Submitted:
26 May 2025
Posted:
26 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Brief Introduction of MICALs
1.2. Intrinsic Disorder in Proteins
2. Results
2.1. Functional Disorder Analysis of Human MICALs
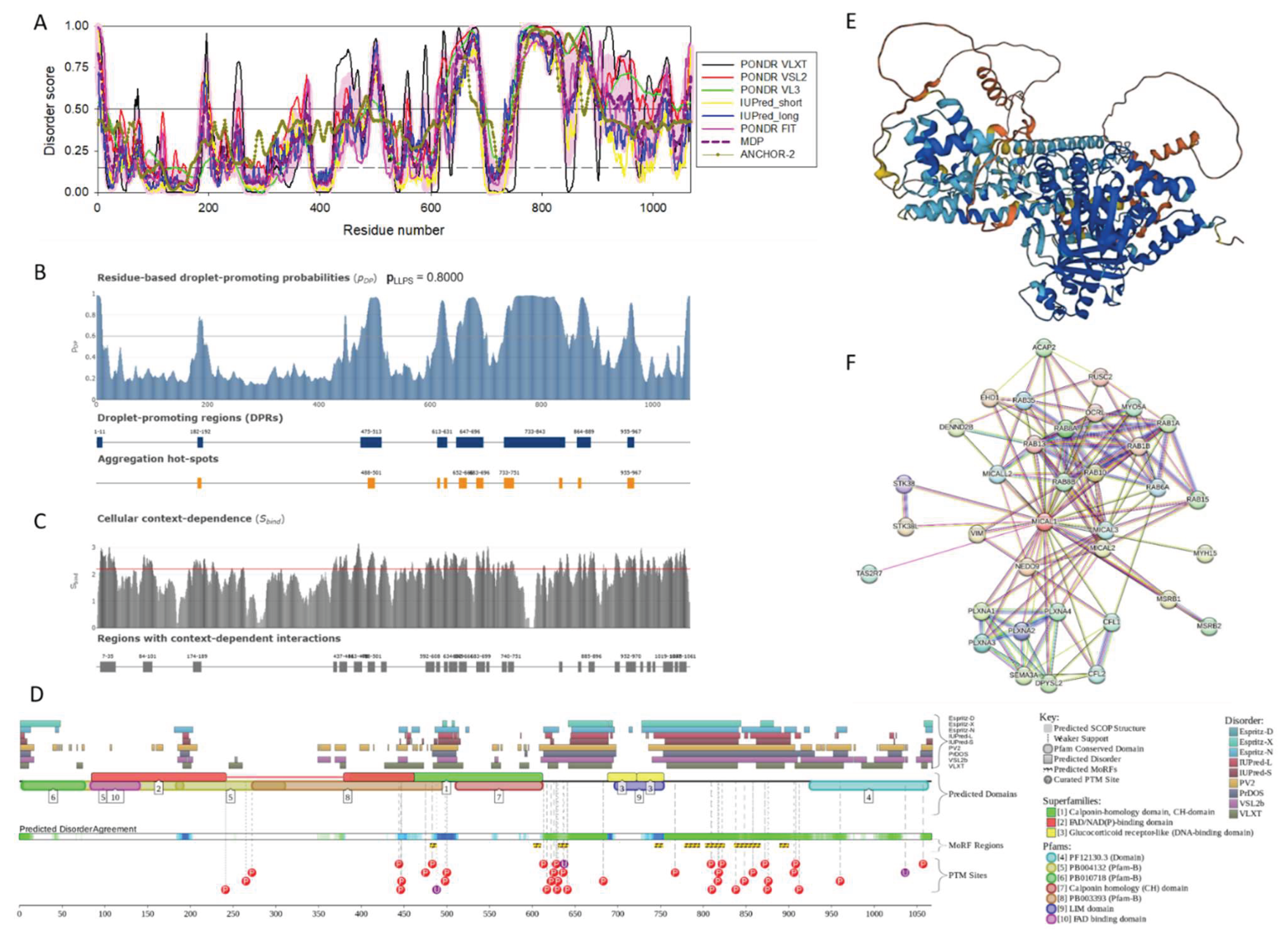
2.1.1. MICAL1 (UniProt ID: Q8TDZ2)
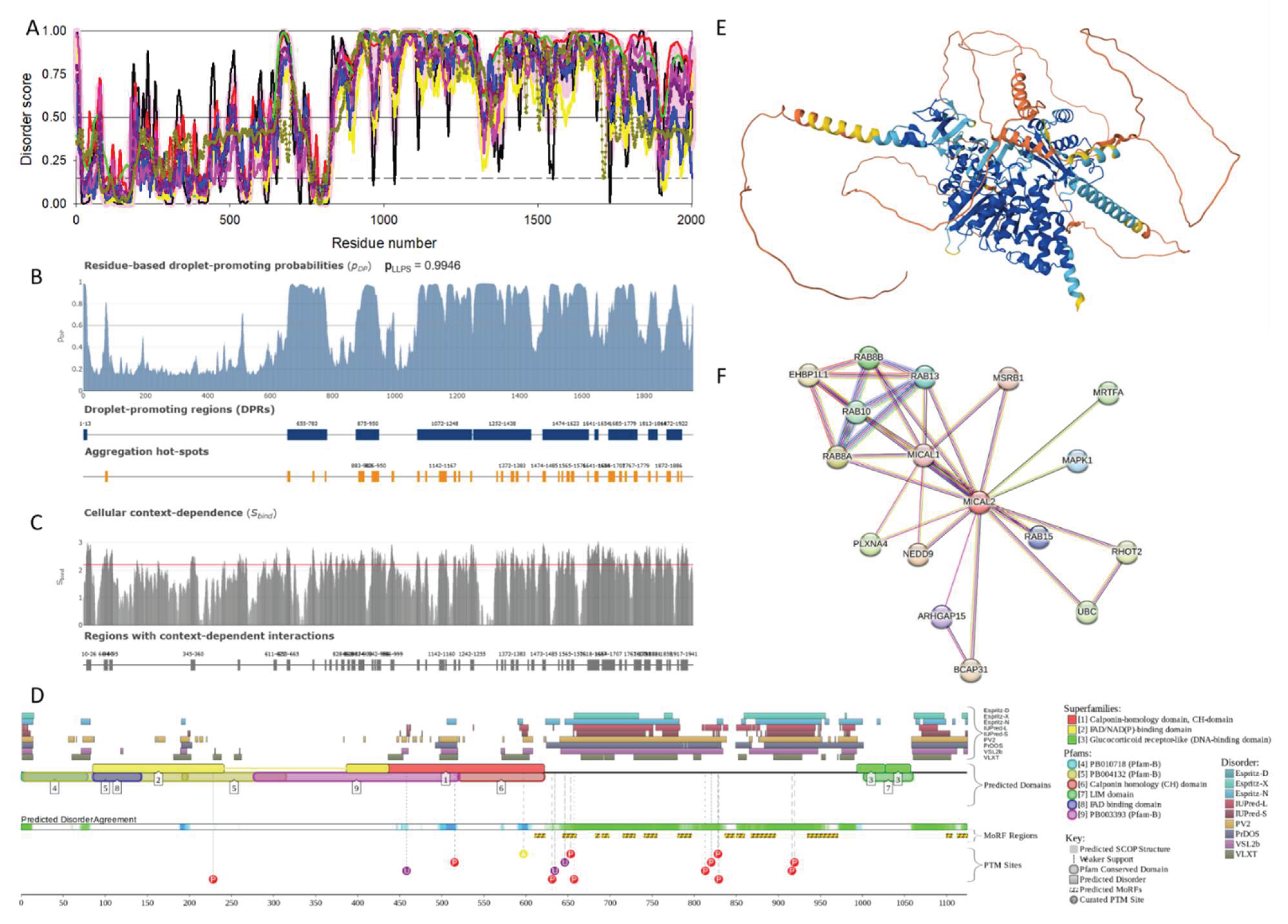
