linked and traceable throughout the entire research lifecycle. Additionally, IntegriLAB connects with LabTrace, an Algorand blockchain-based platform that allows researchers to certify their work, providing tamper-proof validation of contributions. The initial release of the application is accessible via this link -
https://www.integrilab.org/.
3.1. System Architecture
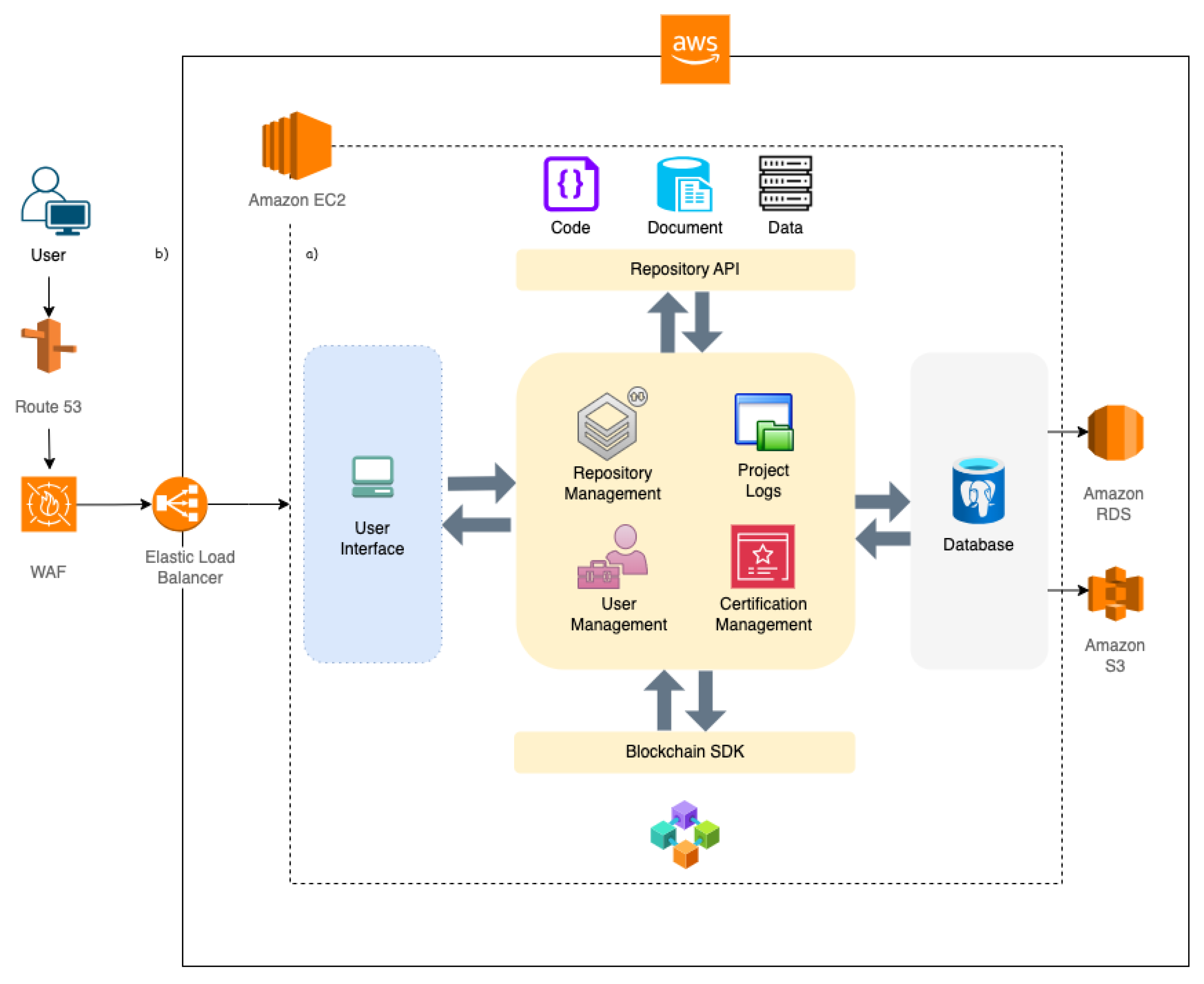
This system follows a three-tier architecture, comprising a frontend user interface, a backend application layer, and a database, as illustrated in the dotted-line diagram in
Figure 3. The frontend offers a user-friendly interface that manages user interactions and sends requests to the backend through RESTful APIs. This interface allows project members and administrators to interact with the system. Users can create and manage projects, connect profiles, track project progress, and engage with repositories.
