1. Introduction
Coronary artery disease (CAD), also known as coronary heart disease or ischemic heart disease, is a chronic condition caused by the buildup of atherosclerotic plaques in the coronary arteries. This accumulation reduces blood flow to the heart muscle and can lead to serious complications such as angina pectoris, acute myocardial infarction, heart failure, or sudden cardiac death. Major risk factors include hypertension, dyslipidemia, smoking, diabetes, obesity, physical inactivity, advanced age, and a family history of cardiovascular disease. Despite advances in diagnosis and therapy, CAD remains one of the leading causes of mortality worldwide [
1].
To improve early detection and treatment planning, image processing has become a critical tool in the clinical management of CAD. Advanced imaging modalities such as coronary computed tomography angiography (CCTA), magnetic resonance imaging (MRI), intravascular ultrasound (IVUS), and optical coherence tomography (OCT) provide high-resolution visualizations of coronary anatomy and pathology. However, extracting clinically meaningful insights from these modalities requires robust image processing techniques capable of handling large volumes of complex data.
For example, CCTA is a non-invasive method widely used to visualize coronary arteries and assess atherosclerotic plaque burden and luminal stenosis. Image processing techniques such as segmentation, 3D reconstruction, and coronary tree extraction are essential for delineating vessel boundaries and identifying regions of interest. Automated or semi-automated segmentation algorithms, including region-growing, active contours, and machine learning-based models, have been developed to improve efficiency and reproducibility in clinical workflows [
2,
3].
Similarly, in IVUS and OCT, which offer microscopic-level resolution, image processing techniques play a key role in characterizing plaque composition and vessel wall morphology. Methods such as speckle noise reduction, edge detection, and texture analysis are employed to distinguish between fibrous, lipid-rich, and calcified plaques. In addition, elastography and deep learning-based techniques are increasingly being integrated to enhance tissue characterization and risk assessment [
4].
A central component of many of these imaging applications is segmentation, a foundational step in medical image analysis that is particularly crucial for delineating anatomical structures such as the coronary artery lumen, vessel wall, and atherosclerotic plaques. Accurate segmentation enables the quantitative assessment of vascular morphology, plaque burden, and luminal narrowing—metrics that are critical for guiding therapeutic decisions such as stent placement or coronary artery bypass grafting [
5]. Traditional segmentation techniques, including thresholding, region growing, edge detection, and active contour models, rely on predefined features and intensity gradients. Although useful in ideal conditions, they often lack robustness in noisy or complex coronary imaging data [
6], and their reliance on manual input introduces inter-operator variability [
7].
To overcome these limitations, deep learning (DL) has emerged as a transformative approach in image segmentation. Architectures like U-Net, introduced by Ronneberger et al. [
8], use encoder–decoder structures with skip connections to preserve spatial context while enabling detailed segmentation. Variants such as ResUNet, Attention U-Net, and 3D U-Net have further improved performance in cardiovascular imaging applications, showing high accuracy in segmenting coronary arteries, lumen borders, and plaques [
9] [
10,
11]. In modalities such as CCTA and IVUS, these networks have also supported the characterization of plaque tissue types [
12,
13], while in IVUS and OCT, they have enabled detailed analysis of vessel wall microstructure [
4].
Building on these architectural innovations, new DL training strategies (such as multi-task learning and weakly supervised learning) have improved model generalization across imaging modalities and patient cohorts [
14]. These advances reduce the dependence on large annotated datasets and enhance the applicability of DL segmentation in real-world settings. Additionally, DL-based radiomics workflows can extract high-dimensional image features from segmented regions, transforming anatomical structures into predictive biomarkers for personalized risk assessment, including the prediction of major adverse cardiac events (MACE) [
15,
16].
While segmentation remains a critical tool in CAD imaging, the need for faster, real-time analysis has catalyzed interest in object detection frameworks, particularly in dynamic or intraoperative contexts. Emerging models based on the YOLOv8 (You Only Look Once, version 8) architecture are helping redefine CAD analysis by detecting pathological structures directly in imaging data with high speed and accuracy. Unlike pixel-wise segmentation, YOLOv8 uses region-based detection, allowing it to identify features such as stenotic lesions, calcified plaques, and vessel bifurcations in a single pass. Its anchor-free detection head, adaptive loss functions, and robust backbone architecture enable simultaneous multi-feature detection with scalability and computational efficiency.
A notable application of this model is the DCA-YOLOv8, which integrates histogram equalization, Canny edge detection, a Double Coordinate Attention mechanism, and a custom AICI loss function. This framework achieved 96.62% precision and 95.06% recall in detecting coronary artery stenosis from X-ray angiography, showing strong clinical potential [
17]. Another implementation, YOLO-Angio, employs a three-phase pipeline (including preprocessing, YOLOv8-based vessel candidate detection, and logic-based tree reconstruction) and achieved an F1 score of 0.4289 in the ARCADE challenge, demonstrating effectiveness in vascular segmentation from angiography [
18].
Furthermore, studies have shown that preprocessing techniques significantly affect the performance of YOLOv8 in CAD imaging. For example, contrast enhancement, image sharpening, and U-Net-generated binary masks have been found to markedly improve YOLOv8’s detection accuracy in coronary angiograms [
19]. These results highlight the synergistic potential of combining classical segmentation outputs with object detection models to create hybrid frameworks that balance interpretability, accuracy, and speed. As such, YOLOv8 represents a promising advance in AI-driven cardiovascular imaging, bridging the gap between high-throughput analysis and real-time clinical decision-making.
In this work, we present a experimental comparison of the performance of the YOLO (You Only Look Once) architecture across the most recent versions (YOLOv8, YOLOv9, and YOLOv11) for the task of coronary vessel segmentation and stenosis detection. The evaluation is conducted using the ARCADE dataset, which provides a standardized benchmark for coronary artery analysis in X-ray angiography. To further investigate factors influencing model performance, we incorporate image preprocessing techniques in order to assess their impact on the learning process and detection accuracy. The performance of each model will be quantitatively assessed using standard segmentation metrics precision, recall, and F1-score, with the aim of identifying the most robust and generalizable configuration for clinical application in coronary artery disease segmentation.
