1. Introduction
The way intracellular biochemistry is organized relies a lot on compartmentalization, which helps ensure that cellular processes are both specific and efficient [
1]. In addition to the traditional membrane-bound organelles, cells also use a fascinating process called Liquid-Liquid Phase Separation (LLPS) to create membraneless organelles (MLOs), often referred to as biomolecular condensates [
2]. These dynamic structures, which include familiar components like nucleoli, P-bodies, and stress granules, form when certain proteins and nucleic acids condense together. This condensation effectively concentrates molecules, allowing for the modulation of reaction rates and the creation of unique biochemical environments [
3]. The discovery of P granules in Caenorhabditis elegans as liquid droplets provided some of the earliest and most convincing evidence that LLPS plays a crucial role in organizing cellular architecture [
4]. This type of compartmentalization, driven by multivalent interactions, offers a reversible and finely-tuned way to control cellular functions in both space and time [
5].
Viruses, which are obligate intracellular parasites, have developed clever strategies to hijack the host's cellular machinery for their own replication. As our understanding of LLPS as a key organizer of cellular life deepens, it has sparked a surge of interest in exploring its role during viral infections [
6]. It is now increasingly clear that a diverse array of viruses commandeers or manipulates host and viral LLPS processes to orchestrate multiple stages of their replication cycles. These include the formation of specialized "viral factories" or replication compartments, sites for genome packaging, and mechanisms for evading host immune surveillance[
5,
6]. The rapid application of LLPS principles from cell biology to virology suggests that viruses are tapping into highly conserved and fundamental organizational mechanisms of the cell [
7,
8]. This realization implies that many viral structures and processes, previously understood in different terms, may be re-interpreted through the dynamic lens of LLPS, potentially revealing that LLPS is even more pervasive in virology than currently documented.
This review will synthesize the current understanding of how LLPs are important in viral replication, focusing on the formation and functional significance of viral condensates. This piece will take into account how this phenomenon changes our understanding of virus-host interactions and viruses causing disease. In addition, it will look at the new therapeutic opportunities that come from targeting LLPS-driven viral activities, while discussing challenges and exciting research routes in this rapidly changing area. The double nature of LLP, which combines both viral replication and antiviral reactions from the host, creates a complex landscape. This complexity sheds light on the fact that any medical approach to the purpose of LLP must be highly specialized to prevent negative effects on significant host functions, which is a significant obstacle to the development of future medicines.
2. Molecular Principles of LLPS in the Viral Context
The formation of viral condensates is underpinned by biophysical principles analogous to those governing the assembly of cellular MLOs [
9]. Central to this process are multivalent, often weak and transient, interactions between biomacromolecules, predominantly viral proteins and nucleic acids [
10]. Several molecular features of viral components are particularly conducive to driving these interactions. Many viral proteins that are key players in LLPS, such as nucleoproteins and phosphoproteins from various viral families, are enriched in Intrinsically Disordered Regions (IDRs) or Low Complexity Regions (LCRs) [
11]. These regions lack a fixed three-dimensional structure, endowing them with conformational flexibility and exposing multiple sites for weak interactions, including electrostatic forces, hydrophobic interactions, hydrogen bonds, and aromatic interactions like π−π stacking and cation-π interactions [
4]. The multivalency arising from multiple binding domains (e.g., RNA-binding domains, oligomerization domains) or repeated motifs on viral scaffold proteins, and their corresponding interaction partners (clients, including viral genomes), is another critical determinant [
1,
12]. This increased number of potential interaction sites enhances the avidity of molecular associations, promoting the demixing necessary for phase separation. The prevalence of IDRs in viral proteins that drive LLPS is unlikely to be coincidental; it may represent an evolutionarily conserved strategy allowing viruses to efficiently create versatile, multivalent interaction hubs with minimal genetic coding, thereby facilitating functional plasticity and rapid adaptation of interaction networks [
13,
14].
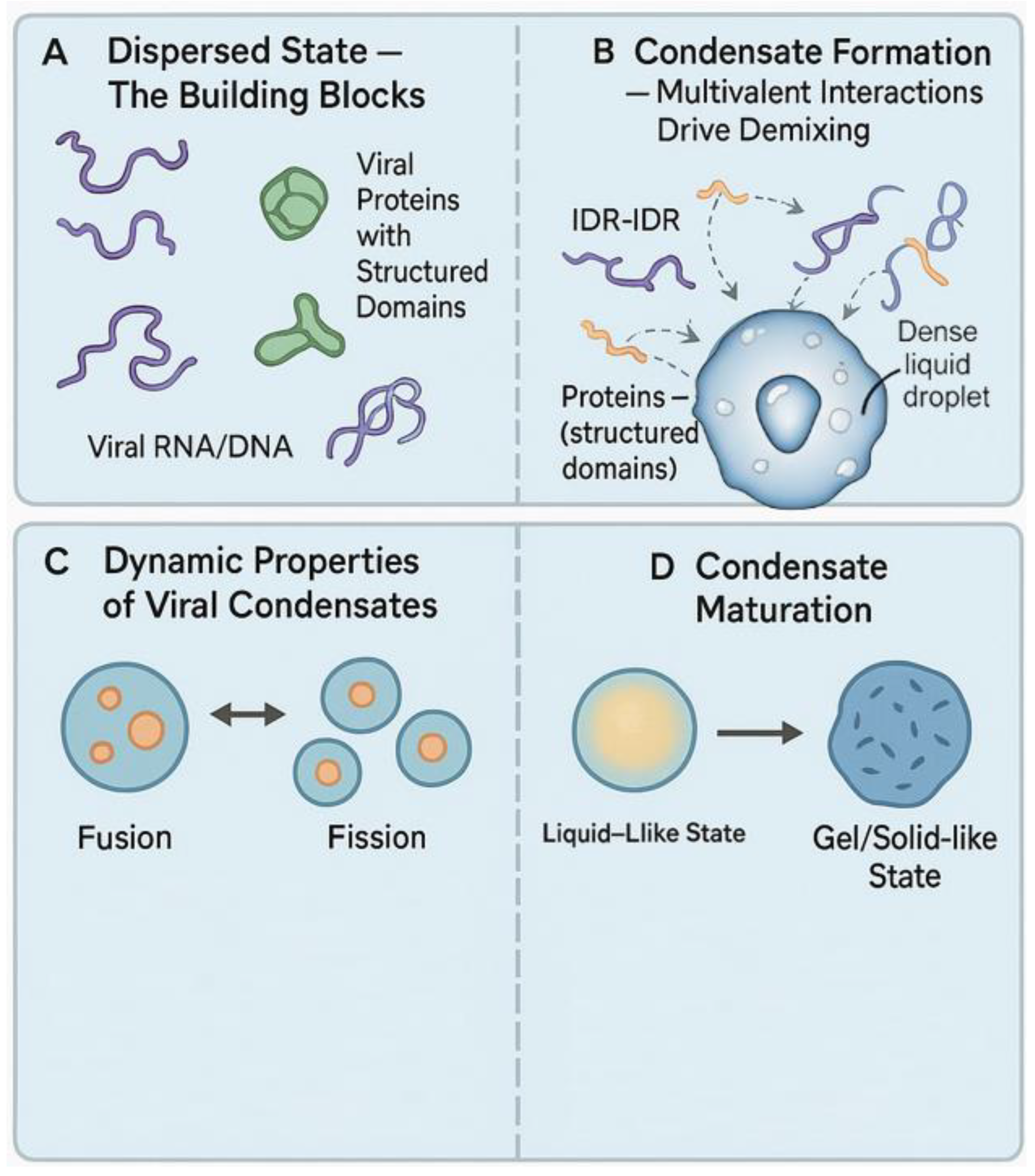
Figure 1.