The backend infrastructure is designed to efficiently process user requests, manage authentication, and oversee data storage. It plays a crucial role in user authentication, project management, and the integration of external services as well as in project certification. The backend communicates directly with the database to facilitate the storage of user and project data. Integration with external repositories is accomplished through the use of custom-developed APIs and open-source solutions for Overleaf (documents), DataLad (data), and GitHub (code). Furthermore, a custom-built SDK is employed to establish a connection with the blockchain platform LabTrace.
The database comprehensively stores all relevant information regarding users, projects, profiles, repository details, and project activities. It effectively manages roles and permissions for different user categories, including Administrator, Principal Investigator (PI), and Member.
3.2. Workflow
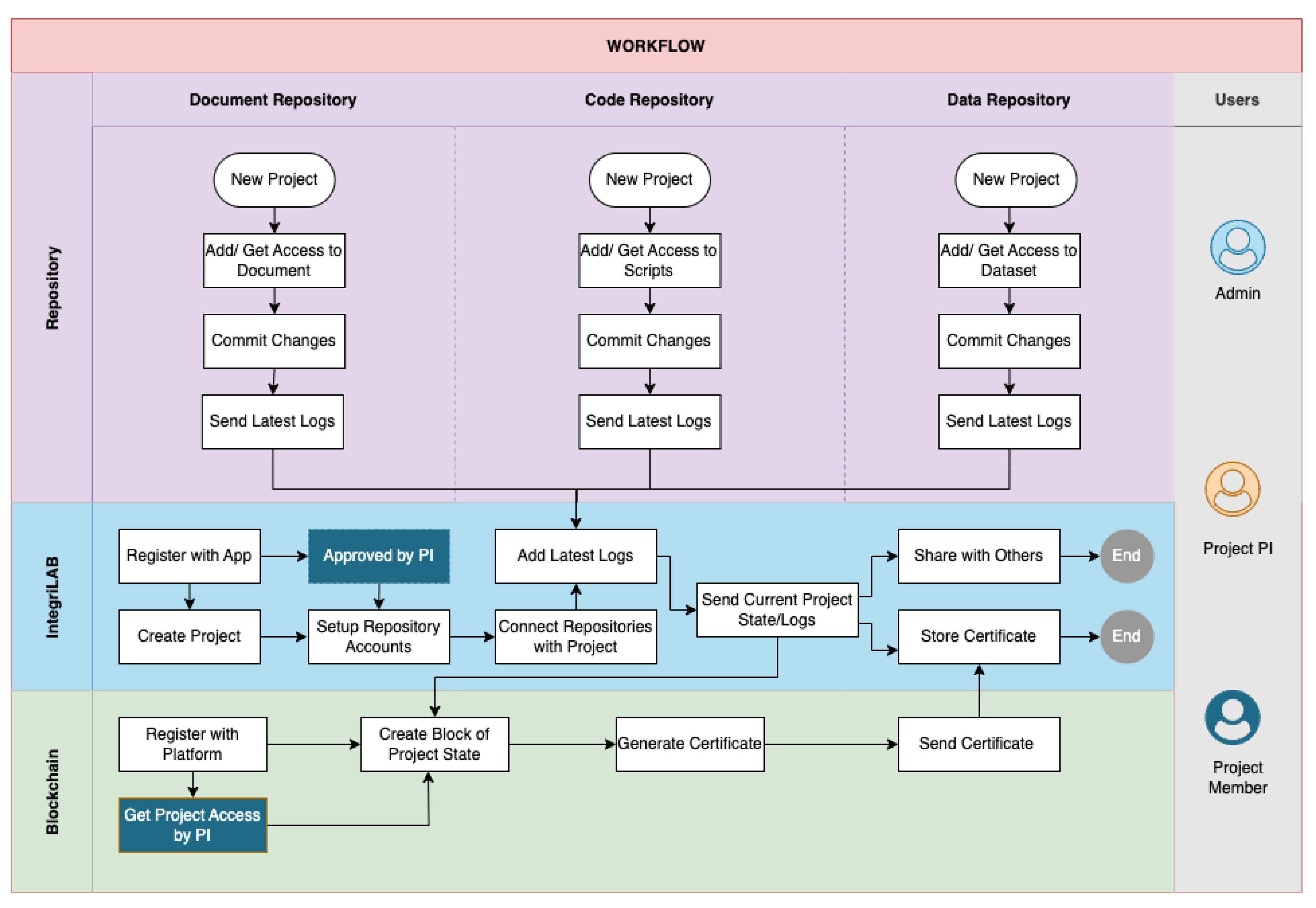
The workflow procedure for IntegriLAB is articulated in
Figure 4. To access the application, users are required to complete a registration process. It is important for users to maintain active accounts on both Overleaf and GitHub, in addition to having a local or remote installation of DataLad for efficient data management. IntegriLAB accommodates three primary user roles: Administrator (responsible for user role Assignment and Entity management), Project Principal Investigator (PI, responsible for project setup and manage user access and permissions), and Project Member (responsible for log into project repository and blockchain validation).