2. Materials and Methods
In this section, we describe the characteristics of the dataset used in our experiments, including its structure, annotation format, and the clinical relevance of its contents. Additionally, we provide a detailed explanation of the image preprocessing technique applied prior to training. Finally, we discuss the different versions of the YOLO architecture (v8, v9, and v11) employed in this study, outlining their structural differences, advancements in detection accuracy and speed, and the rationale behind their selection for the task of coronary vessel classification and stenosis detection.
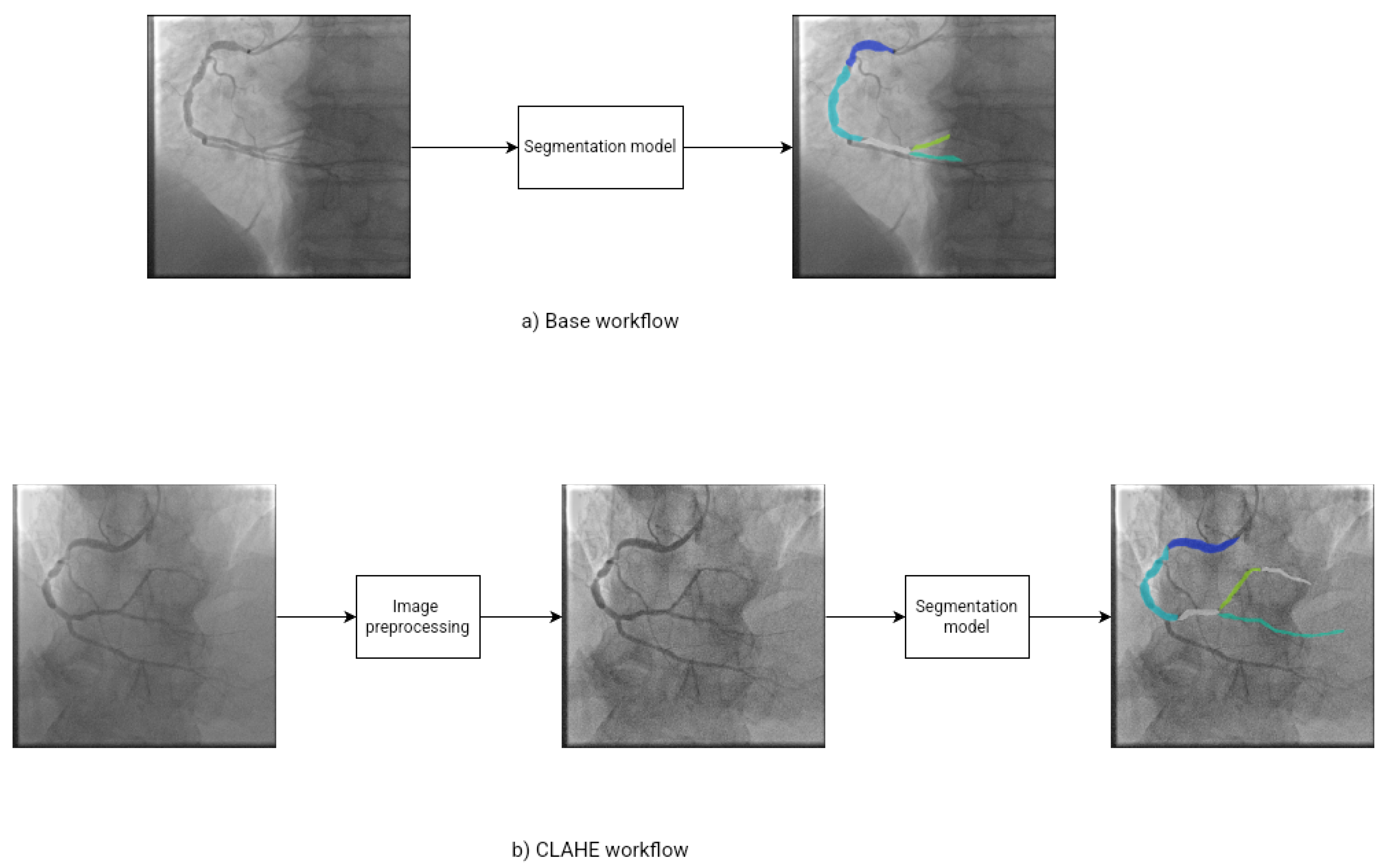
Figure 1 show the workflow of the experiments. We proposed two experimental configuration. In
Figure 1 a) we show the base workflow. We use the base images as input data to segmentation models and obtain the final segmentated image. In
Figure 1 b) show the CLAHE workflow. We apply image preprocessing to enhance the images and help to identify objects to segmentation models, then we use the prepocessing images as input data to segmentions models to obtain the final segmentated images.
2.1. ARCADE Dataset
The Automatic Region-based Coronary Artery Disease Diagnostics using X-ray Angiography Images (ARCADE) [
20] dataset is a publicly available collection designed to support the development, training, and evaluation of AI-based diagnostic tools for coronary artery disease (CAD) through the use of X-ray coronary angiography (XCA) images. It was introduced as part of the ARCADE challenge at the 26th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), with the objective of promoting research in automated detection and interpretation of coronary anatomy and pathology in clinical imaging. The dataset is organized into two primary tasks, each focusing on a critical component of CAD diagnosis:
Coronary artery segmentation: This subset includes 1,200 XCA images, with 1,000 images designated for training and 200 for validation. Each image has been annotated according to the SYNTAX Score methodology, a clinically validated system for evaluating coronary complexity. The annotations divide the coronary tree into 26 anatomically defined segments, enabling fine-grained segmentation of major vessels such as the left anterior descending (LAD), right coronary artery (RCA), and left circumflex artery (LCX), as well as their branches.
Stenosis detection: This subset also contains 1,200 XCA images, similarly split into 1,000 training and 200 validation images. Annotations in this task are designed to localize and label regions affected by atherosclerotic plaques, allowing for the classification of stenosis presence. Each instance highlights pathologically significant lesions that impact coronary blood flow, providing ground truth for evaluating automated diagnostic models.