2.1.2. [F-actin]-Monooxygenase MICAL2 (UniProt ID: O94851)
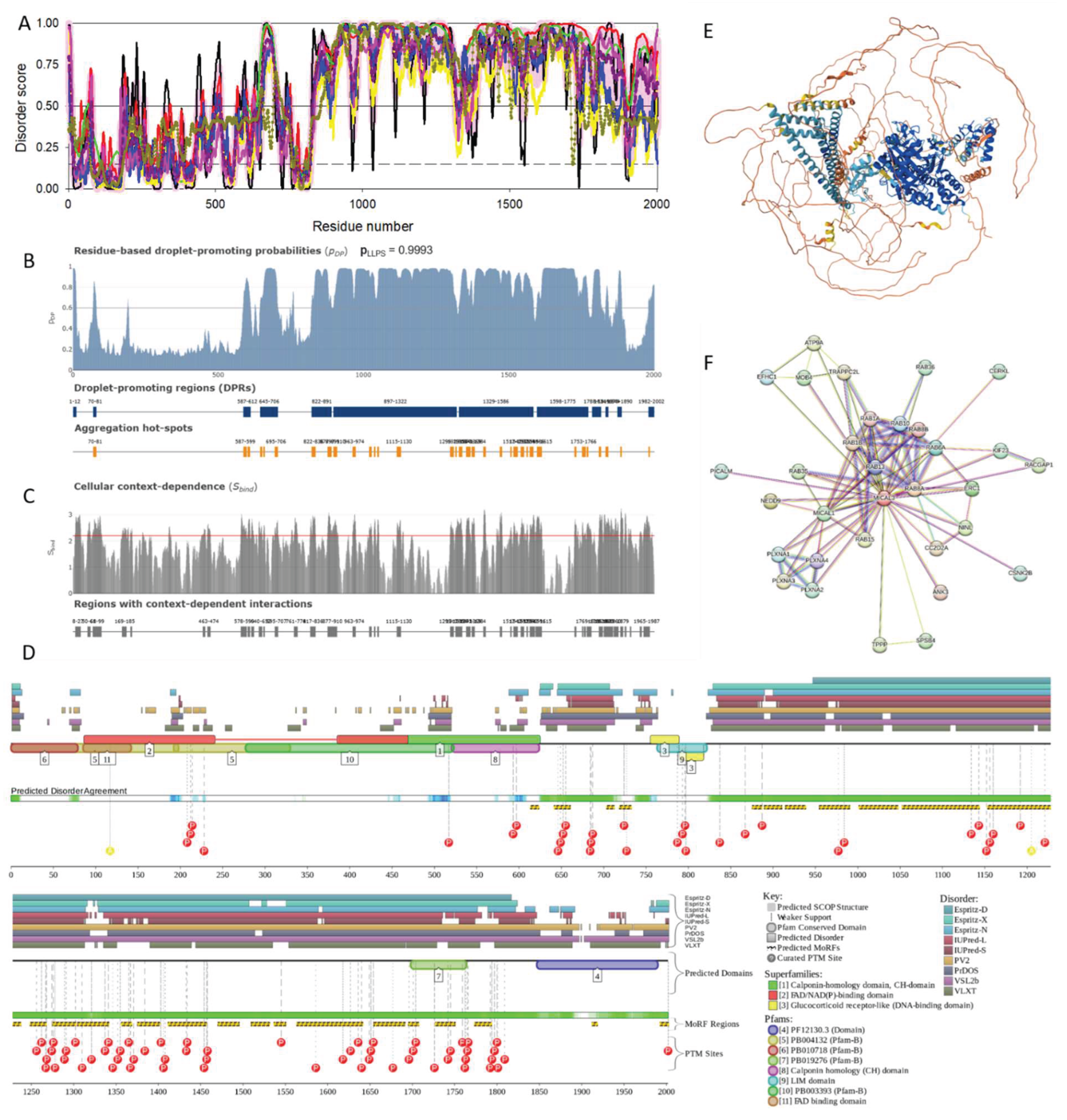