The workflow is initiated by the PI, who is tasked with creating a new project, registering it with LabTrace for blockchain certification, and approving project members upon their registration. Following approval, project members must link their Overleaf, GitHub, and DataLad accounts to IntegriLAB and specify their respective repository names to ensure smooth integration across all tools.
Upon completion of the initial setup, users are able to commit updates from their connected repositories, which may include documentation, datasets, and source code, directly through IntegriLAB. This integration logs details from each repository, thereby promoting a transparent and traceable research process. At any point in the project timeline, users have the option to initiate project certification via LabTrace, which produces tamper-proof digital records that affirm authorship and ownership of research contributions.
3.3. Implementation and Deployment
The IntegriLAB platform is deployed as a web-based application accessible via any modern browser, requiring an internet connection and user registration for access.
Table 1 outlines the complete technology stack employed in its development. The system architecture emphasizes modularity, robustness, and user-centric design, integrating services such as Overleaf, GitHub, DataLad, and LabTrace. The frontend is built using HTML5, CSS3, JavaScript, and Bootstrap to deliver a responsive and intuitive interface optimized for multiple user roles (Administrators, Principal Investigators, and Project Members). The backend is powered by the Django web framework[
30], chosen for its scalability, built-in security features, and seamless integration with third-party services. Django handles all application logic, authentication workflows, and request-response lifecycles.
Overleaf integration is achieved through a custom-built API, enabling real-time synchronization and management of collaborative LaTeX documents directly within the platform. GitHub repositories are accessed using GitPython and RESTful API calls to enable version-controlled source code management. For dataset management, IntegriLAB incorporates DataLad through an in-house developed API, allowing users to link local or remote datasets and synchronize changes within their project workspace.
LabTrace integration introduces blockchain-based certification. A custom SDK enables secure communication with the Algorand blockchain to timestamp and verify project milestones, ensuring tamper-proof records for key contributions.
All persistent data is stored in a PostgreSQL relational database, managed through Django’s Object-Relational Mapper (ORM). This abstracts complex SQL operations and streamlines data access. User roles and permissions are managed via Django’s authentication system, implementing a role-based access control (RBAC) model to enforce secure, hierarchical access to resources based on user roles.
IntegriLAB is deployed on the AWS cloud platform, ensuring scalability, reliability, and robust security, as depicted by the solid-line diagram in
Figure 3. The system’s deployment architecture is dynamic, capable of scaling according to traffic demands and ensuring high availability during peak usage. The application operates on Amazon EC2 instances, offering flexible compute capacity to handle web requests, process backend operations, and manage interactions with both the database and external services. The PostgreSQL database is managed via Amazon RDS (Relational Database Service), which provides automated backups and failover management to ensure continuous uptime. Static assets, including templates, CSS files, research document logs, are stored securely in Amazon S3 (Simple Storage Service). Data in transit is protected with SSL/TLS encryption, safeguarding user data and project-related information. In addition to Django’s built-in security features, AWS Web Application Firewall (WAF) rules are configured to further enhance the overall system security. While the platform is primarily hosted on AWS, deployment on local networks is also possible depending on resource availability. IntegriLAB is a closed-source application, and as such, its source code is not publicly accessible or openly distributed.