In addition to the training and validation sets, Phase 2 of the ARCADE dataset includes test sets containing 300 annotated images for each task, made available for benchmarking model performance in blinded evaluations. All images are standardized to a resolution of
pixels and formatted in a consistent structure suitable for deep learning pipelines. The dataset has been carefully curated and annotated by expert radiologists and cardiologists to ensure clinical validity and labeling consistency. Furthermore, its design aligns with real-world diagnostic workflows, thereby enabling robust testing of machine learning models in scenarios representative of clinical practice. As such, the ARCADE dataset serves as a valuable benchmark for advancing the state-of-the-art in computer-aided diagnosis, with the potential to improve the accuracy, efficiency, and scalability of CAD screening and risk assessment in interventional cardiology. In
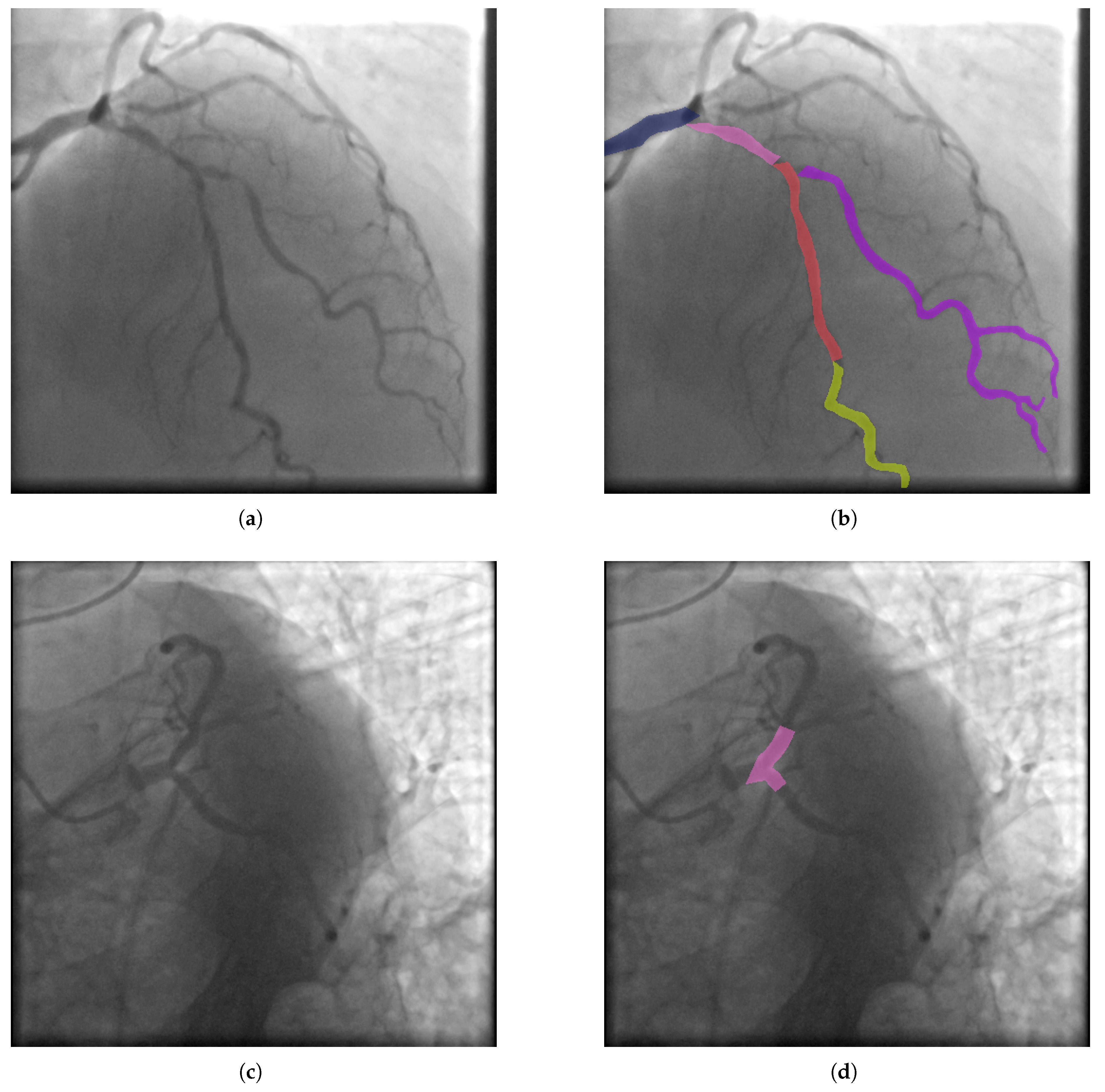
Figure 2 we show an example of vessel and stenosis segmentation in ground truth of dataset.
2.2. Image Preprocessing
In order to ehance the vessel and highlight the object to our segmentation model, we use CLAHE. CLAHE (Contrast Limited Adaptive Histogram Equalization) is an advanced image enhancement technique widely used in medical image processing to improve local contrast and reveal fine structural details in low-contrast or non-uniformly illuminated images. Unlike global histogram equalization, which enhances contrast uniformly across the entire image, CLAHE operates locally by dividing the image into small, non-overlapping regions called tiles and applying histogram equalization to each tile independently. This localized processing preserves edge details and avoids over-amplification of noise.
To further mitigate the risk of noise amplification, CLAHE introduces a contrast-limiting mechanism by clipping the histogram at a predefined threshold, known as the clip limit. The clipped pixels are then redistributed across the histogram, ensuring that areas with uniform intensity do not become oversaturated. After histogram equalization is applied within each tile, bilinear interpolation is used to smoothly combine adjacent tiles, eliminating artificial boundaries and ensuring a seamless transition across the image.
Due to its ability to enhance visibility in regions of low contrast without introducing significant artifacts, CLAHE is particularly beneficial in medical imaging modalities such as X-ray, ultrasound, CT, and coronary angiography, where subtle intensity differences are critical for detecting pathological structures. Its application has been shown to improve both human interpretation and the performance of automated systems for segmentation, classification, and feature extraction [
21,
22].
