1. Introduction
The objective of the European Directive 2001/18/EC on the deliberate release of genetically modified organisms (GMOs) into the environment is to protect human health and the environment in accordance with the precautionary principle [
1]. Since its adoption biotechnology has developed in great pace also with regard to genome editing methods, which are also called new genomic techniques (NGTs) [reviewed by
2]. In recognition of the ability of NGTs of targeted genetic engineering, the European Commission (COM) proposed on July 5, 2023, a
lex specialis to separately regulate certain genetically modified (GM) plants generated with NGTs [
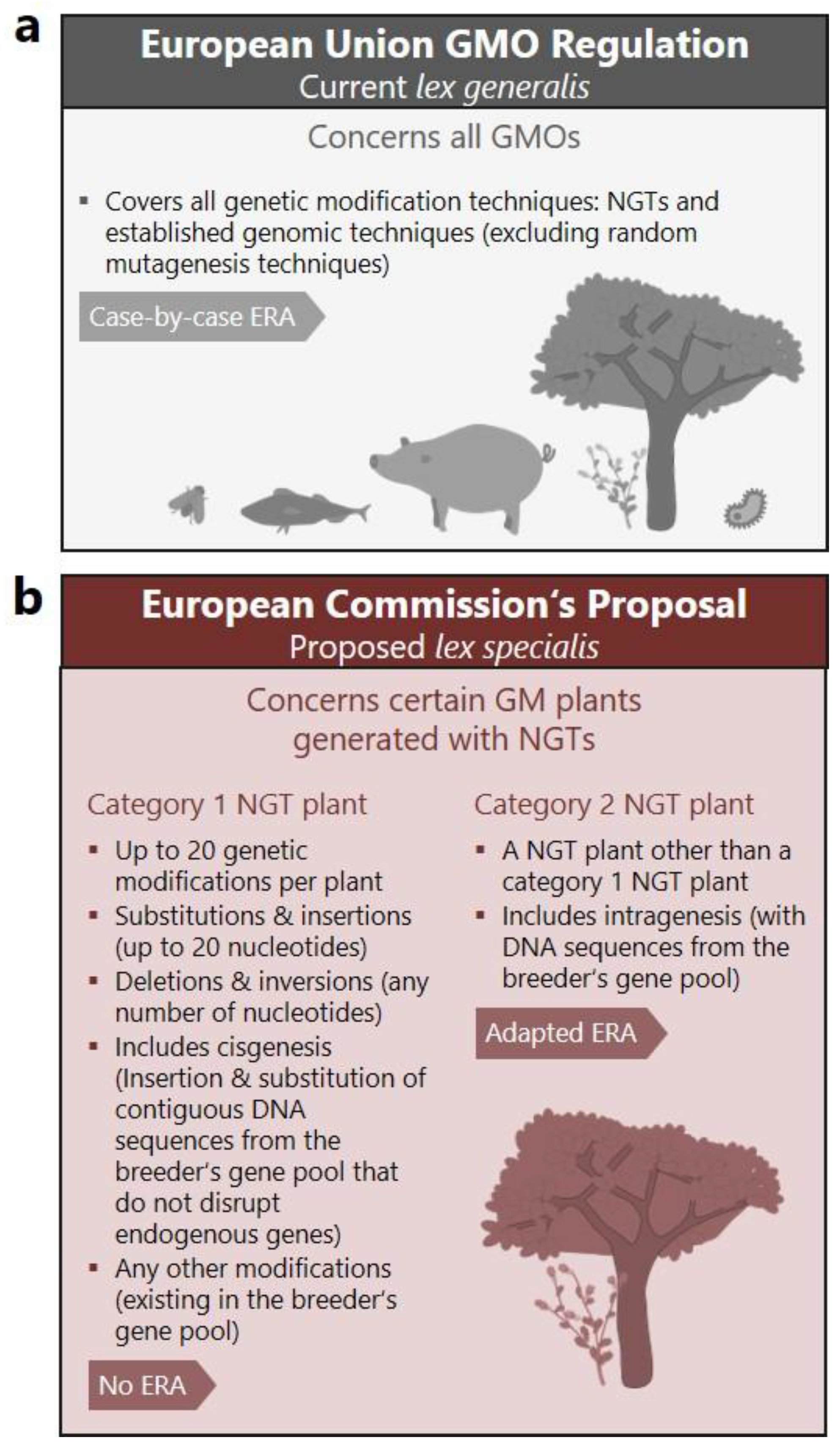
3]. According to the proposal, GM plants generated with NGTs that fulfill the proposed criteria would be assigned to two distinct categories, while all other GM plants would remain regulated as in Directive 2001/18/EC (
Figure 1 and Textbox 1). Meeting the criteria for “category 2 NGT plants” (NGT2) would enable an adapted, possibly reduced risk assessment on a case-by-case basis (
Figure 1b). The proposal goes one step further for GM plants fulfilling the criteria for so-called “category 1 NGT plants” (NGT1) (
Figure 1b). NGT1 plants should receive market access after a simplified, merely technical verification process without risk assessment. The proposal justifies the suggested process with the approach of considering NGT1 plants as
equivalent to conventional plants. This provides that their genetic modifications “could also occur naturally or be produced by conventional breeding techniques”, including random mutagenesis techniques [
3]. The proposal argues that the assumed equivalence to conventional plants makes the mandatory risk assessment of the current GMO regulation obsolete.
Textbox 1
Definitions in the COM proposal
Genetically modified organism (GMO): „[…] an organism, with the exception of human beings, in which the genetic material has been altered in a way that does not occur naturally by mating and/or natural recombination“ (COM/2023/411 final referring to Directive 2001/18/EC, Article 2).