Molecular Principles and Dynamics of Viral Liquid-Liquid Phase Separation (LLPS). (A) Dispersed State – The Building Blocks: In the initial dispersed state, viral proteins and nucleic acids exist individually in the cellular milieu. Viral proteins with Intrinsically Disordered Regions (IDRs) and structured domains are shown, along with viral RNA/DNA strands.(B) Condensate Formation – Multivalent Interactions Drive Demixing: Weak, multivalent interactions—such as IDR-IDR, protein–RNA, and structured domain–protein interactions—facilitate demixing of components into dense, spherical liquid droplets representing viral condensates.(C) Dynamic Properties of Viral Condensates: Viral condensates exhibit liquid-like behavior characterized by fusion (coalescence of smaller droplets into larger ones), fission (splitting into smaller droplets), and dynamic exchange of molecular components between the condensed and dilute phases. (D) Condensate Maturation: Over time, viral condensates can transition from a dynamic, liquid-like state to a more gel-like or solid-like state, reflecting maturation and potentially altered biochemical or biophysical properties.).
Figure 1.
Molecular Principles and Dynamics of Viral Liquid-Liquid Phase Separation (LLPS). (A) Dispersed State – The Building Blocks: In the initial dispersed state, viral proteins and nucleic acids exist individually in the cellular milieu. Viral proteins with Intrinsically Disordered Regions (IDRs) and structured domains are shown, along with viral RNA/DNA strands.(B) Condensate Formation – Multivalent Interactions Drive Demixing: Weak, multivalent interactions—such as IDR-IDR, protein–RNA, and structured domain–protein interactions—facilitate demixing of components into dense, spherical liquid droplets representing viral condensates.(C) Dynamic Properties of Viral Condensates: Viral condensates exhibit liquid-like behavior characterized by fusion (coalescence of smaller droplets into larger ones), fission (splitting into smaller droplets), and dynamic exchange of molecular components between the condensed and dilute phases. (D) Condensate Maturation: Over time, viral condensates can transition from a dynamic, liquid-like state to a more gel-like or solid-like state, reflecting maturation and potentially altered biochemical or biophysical properties.).
Viral nucleic acids, both RNA and DNA, frequently serve as crucial scaffolds or clients in the assembly of viral condensates [
1]. Their polymeric nature, potential for repetitive sequences, and capacity to form complex secondary and tertiary structures provide numerous multivalent binding sites for viral proteins [
4]. Viral RNA has been recognized as a powerful player in the world of liquid-liquid phase separation (LLPS), often boosting the phase separation tendencies of viral proteins. This has been particularly noted with the SARS-CoV-2 Nucleocapsid (N) protein and the HIV-1 Nucleocapsid (NC) protein [
15]. The concentration, specific sequences, and structural shapes of viral RNA can directly affect the properties, makeup, and stability of the resulting condensates [
16,
17]. This indicates that viral RNA isn't just a passive support structure; it's an active player in the architecture and regulation of these processes. The virus might use different RNA segments or specific RNA shapes to finely tune the LLPS process, customizing the condensates for various functions, like replication or packaging, at different points in the infection cycle [
18,
19].
Viral condensation usually shows functions such as dynamic, fluid. They are spherical, thanks to the surface stress that works to reduce surface stress, and they can merge to form large drops or divide into small people. A key feature of their floating nature is a rapid internal installation and exchange of components with the surrounding cellular environment [
11]. Researchers often detect these dynamic symptoms of using biophysical methods such as fluorescence recycling after photobleching (FRAP) [
20]. The physical condition of these viral condenses is not fixed; It can move somewhat more sticky, gel -like or even fasting as a liquid [
21]. These phase changes can be triggered by various factors, such as protein or RNA concentrations, specific post-transplation modifications (PTMS), or how long the transition lasts [
6,
22]. These maturation processes are often carefully regulated and can significantly affect the functionality of the virus. The formation and properties of viral condensate are affected by many elements, including their components, temperatures, pH, ionic power and concentration of PTM -Viral protein. For example, measles virus (MEV) phosphoprotein (P) or SARS-COV-2 N Protein phosplain has been shown to affect the properties of their liquid-elastic phase separation (LLPS) behavior and resulting condensate [
5,
6]. The dynamic physical properties of viral condensation, especially regulated fluid and gel/solid conditions, can serve as an important control mechanism. Early liquid condition can help with the recruitment of components and light enzymatic activities (such as viral genome replica), while later more fixed conditions can protect viral components, improve packaging efficiency or help develop immune response. Reducing these infections and potentially manipulation is an important area for developing new medical strategies.