2.3. YOLOv8
YOLOv8 [
23] is a single-stage, end-to-end deep learning architecture for object detection, instance segmentation, and other visual tasks, developed by Ultralytics as the latest evolution in the "You Only Look Once" (YOLO) family of models. In the context of segmentation, YOLOv8 integrates a lightweight convolutional backbone with a decoupled head architecture, supporting instance-level segmentation masks using a mask prediction branch alongside bounding box regression and classification. It builds upon advancements in previous YOLO versions (e.g., YOLOv5 and YOLOv7) while introducing architectural optimizations such as an anchor-free detection head, dynamic input shapes, and an improved loss function tailored for segmentation accuracy and efficiency.
Unlike classical semantic segmentation networks (e.g., U-Net or DeepLab), YOLOv8 approaches segmentation from an object-centric perspective, predicting polygonal masks for each detected instance using a prototype-based mask representation. This allows YOLOv8 to combine the real-time inference speed of object detectors with the fine-grained mask outputs needed for instance segmentation tasks.
YOLOv8’s segmentation capabilities have been evaluated on benchmarks such as COCO and used in domain-specific tasks, including medical imaging and biomedical instance segmentation, demonstrating competitive performance with significantly lower inference times compared to traditional multi-stage pipelines.
2.4. YOLOv9
Although originally developed as an object detection framework, YOLOv9 [
24] has been extended to support image segmentation tasks through architectural adaptations and the integration of task-specific prediction heads. In the context of semantic and instance segmentation, YOLOv9 incorporates an additional segmentation head parallel to the detection head, enabling pixel-wise classification while maintaining its hallmark real-time processing capabilities. The backbone architecture, enhanced by the Generalized Efficient Layer Aggregation Network (GELAN), enables rich multi-scale feature extraction, which is essential for capturing fine spatial details in segmentation outputs.
Moreover, YOLOv9 introduces the Programmable Gradient Information (PGI) mechanism, a novel training strategy that adaptively emphasizes informative spatial regions during backpropagation. This innovation improves convergence stability and enhances the model’s ability to distinguish subtle object boundaries, making it particularly effective in domains such as medical imaging, where anatomical structures may be small or poorly contrasted. The efficient feature aggregation enabled by GELAN further allows YOLOv9 to perform accurate segmentation in challenging environments, including those involving densely packed or overlapping objects.
When configured for segmentation tasks, YOLOv9 is capable of producing both binary and multi-class masks alongside bounding box predictions, offering a unified approach to detection and segmentation. This dual capability is highly advantageous in real-time applications that demand both object localization and detailed region delineation—such as surgical guidance, autonomous navigation, and pathology identification.
2.5. YOLOv11
YOLOv11 [
25] represents the most recent advancement in the YOLO object detection series, introducing significant improvements not only in detection but also in segmentation capabilities. While primarily developed for real-time object detection, YOLOv11 has been adapted to perform semantic and instance segmentation through the integration of a dedicated segmentation head alongside the standard detection pipeline. This configuration enables the model to generate high-resolution, pixel-level masks in addition to bounding boxes, making it suitable for tasks that require both spatial localization and detailed object delineation.
One of the key innovations in YOLOv11 is the incorporation of a lightweight Vision Transformer (ViT) module into its backbone, which enhances the model’s ability to capture global contextual information across the image. This transformer-based attention mechanism, combined with traditional convolutional layers, improves segmentation accuracy by enabling the network to model long-range dependencies—especially valuable in complex segmentation tasks such as those in medical imaging or remote sensing.
Furthermore, YOLOv11 continues to refine training efficiency through multi-resolution feature fusion and enhanced data augmentation strategies, allowing for better generalization across varying image scales and qualities. The segmentation head in YOLOv11 benefits from precise multi-scale feature aggregation, enabling robust detection of fine structures such as blood vessels, lesions, or boundaries of overlapping objects.
When applied to segmentation, YOLOv11 outputs class-specific segmentation masks at high speed, making it ideal for real-time applications like intraoperative imaging, smart surveillance, and automated visual inspection. Its unified architecture allows simultaneous training of detection and segmentation tasks, leading to improved performance without sacrificing inference speed.
3. Results
We conducted experimentation using the ARCADE dataset to train models for segmenting vessel and train models to segmentate stenosis. The experiments were conducted using a computer with an Intel Xeon W-2133 processor, 32GB of RAM, and an NVIDIA GeForce GTX 1080 graphics card. The system ran on Ubuntu 18.04 operating system, and the CUDA toolkit 10.0 library was utilized.
3.1. Performance Measurements
The models were evaluated using three statistical metrics: precision (
1), recall (
2) and F1-score.
TP corresponds to true positives (there was a object and it was correctly segmentated), FP false positives (there was no object but the model segmentate it as object), and FN false negatives (the model did not segmentate a object in the image). Precision is a measure to know how manny object the model segmentate correctly to all the objects that it segmentates. Recall is a measure to know how many objects the model segmentate compared to the total number of object. F1-score is a measure to combine Precision and Recall to know the average performance of the model in both measures.
3.2. Metric Based Analysis of YOLO Architectures for Segmentation
We trained the YOLOv8, YOLOv9, and YOLOv11 models for 100 epochs using a batch size of 4. The training was performed using the AdamW optimizer with a learning rate of 0.0003 and a momentum value of 0.9. To leverage prior knowledge, we initialized the models with pretrained weights from the COCO dataset.
For model validation, we used YOLO’s built-in val function, evaluating performance with an Intersection over Union (IoU) threshold of 0.5. This means that a predicted region is considered correct if it overlaps with the ground truth by at least 50%, allowing us to assess the segmentation accuracy under standard evaluation criteria.
Table 1 present the result of the different segmentation models trained with base workflow and CLAHE workflow. The experimental results using the ARCADE dataset provide valuable insights into the performance of YOLOv8, YOLOv9, and YOLOv11 in multi-class segmentation tasks, both with and without CLAHE as an image preprocessing technique. Overall, performance across all models and configurations indicates that segmentation quality is highly dependent on the specific class, the number of instances, and the anatomical relevance of each coronary segment.