New genomic technique (NGT): „An umbrella term used to describe a variety of techniques that can alter genetic material of an organism and that have […] been developed since 2001, when the Union legislation on GMOs was adopted“ (COM/2023/411 final, explanatory memorandum).
Established genomic techniques: „Genetic modification techniques developed prior to 2001, when the Union GMO regulation was adopted“ (COM/2023/411 final, explanatory memorandum).
Cis-/intra-/transgenesis: „[…] targeted mutagenesis and cisgenesis (including intragenesis) introduce genetic modifications without inserting genetic material from non-crossable species (transgenesis).“ „The exogenous genetic material can be introduced without (cisgenesis) or with modifications/rearrangements (intragenesis)“ (COM/2023/411 final, explanatory memorandum).
Breeder‘s gene pool: „[…] total genetic information available in one species and other taxonomic species with which it can be cross-bred, including […] embryo rescue, induced polyploidy and bridge crosses“ (COM/2023/411 final, Article 3).
In Annex I of the proposal, criteria of genetic modifications are listed that qualify GM plants as NGT1 plants [
4] (
Figure 1b). The rationale of the NGT1 criteria is to “exclude from category 1 of the proposal NGT plants with complex modifications unlikely to be obtainable by conventional breeding methods” [
5]. Annex I therefore represents the foundation of the entire proposal and the scientific validity and logic of the NGT1 criteria is thus of critical importance. A technical paper provides insights into the scientific considerations of Annex I and formulates justifications for the types, size limits, and numerical limits of genetic modifications considering the genetic variability observed naturally or in conventional breeding [
5]. From here the proposal derives two threshold criteria for Annex I that define that (i) “no more than 20 genetic modifications” and (ii) “substitution or insertion of no more than 20 nucleotides” may be introduced per NGT1 plant (
Figure 1b) [
4]. Importantly, these NGT1 criteria relate exclusively to the molecular type and extent of DNA sequence changes. Thereby, the criteria follow a paradigm in which genetic changes are isolated from their genomic context and functionality. Accordingly, the proposal neither considers intended and unintended effects nor their phenotypic or risk related outcomes.
The aim of this work is to assess the scientific basis of the proposed equivalence criteria in Annex I, focusing on the two threshold criteria mentioned above. A central question is whether basic genetic principles were sufficiently taken into account when drafting Annex I. Taking Annex I as an implementation framework, complex genetic modifications are examined within the scope of possibilities and with a view to developments in the field of artificial intelligence (AI). In this regard, we discuss the basic premise of the proposal that putative NGT1 plants could per se be viewed as equivalent to conventional plants. After all, we question whether the assumed proportional risk constitutes a valid argument for suspending risk assessment in view of the precautionary principle.
2. Genetic Changes: In the Light of Probability, Context, and Combinability
In Annex I of the proposal, the first criterion that is supposed to define equivalence to conventional plants is that a NGT1 plant should differ “from the recipient/parental plant by no more than 20 genetic modifications […] in any DNA sequence sharing sequence similarity with the targeted site” [
4] (
Figure 1b). This criterion classifies all specified genetic changes anywhere in the genome as equally likely to occur. This is because this threshold criterion was derived on the basis of general mutation rates [
5]. The mutation rate, however, reflects neither the different mutation types nor the question whether mutations remain silent or promote biological functions. As the criterion also does not include any distance rule, it would theoretically be possible to place up to twenty modifications next to each other. From our point of view, there are some biological and statistical aspects that need to be addressed in this context.
First of all, the proposal uses the mutation rate as a basis for a defined threshold value of twenty. From a statistical point of view, however, the variables of a general mutation rate deviate strongly from the variables associated with the actual criterion. The proposals initial scientific rationale relied on mutation rates per generation in conventional breeding approaches producing random mutations (mostly single nucleotide polymorphisms). In contrast, the final criterion relies on the probability of specific genomic locations and specific genetic modifications in an infinite number of generations. That would be like equating the one hundred percent probability of numbers being drawn in the lottery with the much lower probability of the selected numbers being drawn. If one were to calculate the statistics for Annex I, one would conclude that the probability of the spontaneous occurrence of twenty very specific genetic changes would require a statistically unrealistic number of independent events and breeding generations. The proposed threshold value to define equivalence is thus based on a statistically untenable equation.
While statistical approaches assume an even distribution, mutations are not evenly distributed throughout the genome [
6,
7]. There are various biological influences that affect the likelihood of mutations in an unidirectional but unpredictable manner [reviewed by
8]. Depending on selective pressure and structural constraints, genomic regions exhibit different mutation rates, which can be observed for example between coding and non-coding regions or between housekeeping and immune system genes. The probability of multiple mutations occurring directly in the same gene or between linked gene groups is comparatively lower. Even taking into account powerful pre-breeding random mutagenesis approaches (like TILLING) that theoretically can promote mutations in every gene of interest, the number and outcome of mutations per gene would be still restricted considering technical limitations like lethality and phenotypic drawbacks [reviewed by
9]. Using general mutation rates for the scientific reasoning therefore neither captures the genetic bias nor the unpredictable progression of incremental mutations.
Furthermore, selective breeding relies on recombination to combine desired or separate unwanted mutations. During recombination pieces of DNA are broken and recombined to produce new combinations of alleles. Recombination occurs rarely within the same gene or among genes in close proximity [reviewed by
10,
11]. Also, the probability of intragenic recombination is influenced by several factors and varies, e.g. between conserved and highly variable genes. The principles of linkage drag and linked genes represent a common drawback in conventional breeding, often requiring many generations before a linkage is resolved. In some crops significant parts of the genome are even inaccessible by breeding [
12,
13]. The greater the number of genes underlying a desired trait, the more difficult it becomes to combine all favorable alleles in the same genotype. Thus, certain combinations of genetic modifications can exceed the limits of intragenic recombination in conventional breeding if e.g. drawn from different breeding lines.