3. LLPS Across the Viral Replication Cycle: New Mechanistic Insights
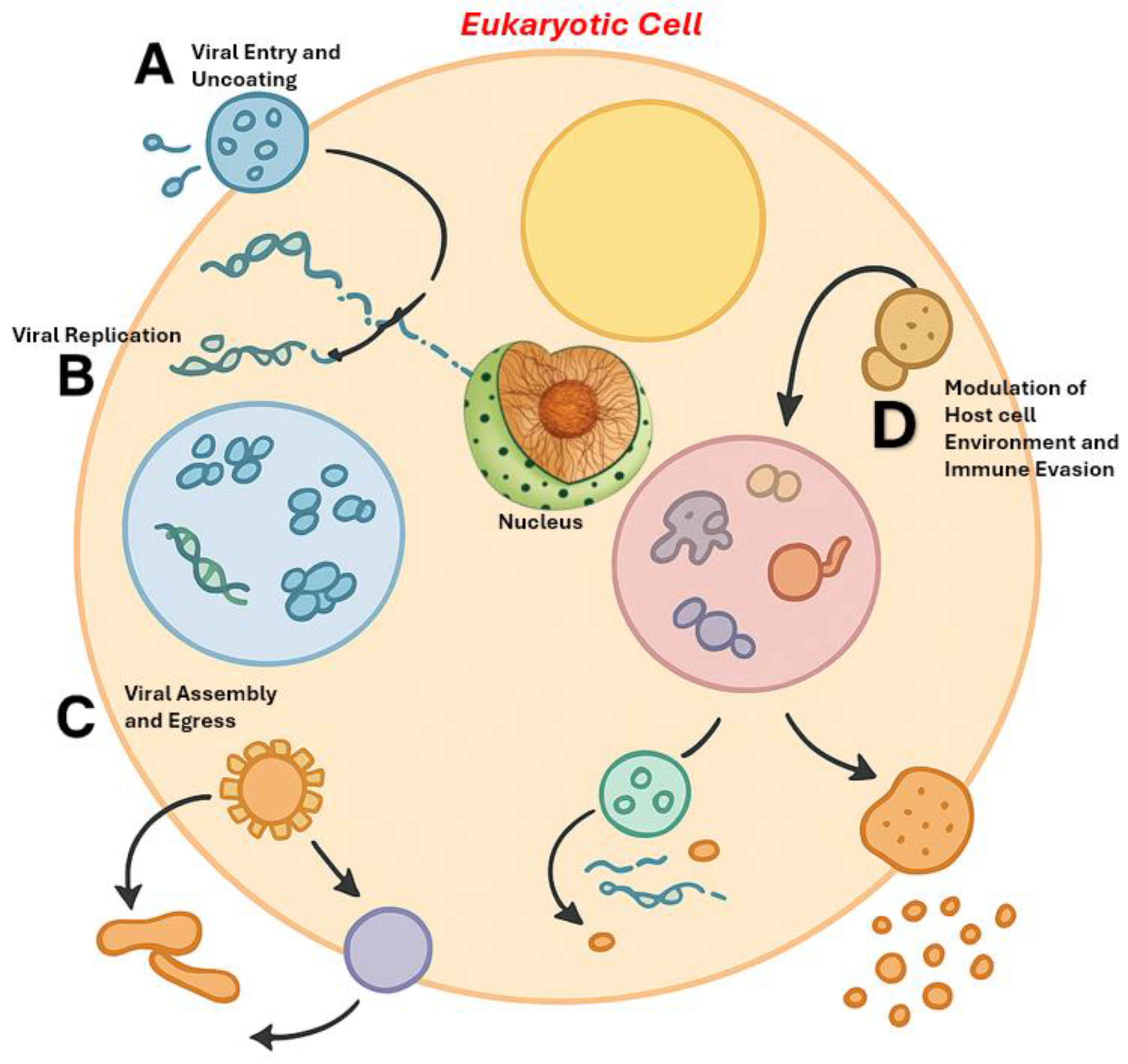
The principles of LLPS prove instrumental in understanding various stages of viral replication, from the initial entry and uncoating events to the final assembly and egress of new virions. Viruses across different families have evolved to harness LLPS, creating specialized compartments that optimize specific steps of their life cycle. The diversity of viral proteins and viral families that employ LLPS for similar functional outcomes, such as creating replication sites or packaging genomes, points towards convergent evolution, where distinct viral lineages have independently "discovered" and utilized LLPS as a highly effective organizing principle.
3.1. Viral Entry and Uncoating
Emerging evidence suggests that LLPS concepts are relevant even at the earliest stages of viral infection, specifically during the disassembly of incoming viral particles and the release of their genetic material into the host cell. For Influenza A Virus (IAV), host factors known to regulate cellular LLPS, such as Transportin-1 (TNPO1), are reportedly hijacked to facilitate the uncoating process. TNPO1 is thought to promote the decondensation of IAV viral ribonucleoprotein (vRNP) complexes, implying that the incoming viral core itself may possess condensate-like characteristics that require dissolution for successful infection [
23]. A similar role for TNPO1 has been implicated in HIV-1 uncoating, suggesting a common strategy where viral capsids or cores might leverage host cell condensate regulators to disassemble. The HIV-1 Gag polyprotein, particularly its nucleocapsid (NC) domain, is known to undergo LLPS [
6,
23]. While often studied in the context of assembly, this intrinsic property could also define the state of Gag within the mature virion and influence its uncoating upon entry into a new cell.
3.2. Genome Replication and Transcription: The Viral Replication Compartment (VRC) as a Biomolecular Condensate
A defining feature of many viral infections is the formation of VRCs, often referred to as viral factories or inclusion bodies. These structures are now widely recognized as MLOs formed through LLPS [
6]. By concentrating viral enzymes, genomic templates, and co-opted host factors, VRCs create a privileged and optimized microenvironment for viral nucleic acid synthesis. These compartments can also serve to shield viral components from host antiviral defense mechanisms.
3.2.1. RNA Viruses
3.2.1.1. Paramyxoviruses
Viruses like Measles Virus (MeV) and Respiratory Syncytial Virus (RSV) form prominent cytoplasmic inclusion bodies (IBs) that are the primary sites of viral RNA replication [
24]. For MeV, these IBs are driven by the multivalent interactions of the viral nucleoprotein (N) and phosphoprotein (P), with their C-terminal intrinsically disordered regions (N-IDR) and the P protein's XD domain (P-XD) playing critical roles [
25,
26]. The assembly, size, and material properties of MeV IBs are further modulated by PTMs, such as the phosphorylation of P, and interactions with host factors like WDR5 and the dynein motor complex [
7,
27]. These IBs not only concentrate the viral replication machinery but can also mature from a dynamic liquid state to a more stable gel-like state over time and may sequester host immune factors [
6]. RSV similarly forms IBs through N and P protein LLPS, which also function in sequestering host antiviral molecules.
3.2.1.2. Coronaviruses
When it comes to SARS-COV-2, N protein plays an important role in liquid-to-foot step paration (LLP). It does interaction with viral RNA and different types of hosts, such as G3BP1, condensed [
28]. The presence of RNA increases the LLP for the N protein, and this process is nice by phosphorus [
29]. This regulation is considered to accommodate the properties of condensate for different tasks: a more fluid position for transcription and replication, and gel for effective genome packaging. These condensed viral replications driven by N protein are important, and hosts can also interfere with the formation of granules and help immunity [
6].
3.2.1.3. Other Negative-Sense RNA Viruses
Viruses such as rabies virus (rabv) and Ebolavirus (ebov) form separate inclusive bodies, such as negligent bodies for rabv, which shows the same properties as liquid organelles[
30]. These are typically driven by the N and P proteins and serve as specialized sites for viral transcription and replication, while also contributing to immune evasion by sequestering key host factors [
31,
32].
3.2.1.4. Influenza A Virus (IAV)
Although IAV replicates its genome in the nucleus, distinct cytoplasmic viral inclusions with liquid-like characteristics are formed post-replication. These condensates involve viral vRNPs and the host protein Rab11a. Current understanding suggests these cytoplasmic condensates are primarily involved in facilitating vRNP trafficking, genome segment assembly, and packaging, rather than de novo RNA synthesis [
6].
3.2.2. DNA Viruses
3.2.2.1. Herpesviruses
Members of this family, including Herpes Simplex Virus-1 (HSV-1), Human Cytomegalovirus (HCMV), and Kaposi's Sarcoma-associated Herpesvirus (KSHV), utilize LLPS to establish nuclear compartments for viral DNA replication [
33,
34]. For instance, the KSHV Latency-Associated Nuclear Antigen (LANA) forms LANA-associated nuclear bodies (LANA-NBs) through LLPS, which are important for maintaining viral latency [
35,
36,
37]. Viral proteins such as HCMV UL112-113 and HSV-1 ICP4 can induce LLPS to form VRCs that recruit the viral DNA polymerase and essential accessory factors, creating optimized sites for genome amplification [
38]. Furthermore, Murid Herpesvirus 68 (MHV-68) has been shown to form cytoplasmic virion assembly compartments (VACs) that exhibit liquid-like properties, driven by the viral tegument protein ORF52 [
8].