For the stenosis class, which represents a critical target in CAD diagnosis, YOLOv9 achieved the highest F1-score 0.417 under the base condition, outperforming both YOLOv8 0.362 and YOLOv11 0.393. However, when CLAHE preprocessing was applied, none of the models demonstrated a consistent improvement; in fact, YOLOv9’s F1-score declined to 0.385, while YOLOv11 dropped slightly to 0.376. These results suggest that although CLAHE may enhance local contrast, it does not necessarily improve the detection of complex pathological structures such as stenoses—possibly due to the increased noise or artificial edges introduced during preprocessing.
In terms of overall vessel segmentation, performance remained relatively stable across models and conditions, with F1-scores consistently around 0.50. YOLOv9 base: 0.500 and YOLOv11 base: 0.510 showed slight advantages over YOLOv8, but CLAHE preprocessing did not significantly enhance results, indicating that vessel detection is already well-optimized in these models and less sensitive to contrast enhancement techniques.
When examining individual vessel segments, large and clearly defined classes (e.g., classes 1 to 6) exhibited consistently high F1-scores across all models (often >0.70), with only minor differences between base and CLAHE conditions. For instance, class 1 achieved F1-scores up to 0.77 with YOLOv11 and CLAHE, confirming the robustness of all three models in segmenting major coronary structures. In contrast, smaller or less frequent segments, such as classes 12, 14a, and 16c, had notably lower F1-scores, often below 0.30. These low values reflect the inherent difficulty in detecting small, irregularly shaped regions, particularly in datasets with limited instances.
Notably, CLAHE preprocessing produced mixed effects across models and classes. While it slightly improved F1-scores in some low-contrast regions for YOLOv8 and YOLOv9 (e.g., classes 9a and 16a), it also led to decreased performance in others, including stenosis and class 13. YOLOv9, which incorporates the PGI training mechanism, appeared especially sensitive to preprocessing: its performance improved in cases where edge contrast was enhanced but tended to decline when CLAHE introduced non-informative artifacts or excessive contrast normalization.
YOLOv11 consistently demonstrated strong performance across most classes under both preprocessing conditions, indicating higher generalizability and robustness. For example, class 11 (a region with 119 instances) achieved an F1-score of 0.849 base and 0.842 CLAHE, showing minimal variation and suggesting that YOLOv11 is less reliant on preprocessing enhancements to achieve optimal performance.
3.3. Visual Analysus of YOLO Mode Predictions
To complement the quantitative evaluation of model performance, this section presents a visual analysis of the segmentation results produced by the different YOLO-based architectures. Visual inspection of model predictions provides valuable insight into the spatial accuracy and morphological coherence of the segmented structures, particularly in clinical tasks such as coronary vessel delineation and stenosis detection. By comparing the outputs of each model with the annotated ground truth under different preprocessing conditions, it is possible to assess the models’ ability to generalize across image variations and to identify specific strengths or limitations that may not be fully captured by numerical metrics alone.
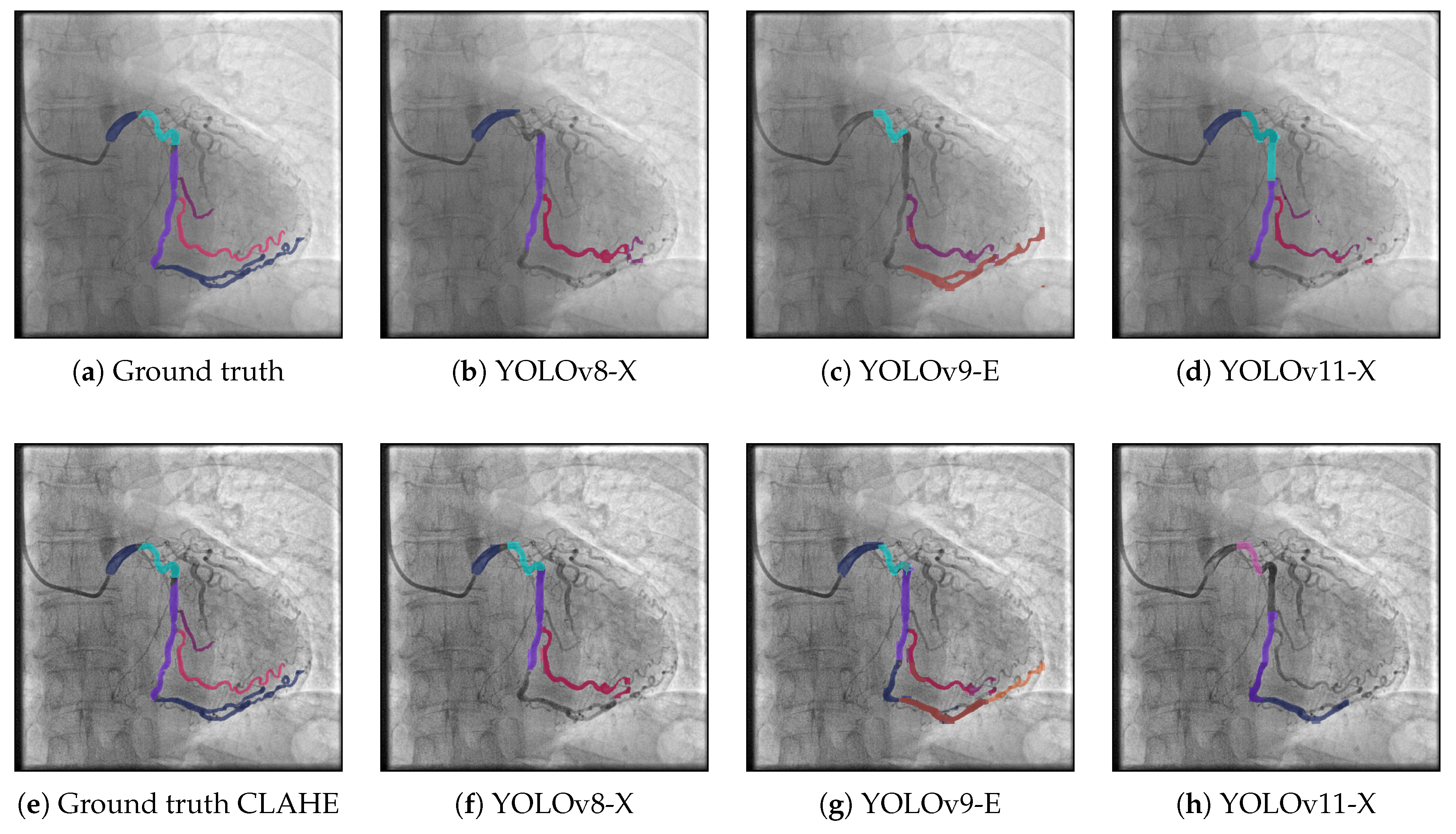