In summary, the statistical likelihood and biological principles, namely genomic context and recombination potential, have not been adequately addressed in the proposal, particularly in Annex I. When both statistical constraints and genetic biases are taken into account, it becomes evident that the space of possible genetic changes is severely limited. As a result, many conceivable combinations are unlikely to arise within realistic evolutionary timescales, even with prolonged crossing or mutagenesis. A pure threshold value would neither capture nor consider the genetic context.
3. NGTs: From Random Mutations to Precision Design
The frequency, type, and length of mutations in plant genomes varies depending on the source of DNA damage, the DNA repair mechanism, the plant type, or plant genome size [reviewed by
8]. In its technical report, the COM concludes for example that “insertions of random sequences were reported to be much smaller” and “insertions of more random sequences are typically of a length of less than ten nucleotides” [
5]. From these findings, another criterion for Annex I was derived, which allows “substitutions or insertions of no more than 20 nucleotides” [
4] (
Figure 1b). Although the word “random” is used several times in the argumentative derivation for the proposal [
5], it is neither defined in more detail nor included in the final criterion. In the following sections, we therefore discuss why the principle of randomness plays a central role in the breeding comparison context and how NGTs can undermine it.
The term "random" generally refers to something that occurs without specific pattern, purpose, or predictable outcome. As a nucleotide does not appear alone in the genome, the genetic context determines whether a mutation appears random or fits into a pattern with biological meaning. In a protein-coding sequence, for example, each nucleotide triplet has a predictable outcome and fulfills the purpose to code for a specific amino acid. The for Annex I considered argument that the “theoretical probability that a random sequence” smaller than 21 nucleotides “may already occur elsewhere in the genome” [
5] has no relation to whether this sequence appears random or fits into a pattern at another position in the genome. Even if insertions of up to twenty random nucleotides were detected somewhere in the genome at some point, this does not mean that an insertion of a very specific sequence at a very specific location is equally likely. That would be like equating the probability of drawing random numbers in the lottery with selected numbers being drawn in an exact order (e.g. 1, 2, 3, 4, 5, 6 and 7). Insertion of specific nucleotides can only be described as precision design. Thus, also this second criterion represents a statistical misinterpretation and an unrealistic assumption of what can actually be achieved in conventional breeding scales.
In contrast to Annex I that makes no methodological distinction, we see a crucial difference in which NGT method is used and for what purpose. NGTs, such as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated system (Cas), cut the DNA with nucleases to generate double-stranded breaks (DSBs) at targeted sites in the genome. DSBs are repaired predominately by non-homologous end joining mostly resulting in small random nucleotide insertions, substitutions, and deletions [reviewed by
14]. This application is generally also known as a site-directed nucleases 1 (SDN1), which requires follow-up selection procedures [reviewed by
15]. Furthermore, SDN2 and SDN3 approaches use homologous donor DNA sequences for an error-prone homology-directed repair of DSBs, which enables the introduction of predefined nucleotides and long inserts. Instead of nucleases and donor DNA sequences, base- and prime-editing CRISPR/Cas applications involve nickases and reverse transcriptases [reviewed by
2]. Without DSBs the error prone DNA machinery is bypassed by these methods to enable pre-determined nucleotide insertions. Since then, NGTs can also be used independently of random repair mechanisms and enable the precise work in translational frames with pre-designed sequences. Thus, on the one hand, genome editing applications like SDN1 artificially resemble the randomness of DNA errors, which are targeted but still require post-selection. On the other hand, advanced prime editing techniques enable targeted non-random sequence insertions that implement designated biological functions.
The precision of NGT prime-editing has been highlighted for protein labeling, which can be used for purification or tracking. In rice, a classic epitope tag consisting of six histidine amino acids (polyhistidine) that are encoded by eighteen nucleotides has been precisely inserted upstream of a stop codon [
16]. This tag is not achievable through conventional breeding in relevant time frames due to the lack of selective pressure, absence of a visible phenotype, and low probability of six consecutive identical triplets. Furthermore, a precise insertion is essential to avoid disrupting protein function while enabling epitope-specific antibody binding. Prime-editing, thus, makes tagging of any protein of interest feasible, a task unthinkable with conventional breeding. Nevertheless, such a GM plant would receive the NGT1 status as it fulfills the Annex I criteria.