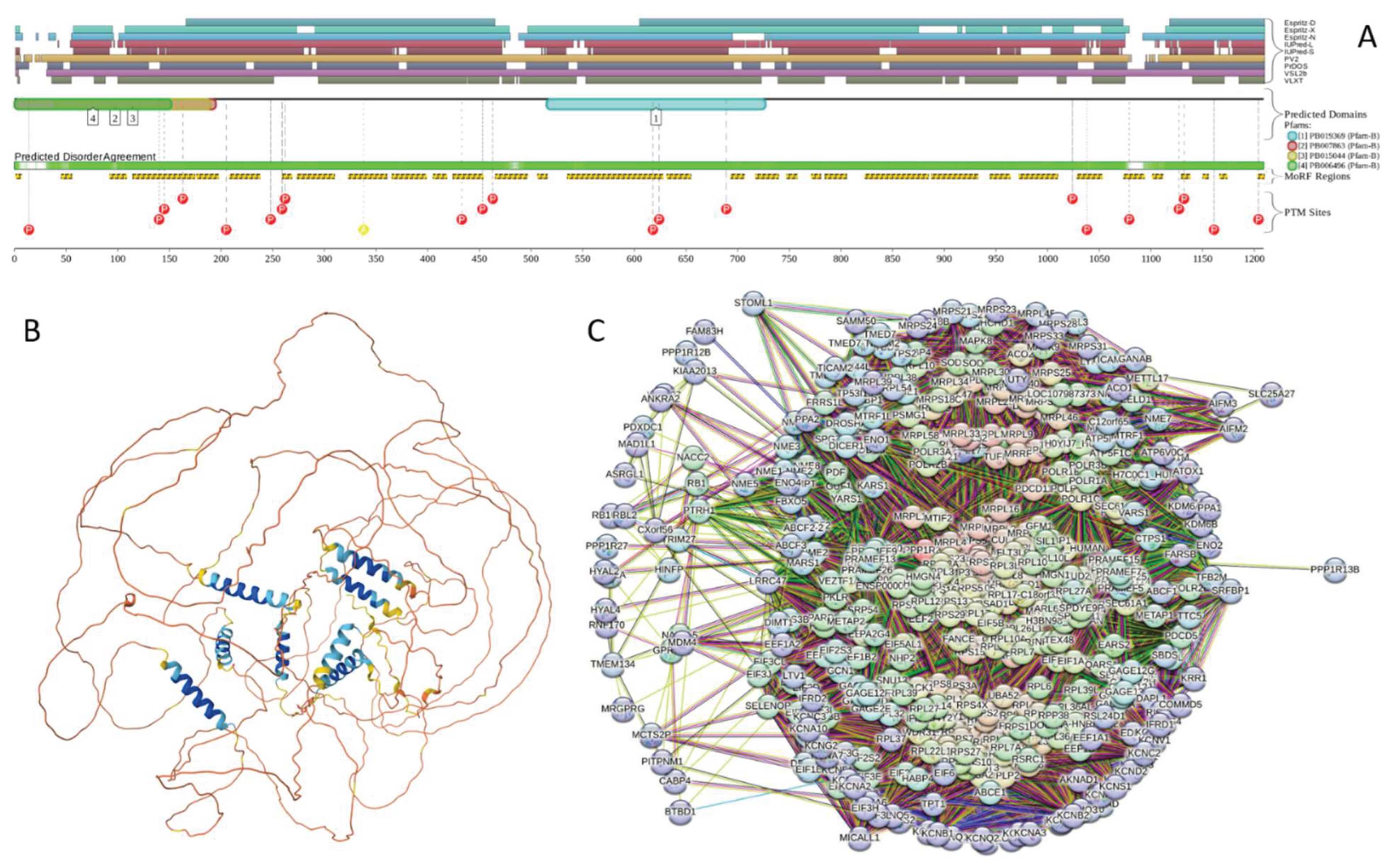
2.1.3. [F-actin]-Monooxygenase MICAL3 (UniProt ID: Q7RTP6)
2.1.4. Human MICAL-Like