Figure 3 presents segmentation results on the ARCADE dataset using three different YOLO-based models under two distinct workflows: the base pipeline (top row) and a pipeline enhanced with CLAHE preprocessing (bottom row). In the base workflow, YOLOv8-X (
Figure 3(b)) captures the main vascular structures but misses several finer branches, particularly in distal regions. YOLOv9-E (
Figure 3(c)) exhibits enhanced sensitivity in detecting vessel structures, particularly in challenging or low-contrast regions. Notably, it is the only model among the three that successfully identifies the vessel segment located in the bottom-center region of the image. However, despite this correct localization, the segment is misclassified, indicating limitations in the model’s class discrimination capabilities. Moreover, YOLOv9-E tends to produce a higher number of false positives compared to the other models, which negatively impacts the overall segmentation precision. This suggests that while the model is effective in capturing subtle vascular structures, further refinement is needed to improve its specificity and reduce erroneous detections.YOLOv11-X (
Figure 3(d)) shows the most faithful reproduction of the ground truth among the base models, offering better continuity and coverage of vessel bifurcations. When applying CLAHE preprocessing, a noticeable improvement is observed across all models. YOLOv8-X (
Figure 3(f)) exhibits enhanced vessel continuity and more complete segment detection compared to its base counterpart. But continue to misclassified some zones that was correctly segmented. If the classifier of the YOLOv9-E could be improve, this can improve the metrics. Notably, YOLOv11-X (
Figure 3(h)) delivers the most accurate segmentation overall, closely aligning with the annotated ground truth and effectively capturing complex vascular morphologies. These findings suggest that CLAHE significantly enhances model performance by improving vessel visibility in angiographic images, particularly in cases where vessel contrast is low or spatial resolution is limited.
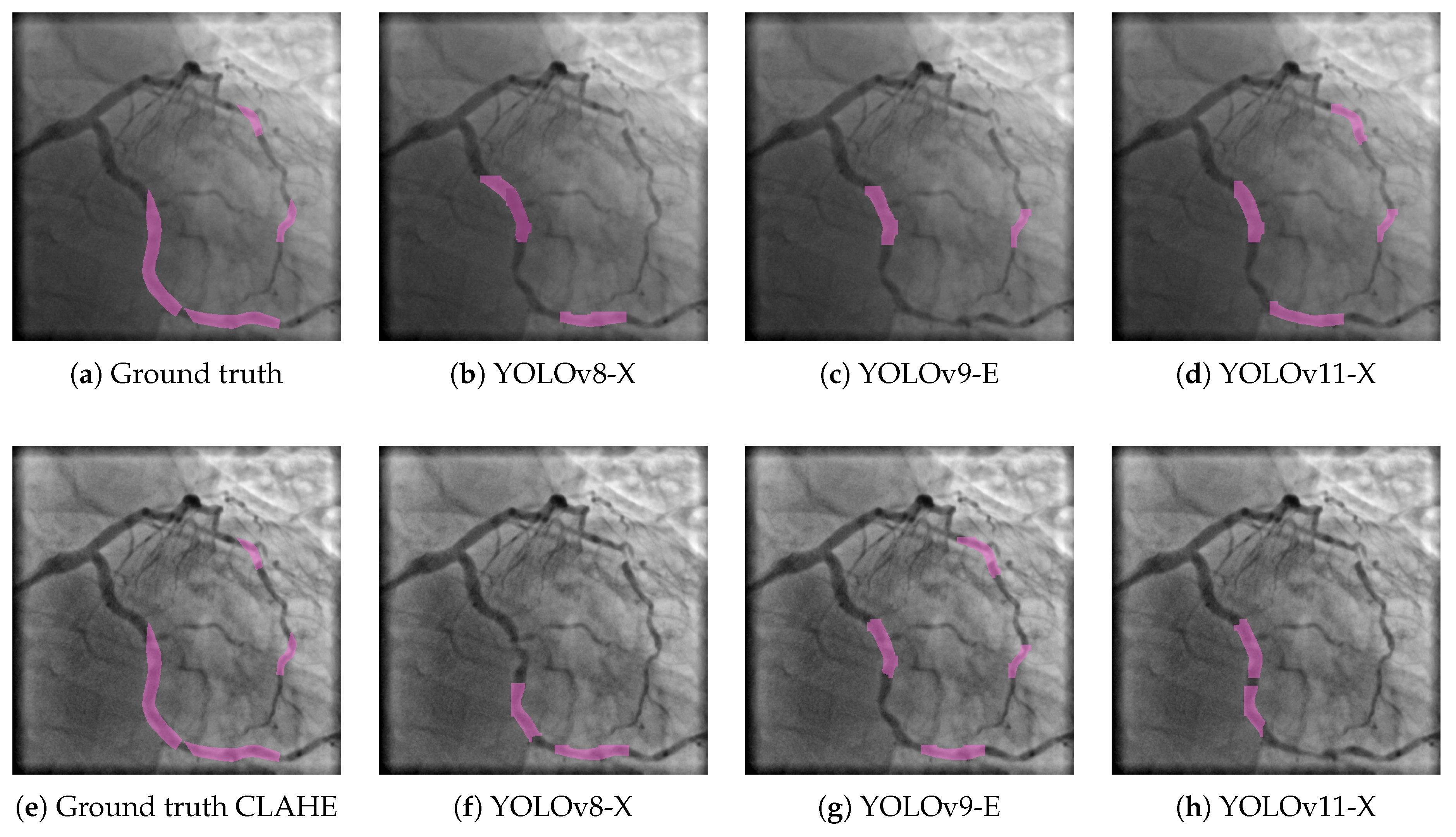
A similar trend is observed in the results for the stenosis class, as shown in
Figure 4. Here, the three YOLO-based models were also evaluated under both the base and CLAHE-enhanced workflows. In the top row, YOLOv8-X (
Figure 4(b)) detects stenotic regions with moderate accuracy but misses portions of the ground truth, resulting in partial segmentation. YOLOv9-E (
Figure 4(c)) exhibits lower sensitivity, capturing only a small fraction of the stenotic segments. YOLOv11-X (
Figure 4(d)) improves upon previous models by identifying a greater extent of the target regions, yet still fails to fully match the reference annotations shown in
Figure 4(a). When CLAHE preprocessing is applied, a marked improvement is observed across all models. YOLOv8-X (
Figure 4(f)) shows increased alignment with the ground truth (
Figure 4(e)), suggesting enhanced contrast allows for better localization of stenotic lesions. YOLOv9-E (
Figure 4(g)) achieves more complete and accurate segmentations compared to its base counterpart, while YOLOv11-X (
Figure 4(h)) delivers the most precise and continuous detection, closely resembling the ground truth. These results confirm the positive impact of CLAHE preprocessing in improving segmentation performance, not only for vascular structures but also for more localized pathological features such as stenosis. The consistent benefit across both classes and all model variants reinforces the robustness of CLAHE as a preprocessing strategy in angiographic image analysis.