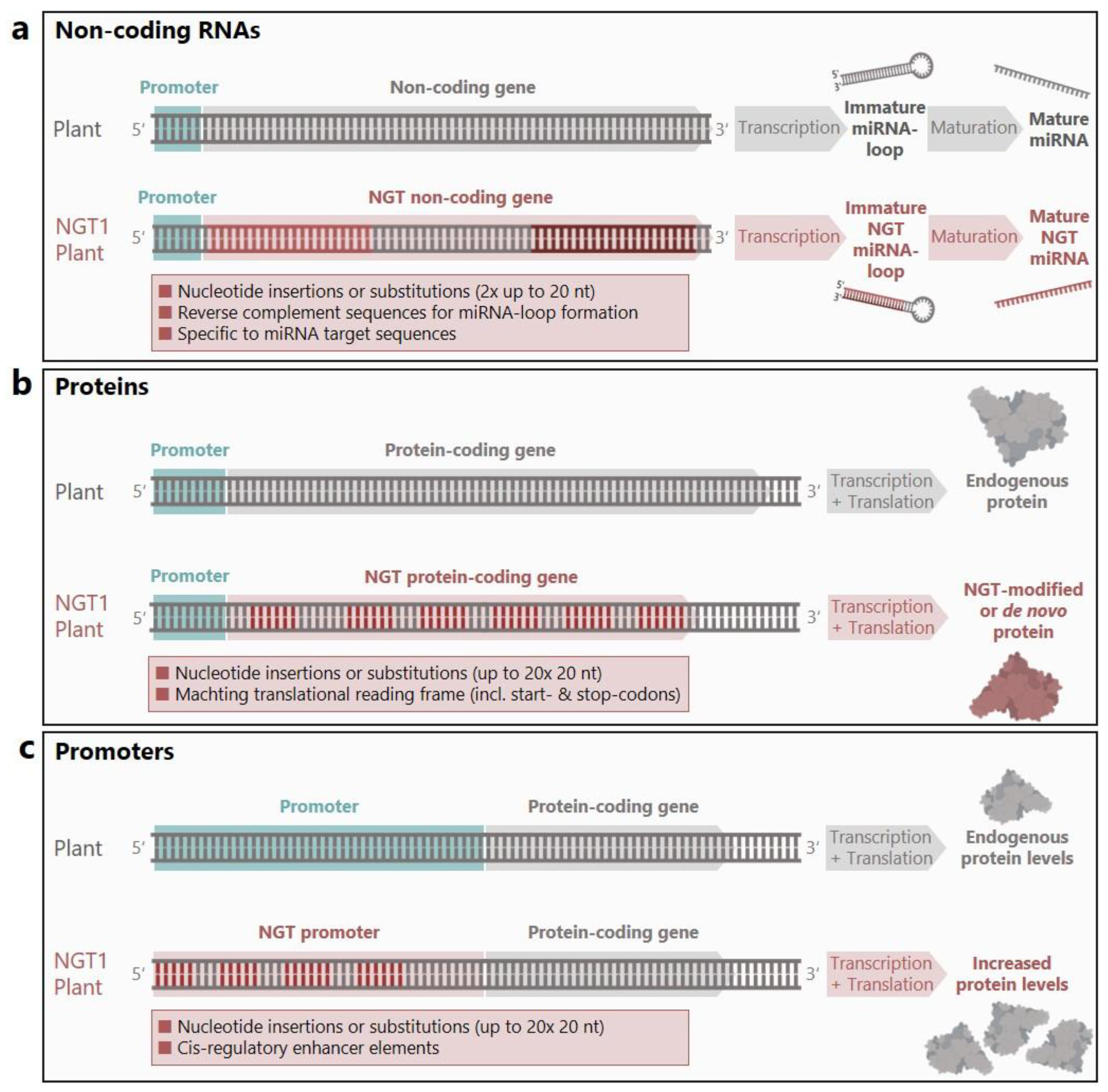
Another evident strength of NGTs like CRISPR/Cas lies in the orchestration of multiple targeted mutagenesis applications. The so-called NGT multiplexing allows the stepwise or simultaneous insertion of several mutations, even in close proximity [reviewed by
17]. This is crucial for plants with gene redundancies, which occur due to polyploidy, gene duplications, and large gene family groups [reviewed by
18]. In addition, some genetic engineering interventions require precise coordination of multiple genome modifications (
Figure 2). For example, the idea of triggering the mechanisms of RNA interference (RNAi) in plants relies on such precision. RNAi is influenced by various non-coding RNAs, including micro RNAs (miRNAs) that regulate gene expression by suppression [reviewed by
19]. With a typical length of 20-24 nucleotides, the recognition sequences of miRNAs represent attractive candidates for NGT applications to control the expression of any gene of interest [reviewed by
20]. Artificial miRNAs clearly demonstrate the strong effect of intentional sequence replacements in contrast to random mutations. The matching sequences enable first the formation of a complementary miRNA hairpin loop and second the specific binding between the mature miRNA and its target (
Figure 2a). If this concept is pursued further, redirected miRNAs could not only act in plants, but also affect insects in an insecticide-like manner [reviewed by
21,
22].
In summary, it is fair to say that despite enormous technical innovations in the field of conventional breeding, plant breeders have to be satisfied with the result of imprecise genome repairs. Besides the necessary selection to achieve desired traits, additional mutations may even be required for functionality. In contrast, NGTs enable precision not only with regard to the target site, but also with regard to the genetic modification itself. NGT interventions are intended to be minimally invasive with maximum efficiency in terms of direct functionality. A mere threshold value makes no differentiation in functionality.