3.4. Data Security and Privacy
IntegriLAB implements a comprehensive security framework to safeguard data and ensure robust user authorization. The application only stores metadata (timestamps, authorship, file names) related to the studies, not the data’s substantive content which reduces risk of exposure and simplifies compliance. Data is encrypted both at rest and in transit, utilizing Amazon RDS encryption[
31] for stored data and SSL/TLS protocols for secure communication. Role-Based Access Control (RBAC) governs user permissions, restricting access to resources based on predefined roles such as Administrator, Principal Investigator, and Member. User authentication is managed through Django’s built-in system, supporting secure registration, login, and password management. Additionally, AWS Web Application Firewall (WAF) rules are configured to protect against common web exploits, enhancing the overall security posture.
3.5. System Validation
Comprehensive testing of IntegriLAB was conducted across multiple levels to ensure functionality, reliability, and user satisfaction. Functional testing validated core operations such as project creation, user registration, repository setup, and certification flow. It confirmed that Principal Investigators (PIs) could approve team members and configure repositories, while team members were able to commit changes and request certification. Integration testing focused on seamless data exchange between IntegriLAB and external platforms like Overleaf, GitHub, DataLad, and LabTrace, ensuring that updates made in any system were accurately reflected across all others. Performance testing evaluated the platform’s responsiveness under load, especially with large datasets and frequent commits, and tested the efficiency of syncing with integrated repositories. Security testing verified robust authentication and authorization for all user roles, assessed the integrity of blockchain certification via LabTrace. Lastly, User Acceptance Testing (UAT) involved real project teams using the platform in a controlled environment to assess usability and workflow alignment with user expectations, with feedback informing iterative improvements.
4. Case Study