A key feature shared by many RNA viruses is the crucial role of viral nucleoproteins (N or NC) and often phosphoproteins (P) in driving liquid-liquid phase separation (LLPS) for replication and assembly. These proteins usually have intrinsically disordered regions (IDRs) and multiple binding sites for RNA and other proteins, which makes them perfect multivalent scaffolds for forming condensates. The interaction between N/NC and RNA seems to be a fundamental building block for creating viral structures based on LLPS.
3.3. Viral Assembly and Egress
LLPS is becoming more and more acknowledged as a crucial organizing principle for gathering viral structural components, which in turn helps streamline the assembly of new virions.
3.3.1. Retroviruses
In HIV-1, the Gag polyprotein, particularly its NC domain, plays a vital role in assembling the virus. This process involves the careful packaging of the dimeric viral genomic RNA (gRNA) and the clustering of Gag proteins at the assembly sites, which are usually found at the host cell's plasma membrane [
39,
40]. Research has shown that the HIV-1 NC protein can undergo liquid-liquid phase separation (LLPS), a process that relies on its zinc finger (ZnF) domains and is affected by RNA and various cytosolic factors [
23]. This phase separation driven by NC is thought to be crucial for concentrating Gag and gRNA, which helps in positioning the gRNA, directing it to assembly sites, and ensuring it gets efficiently packaged into new virions. Disruption of NC LLPS, for example, through Zn
$ chelation or mutagenesis of the ZnF domains, leads to impaired Gag expression, reduced virus release, and mislocalization of the vRNA [
41]. The plasma membrane itself can act as a two-dimensional scaffold, promoting the condensation of Gag. As mentioned earlier, the cytoplasmic condensates containing vRNPs and Rab11a are believed to function as sites where vRNPs are concentrated for the assembly of the segmented viral genome and for their subsequent transport to the plasma membrane for budding and egress [
6].
The material state of these viral condensates, whether they remain highly liquid or transition to more gel-like or even solid states, appears to be functionally tuned for specific stages of the viral lifecycle. For example, highly dynamic, liquid VRCs might be optimal for the rapid exchange of components needed during active replication, whereas more stable, gel-like states could be advantageous for protecting viral genomes during packaging or for sequestering components [
6,
42]. This implies a sophisticated level of viral control over the biophysical properties of these condensates.
3.4. Modulation of Host Cell Environment: Immune Evasion and Stress Response Interference
Viruses strategically employ LLPS not only to build their functional compartments but also to manipulate host MLOs and innate immune signaling pathways, frequently as a means of evading antiviral responses.
3.4.1. Interference with Stress Granules (SGs)
SGs are dynamic MLOs that form in host cells in response to various stresses, including viral infection. They can sequester cellular and viral mRNAs and proteins, often exerting antiviral effects. Consequently, many viruses have evolved sophisticated mechanisms to prevent SG formation or to co-opt SG components for their benefit. For example, the SARS-CoV-2 N protein can sequester the core SG protein G3BP1, thereby suppressing SG assembly and dampening innate immune responses [
6]. The main protease of SARS-CoV-2, known as nsp5, has been shown to interact with G3BP1, which inhibits its liquid-liquid phase separation (LLPS) and, consequently, the formation of stress granules (SG). In a similar vein, the VP35 protein from the Ebola virus can interfere with the aggregation of SG proteins, while the LLPS driven by HIV-1 NC has been associated with the virus's ability to prevent SG assembly [
41].
3.4.2. Sequestration of Immune Factors
Viral condensates, like VRCs or IBs, can serve as molecular sinks, trapping important host antiviral signaling molecules and thus suppressing or reducing the host's immune responses [
43]. For example, the RSV N protein has the ability to sequester MAVS, MDA5, and the NF-κB subunit p65 within viral IBs. Similarly, the RABV P protein is known to capture STAT proteins, which inhibits JAK-STAT signaling. Additionally, the KSHV protein ORF52 can create condensates with cytoplasmic dsDNA, which competitively hinders DNA-induced phase separation and the activation of the cGAS-STING pathway [
44].
3.4.3. Hijacking Host Factors for Viral Benefit
Beyond simply sequestering factors, viruses can also recruit specific host proteins into their condensates to directly aid viral replication or to block interferon (IFN) responses. Examples include RABV recruiting Hsp70 into Negri bodies and EBOV NP recruiting CAD into its IBs. The SARS-CoV-2 N protein has been shown to recruit kinases like TAK1 and IKK into its condensates, leading to NF-κB hyperactivation, which can have complex roles in pathogenesis [
6].
The viral co-option of host MLO components (like G3BP1 into viral condensates) or the direct disruption of host MLOs (like SGs) represents more than just competition for resources. It signifies a strategic "re-wiring" of the host cell's LLPS landscape by the virus. This manipulation could have broader, currently underappreciated consequences for overall cellular homeostasis and signaling pathways, extending beyond direct immune evasion.
Table 1.
Key Viral Proteins and Systems Driving LLPS in Viral Replication.
Table 1.
Key Viral Proteins and Systems Driving LLPS in Viral Replication.