3.4. Comparison with State-of-Art Propposals
In order to evaluate YOLO models, we conducted a review of the state of the art to position our proposal relative to existing approaches. For this comparison, we selected the best-performing configurations in terms of F1-score: YOLOv8-X base for vessel segmentation, and YOLOv9-E base for stenosis segmentation. The
Table 2 presents a comparative analysis against various proposals in terms of Precision, Recall, and F1-score.
In the stenosis detection task, our proposal based on YOLOv9-E achieved an F1-score of 0.417, with a precision of 0.367 and recall of 0.484. While this performance remains moderate in the context of the current state of the art, it represents a clear improvement over the baseline YOLOv8-X model reported by Mlynarski et al., which attained an F1-score of 0.400. These results suggest that YOLOv9-E introduces beneficial architectural enhancements that improve learning dynamics and generalization, particularly in challenging pathological scenarios. Nevertheless, additional optimization will be necessary to reach the performance levels demonstrated by more specialized segmentation models.
In the vessel segmentation task, the best-performing method in terms of F1-score is from Tran et al. using YOLOv8-X, achieving an F1-score of 0.520, with relatively balanced precision and recall. The implementation of YOLOv8-X for vessel segmentation yielded an F1-score of 0.513, with a slightly improved precision 0.495 compared to Tran et al., with recall slightly lower at 0.533 compared to 0.560. This result demonstrates competitive performance and confirms the reproducibility and robustness of YOLOv8-X across different experimental setups. Moreover, our results outperform those from Mlynarski et al. for the same model, who reported an F1-score of 0.490, indicating the effectiveness of our preprocessing or training strategies.
4. Discussion
The experimental results in this study highlight the practical strengths of YOLO-based architectures for coronary artery disease imaging. In particular, YOLOv8 and YOLOv9 offer a compelling balance between computational efficiency and segmentation accuracy, which is crucial for clinical applications requiring fast inference times, such as real-time angiography analysis.
While YOLO models showed high precision, they also exhibited limited recall compared to transformer-based and region-based methods. This suggests a tendency to miss subtle or ambiguous structures, particularly in complex imaging scenarios or when plaque boundaries are indistinct. This behavior underscores a fundamental trade-off in object detection frameworks between specificity and sensitivity.
The role of image preprocessing (especially CLAHE) was found to be task and model dependent. Although CLAHE can enhance local contrast and reveal vessel boundaries, it may also introduce non-informative artifacts or edge noise that confound models during training. This effect was particularly evident in the segmentation of small or irregular stenotic regions, where models like YOLOv9-E showed sensitivity to contrast enhancement but no consistent performance gains.
Furthermore, architectural differences among YOLO versions revealed varying degrees of robustness. YOLOv11, for instance, maintained stable performance across most tasks and preprocessing conditions, likely due to its incorporation of transformer-based attention and improved feature fusion mechanisms. In contrast, YOLOv9 benefited from novel training mechanisms like PGI, but remained more sensitive to preprocessing effects.
In summary, the discussion of findings emphasizes that while YOLO-based models are highly promising for CAD segmentation, further improvements in recall and robustness may require architectural enhancements—such as multiscale decoding, attention mechanisms, or task-specific loss functions—to achieve performance levels comparable to more advanced segmentation approaches.
5. Conclusion
This study presents a comparative evaluation of the latest versions of the YOLO architecture (YOLOv8, YOLOv9, and YOLOv11) applied to coronary vessel segmentation and stenosis detection using the ARCADE dataset. The results demonstrate that YOLO-based architectures are viable for medical segmentation tasks in real-time processing environments, showing particularly strong performance in the segmentation of major coronary vessels.
YOLOv8-X proved to be the most effective model for vessel segmentation, achieving an F1-score of 0.513, while YOLOv9-E delivered the best performance for stenosis detection. Although these results are competitive with previous YOLO-based methods, there is still room for improvement, particularly in the detection of subtle pathological features. This may be addressed by incorporating architectural strategies capable of capturing broader spatial context, such as attention mechanisms or region-focused segmentation techniques.
The application of CLAHE preprocessing showed mixed effects on model performance, suggesting that while it may improve local contrast, it can also introduce noise or artificial edges that hinder the detection of complex structures such as stenoses.
Overall, the findings indicate that recent YOLO versions offer an effective and efficient tool for automatic segmentation in coronary angiography, particularly in clinical scenarios where inference speed is critical. Future work could focus on integrating attention mechanisms or hierarchical segmentation strategies to narrow the performance gap with state-of-the-art architectures and improve detection of small or low-contrast pathological regions.