4. AI: A Regulatory Gap in the NGT Proposal
AI has become a transformative, rapidly evolving tool also in the field of genetic engineering [reviewed by
23,
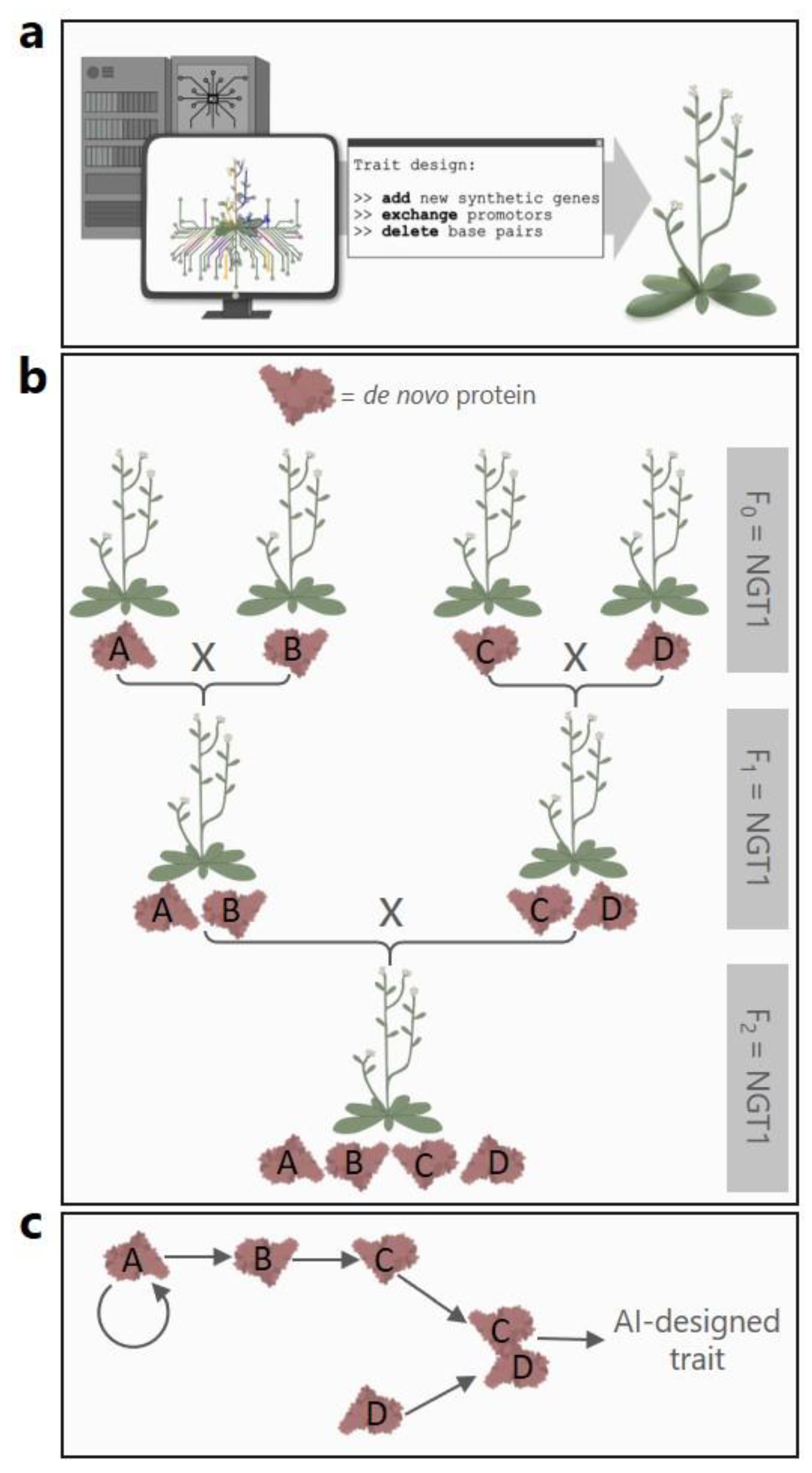
24]. The question of whether NGTs accelerate the breeding of conventional plants within the scope of existing breeding knowledge falls short here. AI-driven tools can analyze vast datasets of plant genomes and phenotypes, thereby identifying new targets for engineering (
Figure 3a). For agricultural purposes, this includes AI-assisted predictions of genetic circuits, optimization of metabolic pathways, and design of novel genes and proteins. The future will show how AI applications would use the leeway of Annex I in the light of big data analyses, algorithms, and machine learning.
For instance, AI leverages the evolutionarily constrained protein sequence space to explore and optimize novel amino acid sequences that are predicted to fold into stable, functional structures for
de novo protein design [reviewed by
23,
25]. NGTs can be used to completely remodel protein functions affecting, for example, the catalytic center of proteins or their transport, solubility, activation, binding, or recognition. More importantly, rather than modifying existing ones,
de novo protein design enables the development of entirely new protein sequences to create molecules with desired properties [reviewed by
26]. Intriguingly, Annex I does not contain any spacing rules for the up to twenty possible genetic modifications per NGT1 plant. Theoretically, a single endogenous nucleotide in between two modifications would be sufficient as a spacer. Thereby several amino acids could be exchanged or introduced in a single protein (
Figure 2b). If twenty changes are possible side by side, we are no longer talking about the insertion (or substitution) of twenty nucleotides. We are talking about up to 400 precisely selected nucleotides only interrupted by individual endogenous nucleotides that could be translated into 140 amino acids. Such precise and coordinated genetic modifications would have to be regarded as complex genetic changes. The process, how, when and where individual sequence components are biotechnologically combined and inserted, is neither specifically considered nor relevant in the proposal. Consequently, the boundaries between supposedly small insertions and sequence constellations that exhibit characteristics of intragenesis and transgenesis are becoming blurred (Textbox 1).
In addition, AI tools find application in synthetic promoter design [reviewed by
27]. Besides the actual genes, the promoter regions as well as enhancing and suppressing
cis-regulatory elements are crucial for the context specific activity of genes. Transcription factor binding sites in plant genomes are approximately 6-10 nucleotides long, easily fitting the Annex I criterion [reviewed by
28] (
Figure 2c). If gene regulation should be regulated by one or more very specific transcription factors, very specific binding sequences must be introduced into the correct genetic context. Such a modified promoter may artificially increase transcription efficiency and strength, adopt expression profiles of responsive tissues, abolish negative feedback regulations, or change promoter induction conditions also in response to external stimuli [reviewed by
27].