Case Study: Enhancing Research Transparency in Schizophrenia Study Using IntegriLAB
IntegriLAB has been applied in a schizophrenia (SCZ) research project investigating dopamine synthesis capacity using [18F] FDOPA PET imaging and radiomics to predict antipsychotic treatment response.[
33] The study involved complex workflows across data storage, code analysis, and documentation, and is currently under peer review. IntegriLAB was integrated with DataLad for version-controlled imaging and feature datasets (CSV format), GitHub for managing Python analysis scripts, and Overleaf for report writing. It recorded metadata—such as authorship, timestamps, and workflow transitions—creating an auditable timeline accessible to the entire team.
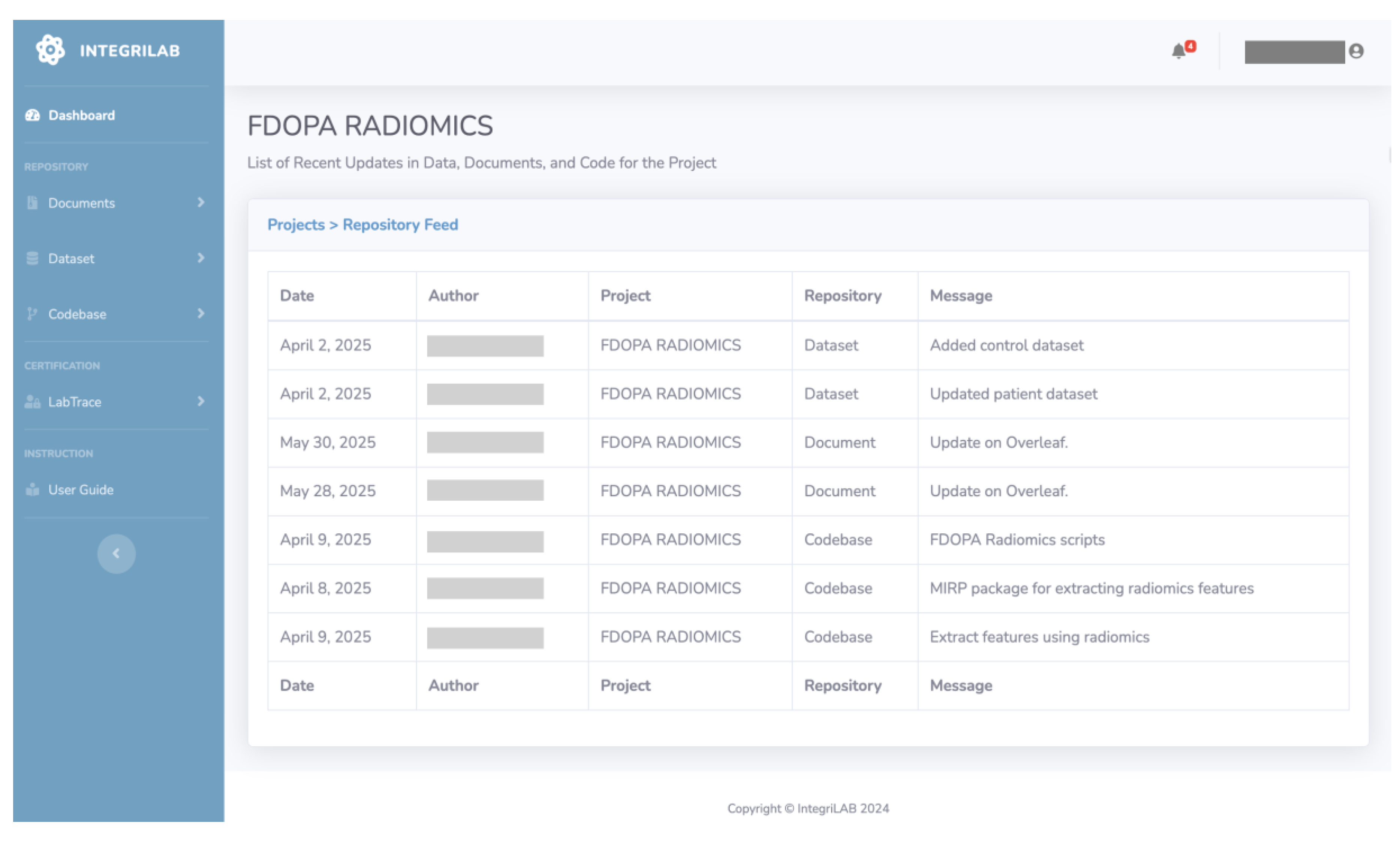
Figure 5 presents the IntegriLAB interface, showing ongoing tasks along with change messages, respective author details, and time logs for each update across the repositories.
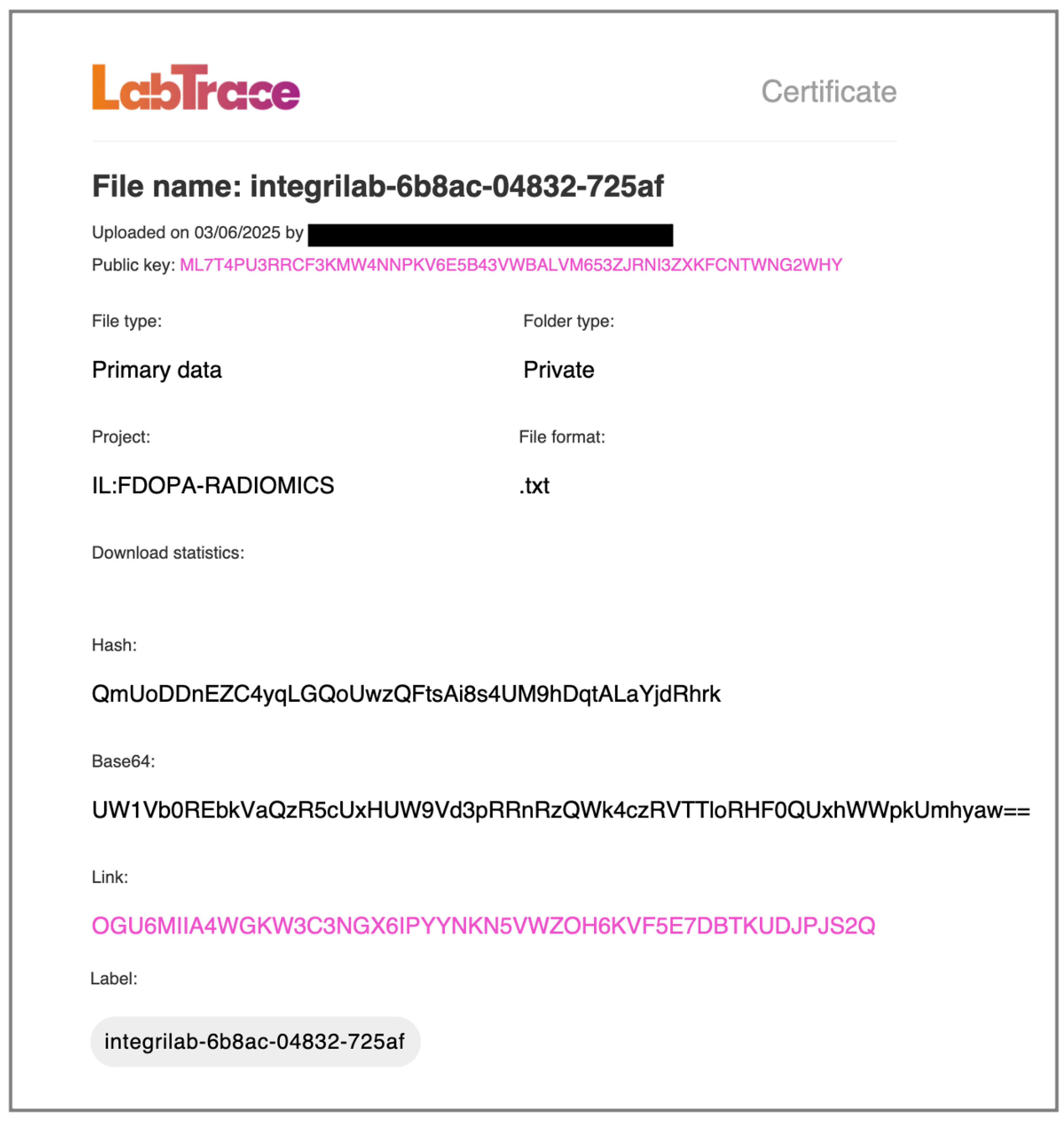
Additionally, IntegriLAB was connected to LabTrace which generated tamper-proof digital certificates for key project milestones. One such certificate, illustrated in
Figure 6, verifies the integrity of a primary data file that stores metadata from the study. The certificate captures project information, author details, and the file’s cryptographic fingerprint (Hash and Base64 encoding), establishes provenance and ownership via Public Key infrastructure, and includes an immutable access token (Link) that links to the certificate’s metadata record on the Algorand blockchain. This implementation enhanced team coordination, centralized oversight, and strengthened the credibility and reproducibility of the research.
5. Discussion
Traditional version control systems like Git play a crucial role in enhancing traceability within scientific research by systematically documenting alterations to scripts and analytical tools. Widely adopted platforms. including Overleaf, Jupyter Notebooks, GitHub, GitLab, and BitBucket, are commonly employed to manage various facets of research workflows. However, the utilization of diverse tools across different stages of a single project can lead to fragmentation, complicating the ability to establish authorship and maintain a cohesive, verifiable record of contributions. Furthermore, it is important to note that version control systems like Git do not provide immutable logging by default, which can result in potential manipulation and raises concerns regarding data integrity.[