| Virus Family / Specific Virus |
Key Viral LLPS Driver(s) |
Critical Domains/Motifs |
Key Interacting Partners (Viral/Host) |
Resulting Condensate / Cellular Process |
Key Regulatory Features |
|
Paramyxoviridae / Measles Virus (MeV) |
N and P proteins |
P-XD (C-term domain of P), N-IDR (C-term intrinsically disordered region of N) [27] |
Viral RNA, host WDR5, host dynein[27] |
Inclusion Bodies (IBs) / Viral Replication Factories, Immune Evasion[6] |
P protein phosphorylation, Liquid-to-gel transition over time [27] |
|
Coronaviridae / SARS-CoV-2[6] |
Nucleocapsid (N) protein |
N-IDRs, RNA-binding domains, Oligomerization domain |
Viral RNA, host G3BP1, host kinases (e.g., SRPK1, GSK3) |
N-RNA condensates / Replication, Transcription, Genome Packaging, SG modulation |
N protein phosphorylation (regulates liquid vs. gel-like state), RNA concentration |
|
Retroviridae / HIV-1 |
Gag polyprotein (specifically Nucleocapsid - NC domain) |
NC Zinc Fingers (ZnF), IDRs in Gag [41] |
Viral genomic RNA (gRNA), host factors (e.g., IP6) [23] |
Gag assembly sites, NC-RNA condensates / RNA packaging, Virion Assembly, SG modulation [23] |
Zn²⁺- [6,27] dependence of NC LLPS, RNA binding[41] |
|
Orthomyxoviridae / Influenza A Virus (IAV) [23] |
Viral Ribonucleoproteins (vRNPs), Nucleoprotein (NP) |
NP oligomerization domains, RNA binding sites[23] |
Host Rab11a, other vRNPs |
Cytoplasmic vRNP inclusions / Genome segment assembly, Trafficking to budding sites[6] 5
|
Interaction with Rab11a, potential modulation by NP PTMs |
|
Herpesviridae / HSV-1, KSHV, HCMV [6] |
ICP4 (HSV-1), LANA (KSHV), UL112-113 (HCMV), ORF52 (MHV-68) |
Various, often IDRs or multivalent domains |
Viral DNA, host replication/transcription factors, viral polymerase |
Nuclear VRCs (HSV-1, HCMV), LANA-NBs (KSHV), Cytoplasmic VACs (MHV-68) / DNA Replication, Latency, Assembly |
Concentration of viral proteins, interaction with viral DNA |
|
Rhabdoviridae / Rabies Virus (RABV) [6,27] |
N and P proteins |
IDRs in N and P, P protein oligomerization domain |
Viral RNA, host STAT proteins, host Hsp70 5
|
Negri Bodies (NBs) / Viral Replication Factories, Immune Evasion |
Protein concentration, interactions between N, P, and L proteins |
4. LLPS: Reshaping Paradigms in Virology
The integration of LLPS principles into virology is not merely an addition of a new mechanism; it is fundamentally reshaping how viral replication, virus-host interactions, and pathogenesis are conceptualized. The traditional view of viral factories, for instance, often depicted them as somewhat static, membrane-associated structures. LLPS provides a more dynamic and flexible framework, re-envisioning these sites as self-assembling MLOs capable of rapid component exchange and responsiveness to cellular conditions. This paradigm explains the membraneless nature of many viral replication compartments, their ability to selectively concentrate necessary viral and host factors, and their capacity to create optimized microenvironments for viral processes without the universal requirement for de novo membrane biogenesis for each distinct compartment [
45]. This shift in understanding offers a more nuanced view of how viruses efficiently organize their replication machinery.
LLPS also introduces new dimensions to the understanding of virus-host interactions. Viruses are now seen not just as entities that interact with individual host proteins in isolation, but as agents that engage with and actively manipulate the host's broader LLPS network and its associated MLOs. This manipulation includes the co-option of host proteins that are integral components of, or regulators for, cellular condensates (e.g., G3BP1 by SARS-CoV-2, Rab11a by IAV, WDR5 by MeV) [
6]. It also involves the disruption of host MLOs, such as SGs, to counteract antiviral responses, or the exploitation of host factors that modulate LLPS processes, like cellular kinases that enact PTMs on viral proteins. This perspective elevates the study of virus-host interactions from a focus on one-to-one molecular engagements to a more systems-level appreciation of how viruses remodel the fundamental organizational principles of the cell. The recognition of LLPS as a key player means that previous observations of viral protein localizations or amorphous "viral inclusions" seen in microscopy studies, whose biophysical nature was often unclear, can now be re-evaluated [
46]. These structures may indeed be functional biomolecular condensates, and re-investigating them with LLPS-specific biochemical and biophysical tools could unlock significant new knowledge from historical data.
Furthermore, the LLPS framework has significant implications for understanding viral pathogenesis, tropism, and evolution. The efficiency with which a virus can establish and utilize LLPS-driven compartments could be a critical determinant of its replication fitness and, consequently, its pathogenic potential. Variations in the host cell's intrinsic LLPS machinery or the differential availability of specific host factors required for the formation or function of viral condensates might contribute to tissue or species tropism. The "biophysical compatibility" of viral proteins with the host cell's phase separation landscape may thus represent a new dimension of viral fitness; viruses whose proteins are more adept at forming functional condensates within a particular host environment may exhibit enhanced replication. Moreover, the inherent flexibility of interactions driven by IDRs, which are common in viral LLPS-driving proteins, could facilitate viral evolution and adaptation. Mutations within IDRs can readily modulate interaction networks and condensate properties, potentially allowing viruses to rapidly adapt to new host environments or to evade host immune pressures [
47]. LLPS, therefore, provides a unifying framework to comprehend diverse viral strategies, such as the formation of replication sites, packaging centers, and immune evasion hubs, which previously might have appeared disconnected. These can now all be viewed as manifestations of a common underlying principle: the creation of specialized, concentrated environments through regulated phase separation.
5. Therapeutic Opportunities Targeting Viral LLPS
The critical and multifaceted roles of LLPS-driven condensates in viral replication, assembly, and immune evasion highlight these structures and their underlying formation mechanisms as attractive and novel targets for antiviral interventions [
6]. The prospect of disrupting or aberrantly modulating these viral condensates offers a new frontier for inhibiting viral propagation or restoring host defenses. The drug ability of LLPS is distinct from traditional enzyme inhibition or receptor antagonism; it involves modulating a complex
process governed by a network of often weak, multivalent interactions. This necessitates innovative screening strategies, such as phenotypic screens that monitor condensate properties directly, and novel medicinal chemistry approaches [
48].
Several strategies for modulating viral condensates are being explored:
5.1. Dissolution or Prevention of Condensate Formation
This can be approached by targeting the multivalent interactions that drive LLPS. Small molecules, peptides, or even antibodies could be designed to block key interaction sites on viral or co-opted host scaffold proteins, or on viral nucleic acids. Another avenue is to alter the properties of scaffold proteins, for instance, by targeting their IDRs/LCRs with compounds that disrupt their necessary conformational ensembles or by using drugs that mimic or block critical PTMs. General LLPS inhibitors, such as 1,6-hexanediol or propylene glycol, have been shown to dissolve condensates in experimental settings, but their lack of specificity and potential cytotoxicity are major limitations for therapeutic use.
5.2. Aberrant Stabilization or Alteration of Material Properties
Instead of dissolving condensates, an alternative strategy is to induce their premature or irreversible gelation or solidification. This could trap essential components or block the dynamic exchange necessary for function, effectively rendering the condensates non-productive [
45]. The compound nucleozin, which targets the IAV NP, acts by "gluing" NP molecules together, leading to more rigid and less dynamic viral inclusions, thereby inhibiting viral replication. Conversely, preventing necessary liquid-to-gel transitions, which might be required for processes like viral genome packaging, could also be disruptive [
49].
5.3. Targeting Specific Components Within Viral Condensates
This involves developing inhibitors against key viral scaffold proteins that are essential for condensate formation or function, such as the SARS-CoV-2 N protein, HIV-1 NC, paramysovirus N/P proteins, or IAV NP. Viral nucleic acid structures that play a role in LLPS could also be targeted. A particularly promising approach involves targeting co-opted host factors that are crucial for the formation or function of viral condensates (e.g., G3BP1, Rab11a, or specific chaperones) [
6]. A refined version of this strategy would be to target the specific
viral interaction surface on these host factors, or the host-factor-binding surface on the viral protein. This could offer enhanced specificity, minimizing disruption to the host factor's normal cellular roles and thereby reducing potential side effects.