Furthermore, AI-assisted machine-learning algorithms have the potential to optimize the outcome and efficiency of multiple interacting signaling nodes that form genetic circuits [
29]. The proposal also stipulates that NGT1 plants that are crossed with each other remain NGT1 plants. This also applies if the breeding product has more than twenty genetic changes. Crossbreeding could thereby produce NGT1 plants that accumulate several
de novo proteins (
Figure 3b). Thus, it would be possible to combine
de novo proteins that act synergistically or as signaling cascades to elicit desired traits (
Figure 3c). In fact, the artificial assembly of multiple regulatory elements and molecules might even be considered
synthetic biology especially when biological pathways and networks are systematically redesigned [reviewed by
24].
In conclusion, the increasing convergence between NGTs, AI, and synthetic biology has not been taken into account in the proposal. The potential of AI to explore and expand the evolutionary sequence space requires a closer look at the genomic context and the synergistic functionality of genetic changes. Mere threshold values do not capture the underlying complexity and impact of AI.
5. Complexity and Risks: Within the Scope of Uniform Regulation
The proposal tolerates risks associated with NGT1 plants by comparing them with risks posed by conventionally bred plants, which are considered within the scope of proportionality [
3]. Scientific findings on potential risks would not be considered in individual cases or retrospectively. The European Food Safety Authority (EFSA) has confirmed in response to an extensive analysis of the Annex I criteria by the French Agency for Food, Environmental and Occupational Health & Safety (ANSES) that the proposals “criteria are not meant to define levels of risk” [
30,
31]. Consequently, risks from NGT1 plants are not ruled out
per se. In view of these risks and taking into account the possible exposure – assuming a significant increase in the cultivation of NGT1 plants in the European Union – the potential hazard is considerable. Furthermore, the risk potential of an NGT plant is not generally linked to the type, size, or extent of the genetic modification. The proposed approach of defining the NGT1 criteria on a molecular basis, independent of the genetic context and functionality, therefore falls short. GM plants can only be adequately assessed on a case-by-case basis, as their properties and risks arise from a complex interplay between the molecular interactome, plant properties, and the receiving environment [reviewed by
32].
The proposed comparative approach would particularly not work if the Annex I criteria do not ensure the desired equivalence to natural and conventionally bred plants. Even if some NGT-generated mutations were highly likely to occur eventually through breeding (e.g. single nucleotide polymorphisms), other NGT1-compatible mutations may not be realized in any relevant time frame of conventional breeding (e.g .
de novo protein design). The more previously unattainable genetic modifications are introduced to achieve desired traits, the more difficult it may become to rule out or predict risks. If modifications under Annex I were to be linked in such a way that the characteristics of intragenesis, transgenesis, or synthetic biology were also fulfilled, associated risks would have to be compared with GMOs and not breeding. Directive 2001/18 thereby identifies diverse risks from GMOs to human health and the environment, including adverse impacts on biodiversity and ecosystems, harmful effects on target and non-target species, and far-reaching changes in agricultural practices [
1]. In case of insect resistant GM plants, including the discussed NGT1 plants with modified miRNAs (
Figure 2a), effects on non-target organisms – especially closely related insects – need to be excluded. Overall, it must be assumed that individual NGT1-compatible plants produced by targeted mutagenesis could have a similar, if not greater, risk potential than plants produced by genetic engineering so far. Probable risks to the environment cannot be excluded if risk assessment (and monitoring) is suspended as foreseen in the proposal for NGT1 plants.
Importantly, AI-assisted design or the novelty of a molecule or system would not disqualify the plant from NGT1 if the Annex I requirements are fulfilled. There is a possibility that there is no evolutionary precedent for de novo designed molecules and networks in a species and that these may be new to nature. In view of the broad application possibilities of NGTs, the general assumption of a “history of safe use” and the specific option for “new to nature” would be mutually exclusive. Despite the lack of scientific data and natural comparability, GM plants categorized as NGT1 would not be subject to an independent risk assessment and could not be rejected if there were concerns. Possible hazards that could emanate from the AI-supported use of NGTs and its innovation potential are neither completely foreseeable nor controllable now or in the future.
In summary, if the scientific validity of Annex I – and consequently the foundation of NGT1 classification – is subject to reasonable doubt, this calls into question the approach of the entire proposal. The general assumptions derived from the equivalence argument, e.g. on a lack of risk and coexistence possibilities, can no longer be regarded as reliable in individual cases. Only a case-specific assessment can keep pace with technological progress, including the use of AI, to ensure a future-proof regulation. Mere thresholds do not capture the complex interactions between molecular changes and risk-related effects on the phenotype and receiving environment.
6. Concluding Remarks and Future Perspectives
The regulatory proposal to exempt certain GM plants generated with NGTs from the current GMO regulatory framework aims “to ensure a high level of protection of human and animal health and the environment” [
3]. In order to meet this objective, the proposed
lex specialis must comply with the precautionary principle on which the current
lex generalis is based [
1]. The approach of equating technological superiority with conventional breeding means using NGTs only within the respective limits. However, the proposal underlies a misconception that all conceivable genetic changes or combinations could occur at some point if crosses or mutagenesis were only carried out long enough. Neither the proposals overall concept nor the Annex I criteria are able to capture the ultimately decisive genomic context that determines functionality and the risk potential of NGT-produced plants. Our results shed light on the possibility that GMO-comparable plants could be produced within the limits of Annex I ”in which the genetic material has been modified in a way that does not occur naturally by mating and/or natural recombination” [
1]. In its current form, the proposed assumption of equivalence is therefore scientifically not justified and not suitable for protection in accordance with the precautionary principle. Our finding would support the 2018 ruling of the European Court of Justice that NGT plants are GMOs to be covered by the European Directive 2001/18/EC [
33].