6]
Therefore, a dedicated platform for managing research workflows is essential to meet the demands of increasingly complex research environments. Researchers should not be burdened with concerns about dataset size or the need to manually track every change. They should be able to store sensitive data securely, whether on local machines or remote servers, without having to upload it to third-party services just to maintain a record of modifications.
The integration of DataLad into IntegriLAB provides a robust solution for tracking dataset changes, supporting data integrity and reproducibility while protecting sensitive or proprietary information. Just as Overleaf facilitates version control for collaborative writing, and GitHub does for code management, IntegriLAB extends these principles across the broader research lifecycle. By incorporating such tools, it maintains detailed logs of research activities, allowing team members to monitor progress, revisit previous states, and restore earlier versions when necessary.
Importantly, IntegriLAB does not require the transfer or storage of confidential data, documents, or code. Instead, it focuses on aggregating and certifying metadata and activity logs from various platforms. This enables researchers to continue using their preferred tools while centralizing workflows to enhance project visibility and team coordination.
While some existing tools already offer logs of operations, others lack robust version control. IntegriLAB adds value not only by bringing these disparate tools into a unified environment but also by using blockchain-based certification to ensure immutability and verifiability, particularly for systems with insufficient logging capabilities. This blockchain-backed approach establishes a tamper-evident timeline of research activities, significantly enhancing cross-platform traceability and reinforcing authorship, attribution, and intellectual property claims.