Author Contributions
Conceptualization, E.D-G. and A.Y-R.; methodology, E.D-G.; software, E.D-G.; validation, E.D-G., A.Y-R., R.M.L-B. and E.L-R; formal analysis, E.D-G., A.Y-R, R.M.L-B and E.L-R; investigation, E.D-G.; resources, A.Y-R., I.F.V-L., R.M.L-B. and E.L-R.; data curation, E.D-G.; writing—original draft preparation, E.D-G.; writing—review and editing, A.Y-R., I.G-A., R.M.L-B and E.L-R; visualization, E.D-G.; supervision, A.Y-R., I.F.V-L., R.M.L-B. and E.L-R; project administration, E.D-G.; funding acquisition, I.G-A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Acknowledgments
The authors would like to acknowledge the support of Secretaría de Ciencia, Humanidades, Tecnología e Innovación (SECIHTI) for the scholarships granted during their graduate studies, as well as the Universidad Autónoma de Sinaloa for its institutional support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Yusuf, S.; Hawken, S.; Ôunpuu, S.; et al. . Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. The Lancet 2004, 364, 937–952. [Google Scholar] [CrossRef] [PubMed]
- Leber, A.W.; et al. . Quantification of obstructive and nonobstructive coronary lesions by 64-slice computed tomography: a comparative study with quantitative coronary angiography and intravascular ultrasound. Journal of the American College of Cardiology 2005, 46, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Zreik, M.; et al. . A recurrent CNN for automatic detection and classification of coronary artery plaque and stenosis in coronary CT angiography. IEEE Transactions on Medical Imaging 2018, 38, 1588–1598. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; et al. . Deep learning for identifying and classifying vulnerable atherosclerotic plaques in intravascular imaging: advances and challenges. Frontiers in Cardiovascular Medicine 2021, 8, 679379. [Google Scholar] [CrossRef]
- Nakamura, S.; et al. . Prognostic impact of quantitative coronary plaque burden assessed by CCTA. European Heart Journal – Cardiovascular Imaging 2019, 20, 601–609. [Google Scholar] [CrossRef]
- Bae, Y.G.; et al. . Segmentation of calcified plaques in IVUS using region growing method. Healthcare Informatics Research 2017, 23, 218–225. [Google Scholar] [CrossRef]
- Yuan, C.; et al. . Coronary artery plaque characterization with CT angiography: limitations and future directions. Current Cardiovascular Imaging Reports 2017, 10, 1–8. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention (MICCAI); 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Chen, S.; Ma, K.; Zheng, Y. Med3D: Transfer learning for 3D medical image analysis. Computer Methods and Programs in Biomedicine 2020, 195, 105618. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention (MICCAI); 2016; pp. 424–432. [Google Scholar] [CrossRef]
- Oktay, O.; et al. . Attention U-Net: Learning Where to Look for the Pancreas. arXiv preprint, arXiv:1804.03999, 2018.
- Yang, G.; Zhang, J.; Liu, J.; et al. . Automatic coronary artery segmentation and calcification detection in CCTA using deep learning. Medical Image Analysis 2021, 67, 101838. [Google Scholar] [CrossRef]
- Zhu, Y.; Xia, Y.; Zhang, Z.; et al. . Deep learning-based IVUS segmentation for plaque characterization. IEEE Transactions on Biomedical Engineering 2021, 68, 1725–1735. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Transactions on Medical Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef]
- Kolossváry, M.; et al. . Machine learning for predicting cardiovascular events using coronary CT radiomics: a multicenter study. Radiology 2019, 292, 188–197. [Google Scholar] [CrossRef]
- Commandeur, F.; et al. . Machine learning for detection of vulnerable plaque from coronary CT angiography-derived radiomics. JACC: Cardiovascular Imaging 2020, 13, 734–745. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, H.; Liu, B.; et al. . YOLOv8-based Coronary Artery Stenosis Detection with Double Coordinate Attention. Sensors 2023, 24, 8134. [Google Scholar] [CrossRef]
- Zhou, Y.; Khanna, A.; Moriconi, F.; et al. . YOLO-Angio: A YOLOv8-Based Coronary Vessel Detection Pipeline for X-ray Angiography. arXiv preprint, arXiv:2310.15898, 2023.
- Mlynarski, D.; Glandut, N.; et al. . Annotated Dataset and Segmentation Masks for X-ray Coronary Angiography Images. Scientific Data 2023, 10, 669. [Google Scholar] [CrossRef]
- Popov, M.; Amanturdieva, A.; Zhaksylyk, N.; Alkanov, A.; Saniyazbekov, A.; Aimyshev, T.; Ismailov, E.; Bulegenov, A.; Kuzhukeyev, A.; Kulanbayeva, A.; et al. Dataset for Automatic Region-based Coronary Artery Disease Diagnostics Using X-Ray Angiography Images. Scientific Data 2024, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Pizer, S.M.; Amburn, E.P.; Austin, J.D.; Cromartie, R.; Geselowitz, A.; Greer, T.; ter Haar Romeny, B.; Zimmerman, J.B.; Zuiderveld, K. Adaptive Histogram Equalization and Its Variations. Computer Vision, Graphics, and Image Processing 1987, 39, 355–368. [Google Scholar] [CrossRef]
- Zuiderveld, K. Contrast Limited Adaptive Histogram Equalization. In Graphics Gems IV; Heckbert, P.S., Ed.; Academic Press Professional, Inc.: San Diego, CA, USA, 1994; pp. 474–485. [Google Scholar]
- Jocher, G.; Chaurasia, A.; Qiu, J. Ultralytics YOLOv8, 2023.
- Wang, C.Y.; Liao, H.Y.M.; Chen, J.W.; Chen, I.H. YOLOv9: Learning What You Want to Learn Using Programmable Gradient Information. arXiv preprint, arXiv:2402.13616, 2024.
- Jocher, G.; Qiu, J. Ultralytics YOLO11, 2024.
- Tran, D.S.; Huynh, A.K.; Huynh, A.D.; Nguyen-Thoi, T. Anatomy-specific two-stage YOLOv8 approach for improved coronary segmentation using the ARCADE dataset. Opt. Continuum 2025, 4, 303–317. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).