Protein 1 (MICAL-L1; UniProt ID: Q8N3F8)
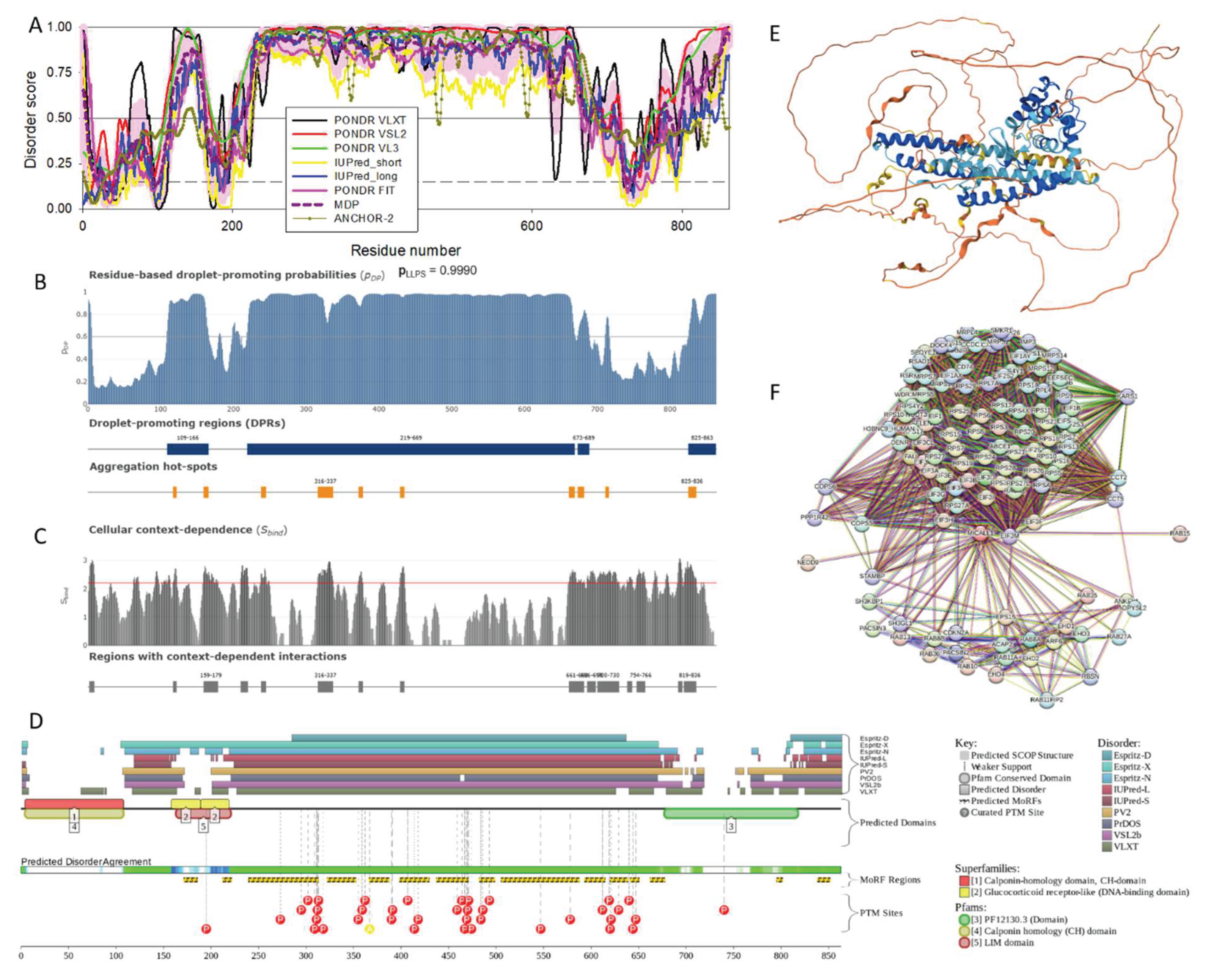
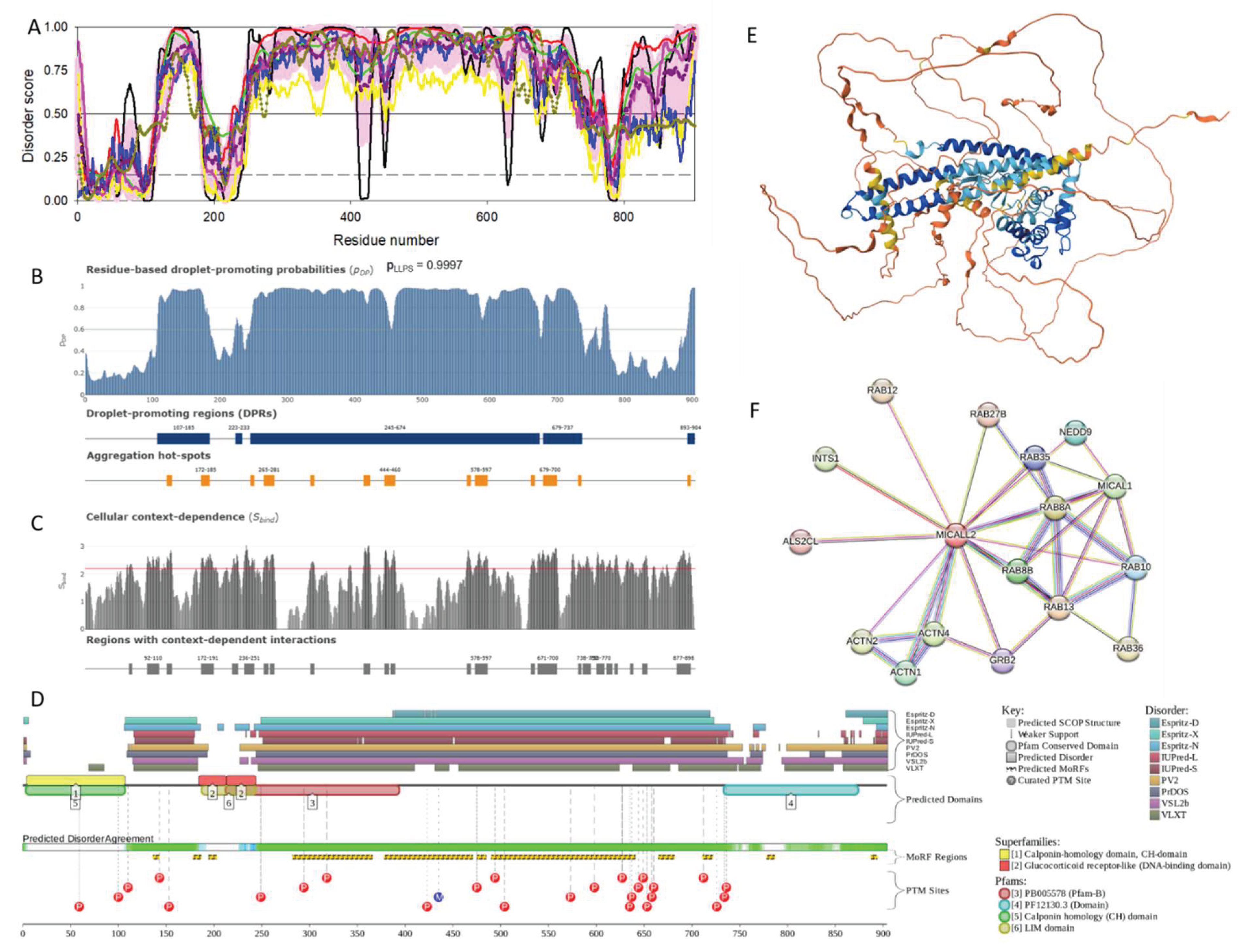