6. Future Works
In the future, integrating Docker technology into IntegriLAB could offer significant advantages by enabling researchers to fully replicate their computational environments—including software dependencies, configurations, and runtime behavior. This would greatly enhance the reproducibility of results. When combined with blockchain-based certification, such integration would provide tamper-proof verification, reinforcing scientific integrity and trust in research outputs.
Additionally, incorporating real-time collaboration tools, such as task assignment, progress tracking, and project milestone management, would further strengthen IntegriLAB’s team coordination capabilities. Expanding support to include popular documentation platforms like Microsoft Word and Google Docs would allow users to continue working within familiar environments while still synchronizing activity logs and metadata with IntegriLAB. To support these enhancements and test scalability, future case studies and proof-of-concept work will explore integration with large-scale data-sharing initiatives such as Psy-ShareD[
35], enabling broader validation and real-world application of the platform across diverse, multi-institutional research environments.
Author Contributions
CRediT: Rubaida Easmin: Conceptualization, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft, Writing –review & editing; Mattia Veronese: Conceptualization, Validation, Writing – original draft, Writing – review & editing; Paul Allen: Supervision, Writing – original draft, Writing – review & editing; Giovanna Nordio: Conceptualization, Validation, Writing – review & editing; Federico Turkheimer: Conceptualization, Supervision, Writing – original draft, Writing –review & editing.
Funding
This work was supported by the National Institute for Health and Care Research (NIHR) Maudsley Biomedical Research Centre (BRC) under Grant NIHR203318.
Acknowledgments
The authors would like to acknowledge Enrico Gava and Marco Centurioni of Shadowy Creators, Web3 Software House, Italy, for their valuable feedback on this research.
Disclosure Statement
There are no relevant financial or non-financial competing interests to report.
References
- Wittek K, Wittek N, Lawton J, Dohndorf I, Weinert A, Ionita A. A blockchain-based approach to provenance and reproducibility in research workflows. 2021 IEEE International Conference on Blockchain and Cryptocurrency (ICBC). 2021; (pp. 1-6). [CrossRef]
- Niso G, Botvinik-Nezer R, Appelhoff S, De La Vega A, Esteban O, Etzel JA, Finc K, Ganz M, Gau R, Halchenko YO, Herholz P. Open and reproducible neuroimaging: from study inception to publication. NeuroImage. 2022;263:119623. [CrossRef]
- Borghi JA, Van Gulick AE. Data management and sharing in neuroimaging: Practices and perceptions of MRI researchers. PloS one. 2018;13(7):e0200562. [CrossRef]
- Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5:11-33. [CrossRef]
- Flywheel. Medical Image Analysis Software. 2022 [accessed 2025 Mar 29]. https://flywheel.io/.
- Bell J, LaToza TD, Baldmitsi F, Stavrou A. Advancing open science with version control and blockchains. 2017 IEEE/ACM 12th International Workshop on Software Engineering for Science (SE4Science). 2017;(pp. 13-14). [CrossRef]
- LabArchives. The Modern Electronic Lab Notebook (ELN). 2022 [accessed 2025 Mar 08]. https://www.labarchives.com/.
- Schröder M, Biskup T. LabInform ELN: A lightweight and flexible electronic laboratory notebook for academic research based on the open-source software DokuWiki. ChemRxiv. 2023. [CrossRef]
- SciNote Blog. How To Choose The Best Electronic Lab Notebook Software. 2024 [accessed 2025 Mar 08]. https://www.scinote.net/blog/electronic-lab-notebook-guide/.
- CARP N, Minges A, Piel M. eLabFTW: An open source laboratory notebook for research labs. J. Open Source Softw.. 2017;2(12):146. [CrossRef]
- Benchling. Cloud-based platform for biotech R&D. 2023 [accessed 2025 Mar 08]. https://www.benchling.com/.
- eLabNext. Your go-to lab platform. 2024 [accessed 2025 Mar 08]. https://www.elabnext.com/.
- Genemod. Lab Management Software: LIMS | Genemod. 2020 [accessed 2025 Mar 08]. https://genemod.net/.
- Riley EM, Hattaway HZ, Felse PA. Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory. Journal of Biological Engineering. 2017;11:1-9. [CrossRef]
- Szczepanik M, Wagner AS, Heunis S, Waite LK, Eickhoff SB, Hanke M. Teaching research data management with DataLad: a multi-year, multi-domain effort. Neuroinformatics. 2024;7:1-1. [CrossRef]
- GitHub. Build and ship software on a single, collaborative platform. 2004 [accessed 2025 Mar 09]. https://github.com/.