Although the analysis of the other Annex I criteria for NGT1 plants (
Figure 1b) would go beyond the scope of this publication, we would like to point out few critical aspects. First, the criteria generally require a more comprehensive scientific justification and clarification of terms in order to enable a substantive assessment, as already pointed out in the analysis by ANSES [
31]. For example, vague terms leave room for interpretation. It is not clear whether a promoter and an open reading frame are regarded as an inseparable “contiguous DNA sequence” or whether a single exon or motif already represents a “contiguous DNA sequence” (
Figure 1b). Secondly, the possibility of combining the different criteria would considerably broaden the spectrum of genetic modifications and their potential risks. For example, it cannot be clearly ruled out that highly expressed cisgenes can be inserted and further modified with up to nineteen specific twenty-nucleotide insertions (e.g. to enable the ubiquitous expression of
de novo proteins). Finally, it should also be noted that the regulatory process is still ongoing and that the final negotiations in the trilogue could also change the Annex I criteria. For example, according to the resolutions passed by the European Parliament on February 7, 2024, the total number of genetic modifications per plant is not to be regulated [
34]. This would mean that advanced NGT multiplexing applications like gluten-reduced wheat [
35] would also fall under NGT1. Threshold values are ultimately political decisions. Whereby in our view, it is not the size of genetic changes but the genetic context and functionality that are decisive in ensuring a high level of protection.
Another critical point is that the adaptation of the GMO regulatory framework could be in place for decades to come and it must therefore take account of technical progress. It is important to discuss which GM plants would currently be subject to the proposed regulation [
22]. However, the proposal would not only affect the currently most common SDN1 applications [
36]. In the future, genetic modifications in plants could be specifically designed so that they meet the proposed criteria for the NGT1 fast track, although this would also bypass the currently mandatory risk assessment. Even if methods for certain complex modifications are currently not available, technical feasibility should only be a matter of time at the current pace of development. It is remarkable that at a time when AI innovations are overcoming the humanly possible, deregulation is being proposed that falls back on old-fashioned breeding principles. As the use of AI in the production of novel biomolecules is not considered in the proposal, the proposals concept risks creating a regulatory gap that could enable so far unknown applications of synthetic biology in agriculture, molecular pharming, and beyond.
Funding
The author(s) declare no financial support was received for the research, authorship, and/or publication of this article.
Author’s Contributions
J.M. conceptualized the manuscript, analyzed the scientific literature, prepared the figures, wrote the original draft, and controlled the writing-review and -editing. S.S. conceptualized the manuscript, analyzed the scientific literature, and contributed to the manuscript writing. ME supervised and contributed to the manuscript writing. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Acknowledgments
We thank Mathias Otto for critical comments regarding the manuscript and Finja Bohle for illustrations.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
| AI: Artificial intelligence |
| ANSES: Agence Nationale de Sécurité Sanitaire |
| Cas: CRISPR-associated system |
| COM: European Commission |
| CRISPR: Clustered Regularly Interspaced Short Palindromic Repeats |
| DSB: Double-stranded breaks |
| EFSA: European Food Safety Authority |
| ERA: Environmental risk assessment |
| GM: Genetically modified |
| GMO: Genetically modified organism |
| miRNA: Micro RNA |
| NGT: New genomic technique |
| NGT1: Category 1 NGT plant |
| NGT2: Category 2 NGT plant |
| nt: Nucleotide |
| SDN1/2/3: Site-directed nucleases 1/2/3 |
References
- European Parliament and Council of the European Union. Directive 2001/18/EC on the deliberate release into the environment of genetically modified organisms and repealing Council Directive 90/220/EEC; 2001.
- Bhuyan SJ, Kumar M, Ramrao Devde P, Rai AC, Mishra AK, Singh PK, Siddique KHM. Progress in gene editing tools, implications and success in plants: a review. Front Genome Ed. 2023;5:1272678. [CrossRef]
- European Commission. Proposal for a REGULATION OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL on plants obtained by certain new genomic techniques and their food and feed, and amending Regulation (EU) 2017/625: COM(2023) 411 final; 2023.
- European Commission. ANNEXES to the Proposal for a REGULATION OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL on plants obtained by certain new genomic techniques and their food and feed, and amending Regulation (EU) 2017/625: COM(2023) 411 final ANNEXES 1 to 3; 2023.
- European Commission. Regulation on new genomic techniques (NGT) – Technical paper on the rationale for the equivalence criteria in Annex I: 2023/0226(COD), 14204/23; 2023.
- Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–4. [CrossRef]
- Weng M-L, Becker C, Hildebrandt J, Neumann M, Rutter MT, Shaw RG, et al. Fine-Grained Analysis of Spontaneous Mutation Spectrum and Frequency in Arabidopsis thaliana. Genetics. 2019;211:703–14. [CrossRef]
- Quiroz D, Lensink M, Kliebenstein DJ, Monroe JG. Causes of Mutation Rate Variability in Plant Genomes. Annu Rev Plant Biol. 2023;74:751–75. [CrossRef]
- Szurman-Zubrzycka M, Kurowska M, Till BJ, Szarejko I. Is it the end of TILLING era in plant science? Front Plant Sci. 2023;14:1160695. [CrossRef]
- Otto SP, Payseur BA. Crossover Interference: Shedding Light on the Evolution of Recombination. Annual Review of Genetics. 2019;53:19–44. [CrossRef]
- Epstein R, Sajai N, Zelkowski M, Zhou A, Robbins KR, Pawlowski WP. Exploring impact of recombination landscapes on breeding outcomes. Proc Natl Acad Sci U S A. 2023;120:e2205785119. [CrossRef]
- Chen Y-Y, Schreiber M, Bayer MM, Dawson IK, Hedley PE, Lei L, et al. The evolutionary patterns of barley pericentromeric chromosome regions, as shaped by linkage disequilibrium and domestication. Plant J. 2022;111:1580–94. [CrossRef]
- Des Danguy Déserts A, Bouchet S, Sourdille P, Servin B. Evolution of Recombination Landscapes in Diverging Populations of Bread Wheat. Genome Biol Evol 2021. [CrossRef]
- Bennett EP, Petersen BL, Johansen IE, Niu Y, Yang Z, Chamberlain CA, et al. INDEL detection, the 'Achilles heel' of precise genome editing: a survey of methods for accurate profiling of gene editing induced indels. Nucleic Acids Res. 2020;48:11958–81. [CrossRef]
- Cardi T, Murovec J, Bakhsh A, Boniecka J, Bruegmann T, Bull SE, et al. CRISPR/Cas-mediated plant genome editing: outstanding challenges a decade after implementation. Trends Plant Sci. 2023;28:1144–65. [CrossRef]
- Li J, Ding J, Zhu J, Xu R, Gu D, Liu X, et al. Prime editing-mediated precise knockin of protein tag sequences in the rice genome. Plant Commun. 2023;4:100572. [CrossRef]
- Abdelrahman M, Wei Z, Rohila JS, Zhao K. Multiplex Genome-Editing Technologies for Revolutionizing Plant Biology and Crop Improvement. Front Plant Sci. 2021;12:721203. [CrossRef]
- Nei M, Rooney AP. Concerted and Birth-and-Death Evolution of Multigene Families. Annual Review of Genetics. 2005:121–52. [CrossRef]
- Bhogireddy S, Mangrauthia SK, Kumar R, Pandey AK, Singh S, Jain A, et al. Regulatory non-coding RNAs: a new frontier in regulation of plant biology. Funct Integr Genomics. 2021;21:313–30. [CrossRef]
- Bravo-Vázquez LA, Méndez-García A, Chamu-García V, Rodríguez AL, Bandyopadhyay A, Paul S. The applications of CRISPR/Cas-mediated microRNA and lncRNA editing in plant biology: shaping the future of plant non-coding RNA research. Planta. 2023;259:32. [CrossRef]
- Liu S, Jaouannet M, Dempsey DA, Imani J, Coustau C, Kogel K-H. RNA-based technologies for insect control in plant production. Biotechnol Adv. 2020;39:107463. [CrossRef]
- Bohle F, Schneider R, Mundorf J, Zühl L, Simon S, Engelhard M. Where does the EU-path on new genomic techniques lead us? Front. Genome Ed. 2024. [CrossRef]
- Winnifrith A, Outeiral C, Hie BL. Generative artificial intelligence for de novo protein design. Curr Opin Struct Biol. 2024;86:102794. [CrossRef]
- Zhang D, Xu F, Wang F, Le L, Pu L. Synthetic biology and artificial intelligence in crop improvement. Plant Commun. 2024:101220. [CrossRef]
- Hayes T, Rao R, Akin H, Sofroniew NJ, Oktay D, Lin Z, et al. Simulating 500 million years of evolution with a language model. Science. 2025:eads0018. [CrossRef]
- Huang P-S, Boyken SE, Baker D. The coming of age of de novo protein design. Nature. 2016;537:320–7. [CrossRef]
- Yasmeen E, Wang J, Riaz M, Zhang L, Zuo K. Designing artificial synthetic promoters for accurate, smart, and versatile gene expression in plants. Plant Commun. 2023;4:100558. [CrossRef]
- Strader L, Weijers D, Wagner D. Plant transcription factors - being in the right place with the right company. Curr Opin Plant Biol. 2022;65:102136. [CrossRef]
- Hiscock, TW. Adapting machine-learning algorithms to design gene circuits. BMC Bioinformatics. 2019;20:214. [CrossRef]
- EFSA Panel on Genetically Modified Organisms. Scientific opinion on the ANSES analysis of Annex I of the EC proposal COM (2023) 411 (EFSA-Q-2024-00178). EFSA Journal 2024. [CrossRef]
- AGENCE NATIONALE DE SÉCURITÉ SANITAIRE (ANSES) de l'alimentation, de l'environnement et du travail. OPINION of the French Agency for Food, Environmental and Occupational Health & Safety on the scientific analysis of Annex I of the European Commission’s Proposal for a Regulation of 5 July 2023 on new genomic techniques (NGTs): Review of the proposed equivalence criteria for defining category 1 NGT plants; 2023.
- Eckerstorfer MF, Grabowski M, Lener M, Engelhard M, Simon S, Dolezel M, et al. Biosafety of genome editing applications in plant breeding: Considerations for a focused case-specific risk assessment in the EU. BioTech 2021. [CrossRef]
- European Court of Justice. Judgement of the Court (Grand Chamber) in Case C-528/16. 2018, July 25.
- European Parliament. Plants obtained by certain new genomic techniques and their food and feed. Amendments adopted by the European Parliament on 7 February 2024 on the proposal for a regulation of the European Parliament and of the Council on plants obtained by certain new genomic techniques and their food and feed, and amending Regulation (EU) 2017/625 (COM(2023)0411 – C9-0238/2023 – 2023/0226(COD)). 2024.
- Sánchez-León S, Gil-Humanes J, Ozuna CV, Giménez MJ, Sousa C, Voytas DF, Barro F. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J. 2018;16:902–10. [CrossRef]
- Parisi C, Rodríguez-Cerezo E. Current and future market applications of new genomic techniques. Publications Office of the European Union 2001. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).