5.5. Modulating Post-Translational Modifications (PTMs)
Since PTMs like phosphorylation can critically regulate the LLPS behavior of viral proteins and the properties of the resulting condensates, targeting the host or viral enzymes (kinases, phosphatases) responsible for these modifications is a viable strategy. For example, activators of the host kinase SRPK1, which phosphorylates the SARS-CoV-2 N protein, have been proposed as potential antivirals because N phosphorylation can alter its LLPS behavior and interaction with RNA [
50].
Preclinical progress has been made for several viruses:
SARS-CoV-2 N protein: Strategies include the use of small molecules like gallic catechin gallate (GCG) from green tea, which can inhibit N protein LLPS. Interference peptides (NIP I-V) have been designed to disrupt N protein interactions essential for LLPS. Furthermore, modulating N protein phosphorylation via kinase activators (e.g., SRPK1) or inhibitors is an active area of investigation, as mutations in N protein IDRs are known to affect LLPS propensity [
6].
HIV-1 NC protein: The ZnF domains of NC are critical for its LLPS activity. Zn chelators or mutations in the ZnF domains disrupt NC-LLPS and significantly impair viral replication, identifying the ZnF motif as a druggable target for pan-retroviral therapies [
41].
Influenza A Virus NP: As mentioned, nucleozin induces NP oligomerization, hardens viral inclusions, and shows antiviral activity in preclinical models [
49]. Other small molecules targeting NP's nuclear localization signals or critical salt bridges also inhibit viral replication [
51].
Paramyxoviruses (e.g., MeV): Mutations in the MeV P protein that affect its LLPS capability have been shown to reduce viral transcript levels [
23]. Inhibitors of CK2, a kinase that phosphorylates P, can decrease the size of MeV IBs [
27].
Many current examples of LLPS modulators are either general compounds with known cytotoxicity (like 1,6-hexanediol) or molecules that were repurposed after their effects on LLPS were discovered. There is a significant need for the de novo design and discovery of highly specific modulators of viral LLPS. Compounds like nucleozin and GCG represent promising starting points, but a more systematic approach to identify and develop such agents is crucial for translating the therapeutic potential of targeting viral LLPS into clinical reality.
Table 2.
Therapeutic Strategies Targeting LLPS in Viral Infections.
Table 2.
Therapeutic Strategies Targeting LLPS in Viral Infections.
| Therapeutic Strategy |
Molecular Target Class |
Type of Modulator |
Specific Viral Example(s) & Protein(s) |
Potential Antiviral Effect |
Key Challenges / Current Status |
| Disrupt Condensate Formation/Integrity [6,41] |
Viral IDRs/LCRs, Viral Oligomerization Domains, Viral-Host Protein Interfaces |
Small Molecule (dissolver, binder), Peptide (interface blocker, IDR binder), Antibody/Nanobody |
SARS-CoV-2 N (NIP I-V, GCG) ; HIV-1 NC (Zn$^{2+}$ chelators) |
Inhibit VRC formation, Block RNA packaging, Disrupt viral assembly |
Specificity, Delivery, Off-target effects, Preclinical/Early research |
| Modulate Material Properties[49] |
Viral Scaffold Proteins (e.g., NP, N) |
Small Molecule (e.g., "glues", stabilizers) |
IAV NP (Nucleozin) ; SARS-CoV-2 N |
Impair condensate dynamics, Prevent necessary phase transitions (e.g., liquid-to-gel) |
Achieving precise modulation, Understanding functional consequence of altered properties |
| Inhibit Scaffold Protein Function[27,51] |
Key viral proteins driving LLPS (e.g., N, NC, P) |
Small Molecule Inhibitor, Peptide, Nucleic Acid Aptamer |
IAV NP (NLS-binding compounds) ; MeV P |
Reduce viral replication/transcription, Inhibit viral factory function |
Identifying druggable pockets on IDP-rich scaffolds, Specificity |
| Inhibit Client Recruitment [6] |
Interaction sites between viral scaffolds and essential viral/host clients |
Small Molecule, Peptide |
SARS-CoV-2 N and G3BP1 |
Prevent concentration of essential factors (e.g., polymerase, RNA) in VRCs, Restore host defenses |
High specificity is required to avoid disrupting host protein functions |
| Modulate PTMs Regulating LLPS[27] |
Cellular/Viral Enzymes (e.g., kinases, phosphatases) modifying viral LLPS proteins |
Small Molecule Enzyme Inhibitor/Activator |
SARS-CoV-2 N (SRPK1 activators) ; MeV P (CK2 inhibitors) |
Alter condensate properties, Inhibit/promote specific viral processes dependent on PTM state |
Specificity for viral targets, Understanding complex PTM codes, Potential host cell toxicity |
6. Challenges and Future Perspectives
While the study of LLPS in virology has opened exciting new frontiers, significant challenges remain, both in fundamental understanding and in translating these discoveries into effective antiviral therapies. Overcoming these hurdles will require continued interdisciplinary collaboration and technological innovation.
One set of challenges pertains to the technical and methodological aspects of studying viral LLPS. Characterizing the dynamic, often transient, and heterogeneous nature of viral condensates in situ within infected cells and in vivo in animal models remains a considerable task. Advanced microscopy techniques like FRAP, FLIP, FCS, SPT, and cryo-electron tomography (cryo-ET) are providing valuable insights, but their application in the context of viral infection requires sophisticated instrumentation and expertise [
45]. A persistent concern is the need to rigorously distinguish bona fide, functionally relevant LLPS events from non-specific protein aggregation or artifacts, particularly when relying on overexpression systems [
11]. Furthermore, there is a pressing need for more refined tools to probe the precise biophysical properties of MLOs within the complex environment of a living cell and to comprehensively define their molecular composition and dynamic changes during infection [
46].
In the realm of therapeutic development, achieving specificity for viral condensates over essential host MLOs is a paramount challenge. Viruses often utilize the same "molecular language"—such as IDRs, multivalency, and RNA-binding motifs as the host cell to drive phase separation. This shared mechanistic basis means that broad-spectrum LLPS disruptors (e.g., 1,6-hexanediol) are likely to cause significant off-target effects and cytotoxicity [
6]. Therefore, successful antiviral strategies will likely need to target unique viral components within the condensates, highly specific virus-host interaction interfaces that are not replicated in host MLOs, or unique regulatory features of viral LLPS. The delivery of LLPS-modulating agents, especially larger molecules like peptides or nucleic acids, to the correct subcellular compartments where viral condensates form also poses a considerable hurdle. The "double-edged sword" nature of LLPS, where it plays roles in both viral processes and host immunity, further complicates therapeutic design, demanding interventions that can selectively inhibit viral exploitation of LLPS without compromising beneficial host responses. The dynamic nature and potential for redundancy within LLPS systems, where multiple weak interactions contribute to condensate stability, might also render single-molecule inhibitors less effective. This suggests that combination therapies targeting different components or aspects of viral condensate formation, stability, or maturation could prove more efficacious.