- Overleaf. Online LaTeX Editor. 2014 [accessed 2025 Mar 09]. https://www.overleaf.com/.
- LabTrace. LabTrace. 2023 [accessed 2025 Mar 09]. https://labtrace.io/.
- National Academies of Sciences, Medicine, Policy, Global Affairs, Board on Research Data, Information, Division on Engineering, Physical Sciences, Committee on Applied, Theoretical Statistics, Board on Mathematical Sciences. Reproducibility and replicability in science. National Academies Press. 2019. [CrossRef]
- Gorgolewski KJ, Poldrack RA. A practical guide for improving transparency and reproducibility in neuroimaging research. PLoS biology. 2016;14(7): e1002506. [CrossRef]
- Baker, M. 1,500 scientists lift the lid on reproducibility. Nature. 2016; 533(7604):452-454. [CrossRef]
- Botvinik-Nezer R, Holzmeister F, Camerer CF, Dreber A, Huber J, Johannesson M, Kirchler M, Iwanir R, Mumford JA, Adcock RA, Avesani P. Variability in the analysis of a single neuroimaging dataset by many teams. Nature. 2020;582(7810):84-88. [CrossRef]
- Zhao C, Jarecka D, Covitz S, Chen Y, Eickhoff SB, Fair DA, Franco AR, Halchenko YO, Hendrickson TJ, Hoffstaedter F, Houghton A. A reproducible and generalizable software workflow for analysis of large-scale neuroimaging data collections using BIDS Apps. Imaging Neuroscience. 2024;2:1-9. [CrossRef]
- White T, Blok E, Calhoun VD. Data sharing and privacy issues in neuroimaging research: Opportunities, obstacles, challenges, and monsters under the bed. Human Brain Mapping. 2022;43(1):278-91. [CrossRef]
- Higgins SG, Nogiwa-Valdez AA, Stevens MM. Considerations for implementing electronic laboratory notebooks in an academic research environment. Nature Protocols. 2022;17(2):179-89. [CrossRef]
- LabArchives. Data Storage - General Information About LabArchives. 2023 [accessed 2025 Mar 08]. https://help.labarchives.com/hc/en-us/articles/11724563888276-Data-Storage.
- Blockchain. Blockchain - Wikipedia. 2023 [accessed 2025 Mar 20]. https://en.wikipedia.org/wiki/Blockchain.
- Alzoubi YI, Mishra A. Green blockchain–A move towards sustainability. Journal of Cleaner Production. 2023; 430:139541. [CrossRef]
- Iberdrola. What are green cryptocurrencies and why are they important? - Iberdrola. 2023 [accessed 2025 Mar 20]. https://www.iberdrola.com/sustainability/green-cryptocurrencies.
- Forcier J, Bissex P, Chun WJ. Python web development with Django. Addison-Wesley Professional; 2008.
- Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES, Munafò MR. Power failure: why small sample size undermines the reliability of neuroscience. Nature reviews neuroscience. 2013;14(5):365-76. [CrossRef]
- Amazon. Encrypting Amazon RDS resources. 2020 [accessed 2025 Mar 10]. https://docs.aws.amazon.com/AmazonRDS/latest/UserGuide/Overview.Encryption.html.
- Lindquist, Martin. Neuroimaging results altered by varying analysis pipelines. Nature Publishing Group UK London. (2020): 36-37. [CrossRef]
- Schiulaz A, Nordio G, Giacomel A, Easmin R, Bettinelli A, Selvaggi P, Williams S, Turkheimer F, Jauhar S, Howes O, Veronese M. Radiomic Analysis of Striatal [18F] FDOPA PET Imaging in Patients with Psychosis for the Identification of Antipsychotic Response. Molecular Imaging and Biology. 2025 :1-4. 5 May. [CrossRef]
- Allen P, Zurita M, Easmin R, Bucci S, Kempton MJ, Rogers J, Mehta UM, McGuire PK, Lawrie SM, Whalley H, Gadelha A. The Psychosis MRI Shared D ata Resource (Psy-ShareD). Hoboken, USA: John Wiley & Sons, Inc. 2025. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).