Numerous key questions remain unanswered, paving the way for future research. A deeper mechanistic understanding is required to elucidate the precise molecular grammars including specific amino acid sequences in IDRs, PTM codes, and critical RNA structural motifs that dictate the formation, specific composition, material properties, and regulation of distinct viral condensates. Identifying the full spectrum of host factors that are recruited into or modulate viral LLPS, and determining whether any host factors can specifically discriminate between viral and cellular LLPS, is crucial. How variations in viral LLPS efficiency or condensate properties, potentially arising from viral mutations, correlate with differences in viral replication efficacy, transmission dynamics, and disease severity across different viral strains or host species is an important area for investigation in viral pathogenesis. The functional consequences of the observed dynamic transitions in material properties (e.g., liquid-to-gel) of viral condensates and the mechanisms regulating these transitions need to be fully explored. There is also a significant need for the development of novel high-throughput screening platforms specifically designed to identify modulators of viral LLPS. These could include phenotypic screens based on imaging changes in condensate morphology or dynamics, or target-based screens focusing on multivalency or IDR interactions. A critical component of such screening platforms would be the development of "LLPS reporters" for specific viral condensates that not only indicate presence but also reflect the functional state of the condensate. Recent advancements in structural biology techniques, like cryo-electron tomography (cryo-ET) and integrative/computational modeling, are crucial for getting a clearer picture of the complex architecture of viral condensates. Additionally, delving into the intricate interactions between LLPS-driven viral compartments and traditional membrane-bound organelles, as well as membrane-associated viral replication organelles (VROs), will give us a more comprehensive understanding of how viruses hijack cellular organization [
45].
To wrap things up, diving into the study of LLPS in virology is an exciting journey that’s really changing how we see essential viral processes. It’s also opening up some thrilling new paths for antiviral treatments. Sure, there are significant hurdles to overcome, especially when it comes to ensuring therapeutic specificity and dealing with methodological challenges. However, the chance to target these unique viral microenvironments holds great promise for creating a new wave of antiviral medications. The ongoing collaboration across various fields, bringing together insights from virology, biophysics, cell biology, structural biology, and chemical biology will be crucial in unlocking the full potential of LLPS research as we continue the fight against viral diseases.
Author Contributions
Sanjida and M.I.H. conceptualized the study and prepared the original draft. S.R.A. and M.S.A. reviewed and edited the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors would like to thank all colleagues who contributed indirectly to this work.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhang: J.Z., S. Mehta, and J. Zhang, Liquid–liquid phase separation: a principal organizer of the cell’s biochemical activity architecture. Trends in pharmacological sciences, 2021. 42(10): p. 845-856.
- Zhao, Y.G.; Zhang, H. Phase Separation in Membrane Biology: The Interplay between Membrane-Bound Organelles and Membraneless Condensates. Dev. Cell 2020, 55, 30–44. [CrossRef]
- Lim, J.-Y.; Kim, Y.; Jeon, S.; Jeon, Y.; Kim, W.; Cha, B. Emerging regulatory mechanisms and functions of biomolecular condensates: implications for therapeutic targets. Signal Transduct. Target. Ther. 2025, 10, 1–42. [CrossRef]
- Gomes, E.; Shorter, J. The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294, 7115–7127. [CrossRef]
- Li, J.; Yang, F.; Zahid, A.; Yao, X.; Ismail, H.; Liu, X.; Dou, Z.; Liu, X. Mechanisms and regulation underlying membraneless organelle plasticity control. J. Mol. Cell Biol. 2021, 13, 239–258. [CrossRef]
- Hu, J.; Cui, J.; Jin, S.; Zhang, C.; Yang, S.; Wu, J.; Cai, S.; Guan, X.; Wu, Y.; Shen, W. Molecular mechanisms and cellular functions of liquid-liquid phase separation during antiviral immune responses. Front. Immunol. 2023, 14. [CrossRef]
- Parissi, V.; Li, H.; Ernst, C.; Greb-Markiewicz, B.; Kolonko-Adamska, M.; Man, J.; Ng, B.W.-L. Phase separation in viral infections. Trends Microbiol. 2022, 30, 1217–1231. [CrossRef]
- Ahn, J.-H.; Chung, W.-C.; Song, M.J. Liquid–liquid phase separation drives herpesvirus assembly in the cytoplasm. J. Cell Biol. 2022, 222. [CrossRef]
- Lagaudrière-Gesbert, C.; Gaudin, Y.; Albertini, A.; Glon, D.; Léonardon, B.; Guillemot, A. Biomolecular condensates with liquid properties formed during viral infections. Microbes Infect. 2024, 26, 105402. [CrossRef]
- Li, Z.; Zhang, X.; Zheng, R.; Ma, J. Liquid-liquid Phase Separation in Viral Function. J. Mol. Biol. 2023, 435, 167955. [CrossRef]
- Peng, P.-H., K.-W. Hsu, and K.-J. Wu, Liquid-liquid phase separation (LLPS) in cellular physiology and tumor biology. American journal of cancer research, 2021. 11(8): p. 3766.
- Xue, B.; Habchi, J.; Blocquel, D.; Uversky, A.V.; Uversky, V.N.; Kurgan, L.; Longhi, S. Structural Disorder in Viral Proteins. Chem. Rev. 2014, 114, 6880–6911. [CrossRef]
- Borcherds, W., et al., How do intrinsically disordered protein regions encode a driving force for liquid–liquid phase separation? Current opinion in structural biology, 2021. 67: p. 41-50. [CrossRef]
- Rosen, M.K.; Lin, Y.; Currie, S.L. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 2017, 292, 19110–19120. [CrossRef]
- Lee, K.S. SARS-CoV-2 immunogenicity models inform novel strategies that can improve mRNA vaccines. 2024. [CrossRef]
- Ke, W., Functional nucleic acid nanoparticles and their delivery. 2020, The University of North Carolina at Charlotte.
- Bergeron, H.C., Blocking the RSV G Protein CX3C Chemokine Motif Protects Against RSV Disease. 2023, University of Georgia.
- Grandori, R.; Brocca, S.; Longhi, S.; Uversky, V. Liquid–Liquid Phase Separation by Intrinsically Disordered Protein Regions of Viruses: Roles in Viral Life Cycle and Control of Virus–Host Interactions. Int. J. Mol. Sci. 2020, 21, 9045. [CrossRef]
- Nosella, M., The Effects Of Post-Translational Modification On Intrinsically Disordered Protein Phase Separation And Nucleosome Dynamics. 2024, University of Toronto (Canada).
- Geiger, F., et al., Rotavirus replication factories are complex ribonucleoprotein condensates. Biorxiv, 2020: p. 2020.12. 18.423429.
- Shanker, S.; Estes, M.K.; Ayyar, B.V.; Pollet, J.; Kaur, G.; Prasad, B.V.V.; Stossi, F.; Crawford, S.E.; Kaundal, S.; Anish, R. RNA-dependent RNA polymerase of predominant human norovirus forms liquid-liquid phase condensates as viral replication factories. Sci. Adv. 2024, 10, eadp9333. [CrossRef]
- Sanfeliu-Cerdán, N.; Krieg, M. The mechanobiology of biomolecular condensates. Biophys. Rev. 2025, 6, 011310. [CrossRef]
- Etibor, T.A.; Yamauchi, Y.; Amorim, M.J. Liquid Biomolecular Condensates and Viral Lifecycles: Review and Perspectives. Viruses 2021, 13, 366. [CrossRef]
- Su, J.M.; Wilson, M.Z.; Samuel, C.E.; Ma, D. Formation and Function of Liquid-Like Viral Factories in Negative-Sense Single-Stranded RNA Virus Infections. Viruses 2021, 13, 126. [CrossRef]
- Moncman, C.L.; Leung, D.W.; Creamer, T.P.; El Najjar, F.; Wu, C.; Cifuentes-Munoz, N.; Edmonds, K.; Boggs, K.B.; Ossandón, C.; Roe, M.; et al. Human Metapneumovirus Phosphoprotein Independently Drives Phase Separation and Recruits Nucleoprotein to Liquid-Like Bodies. mBio 2022, 13, e0109922. [CrossRef]
- Gerresheim, G.K.; Dolnik, O.; Biedenkopf, N. New Perspectives on the Biogenesis of Viral Inclusion Bodies in Negative-Sense RNA Virus Infections. Cells 2021, 10, 1460. [CrossRef]
- Zhou, Y.; Su, J.M.; Samuel, C.E.; Ma, D. Measles Virus Forms Inclusion Bodies with Properties of Liquid Organelles. J. Virol. 2019, 93, e00948-19. [CrossRef]
- Yang, Z., et al., Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication. BioRxiv, 2023.
- Cascarina, S.M. and E.D. Ross, Phase separation by the SARS-CoV-2 nucleocapsid protein: Consensus and open questions. Journal of Biological Chemistry, 2022. 298(3): p. 101677. [CrossRef]
- Nikolic, J.; Le Bars, R.; Lama, Z.; Scrima, N.; Lagaudrière-Gesbert, C.; Gaudin, Y.; Blondel, D. Negri bodies are viral factories with properties of liquid organelles. Nat. Commun. 2017, 8, 58. [CrossRef]
- Nevers, Q.; Albertini, A.A.; Lagaudrière-Gesbert, C.; Gaudin, Y. Negri bodies and other virus membrane-less replication compartments. Biochim. Biophys. Acta 2020, 1867, 118831. [CrossRef]
- Darling, A.L.; Oldfield, C.J.; Liu, Y.; Uversky, V.N. Intrinsically Disordered Proteome of Human Membrane-Less Organelles. Proteomics 2017, 18, e1700193. [CrossRef]
- Bosse, J.B.; Brune, W.; Caragliano, E. Herpesvirus Replication Compartments: Dynamic Biomolecular Condensates?. Viruses 2022, 14, 960. [CrossRef]
- RANGE, H. and I. DOSE, PATHOGEN SAFETY DATA SHEET-INFECTIOUS SUBSTANCES.
- Deng, Z.; De Leo, A.; Vladimirova, O.; Hayden, J.; Lieberman, P.M.; Wiedmer, A. Phase separation and DAXX redistribution contribute to LANA nuclear body and KSHV genome dynamics during latency and reactivation. PLOS Pathog. 2021, 17, e1009231. [CrossRef]
- Uppal, T.; Banerjee, S.; Sun, Z.; Verma, S.C.; Robertson, E.S. KSHV LANA—The Master Regulator of KSHV Latency. Viruses 2014, 6, 4961–4998. [CrossRef]
- Li, S.; Raina, K.; Kazemian, M.; Szymula, A.; George, A.; Ramachandran, A.; Van Sciver, N.; Wang, M.; Zhao, B.; Tumuluri, V.S.; et al. Kaposi’s sarcoma herpesvirus latency-associated nuclear antigen broadly regulates viral gene expression and is essential for lytic infection. PLOS Pathog. 2024, 20, e1011907. [CrossRef]
- Bourqui, L.; Anfossi, M.; Michaelsen, K.; Vogt, B.; Fraefel, C.; Georgi, F.; Seyffert, M.; Tobler, K.; Greber, U.F. The HSV-1 Transcription Factor ICP4 Confers Liquid-Like Properties to Viral Replication Compartments. Int. J. Mol. Sci. 2021, 22, 4447. [CrossRef]
- Hanson, H.M.; Willkomm, N.A.; Yang, H.; Mansky, L.M. Human Retrovirus Genomic RNA Packaging. Viruses 2022, 14, 1094. [CrossRef]
- Moog, C.; Boutant, E.; Réal, E.; Zeiger, M.; Mély, Y.; Anton, H.; Klingler, J. How HIV-1 Gag Manipulates Its Host Cell Proteins: A Focus on Interactors of the Nucleocapsid Domain. Viruses 2020, 12, 888. [CrossRef]
- Niu, M.; Monette, A.; Rao, S.; Chen, L.; Gorelick, R.J.; Mouland, A.J. Pan-retroviral Nucleocapsid-Mediated Phase Separation Regulates Genomic RNA Positioning and Trafficking. Cell Rep. 2020, 31, 107520–107520. [CrossRef]
- Charman, M.; Weitzman, M.D. Replication Compartments of DNA Viruses in the Nucleus: Location, Location, Location. Viruses 2020, 12, 151. [CrossRef]
- Van Royen, T., et al., How RSV proteins join forces to overcome the host innate immune response. Viruses, 2022. 14(2): p. 419. [CrossRef]
- Jobe, F.; Simpson, J.; Hawes, P.; Guzman, E.; Bailey, D. Respiratory Syncytial Virus Sequesters NF-κB Subunit p65 to Cytoplasmic Inclusion Bodies To Inhibit Innate Immune Signaling. J. Virol. 2020, 94, e01380-20. [CrossRef]
- Longhi, S.; Galloux, M. Unraveling Liquid–Liquid Phase Separation (LLPS) in Viral Infections to Understand and Treat Viral Diseases. Int. J. Mol. Sci. 2024, 25, 6981. [CrossRef]
- Bosse, J.B.; Brune, W. Viral dew: Phase separation and the formation of viral replication compartments. PLOS Pathog. 2023, 19, e1011145. [CrossRef]
- Nguyen, A., et al., Modulation of biophysical properties of nucleocapsid protein in the mutant spectrum of SARS-CoV-2. Elife, 2024. 13. [CrossRef]
- Mitrea, D.M.; Gomes, B.F.; Mittasch, M.; Klein, I.A.; Murcko, M.A. Modulating biomolecular condensates: a novel approach to drug discovery. Nat. Rev. Drug Discov. 2022, 21, 841–862. [CrossRef]
- Kong, H.; Amada, J.X.; Ng, B.W.-L.; Scheeff, S. Making it hard to replicate. eLife 2023, 12. [CrossRef]
- Sharma, B., et al., Post-translational modifications (PTMs), from a cancer perspective: an overview. Oncogen, 2019. 2(3): p. 12.
- Yang, F., et al., Discovery of a novel specific inhibitor targeting influenza A virus nucleoprotein with pleiotropic inhibitory effects on various steps of the viral life cycle. Journal of virology, 2021. 95(9): p. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).