2.1.5. MICAL-L2 (MICAL-LIKE 2 or JRAB; UniProt ID: Q8IY33)
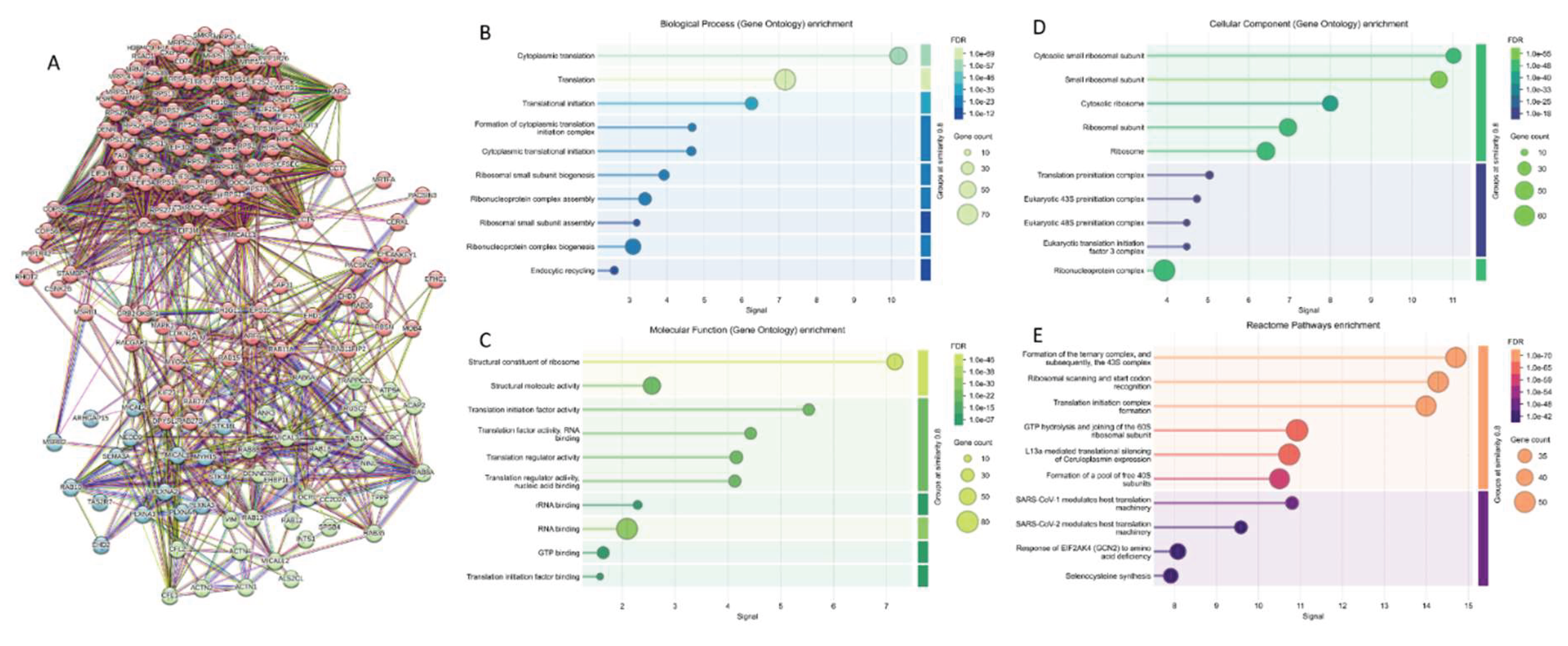
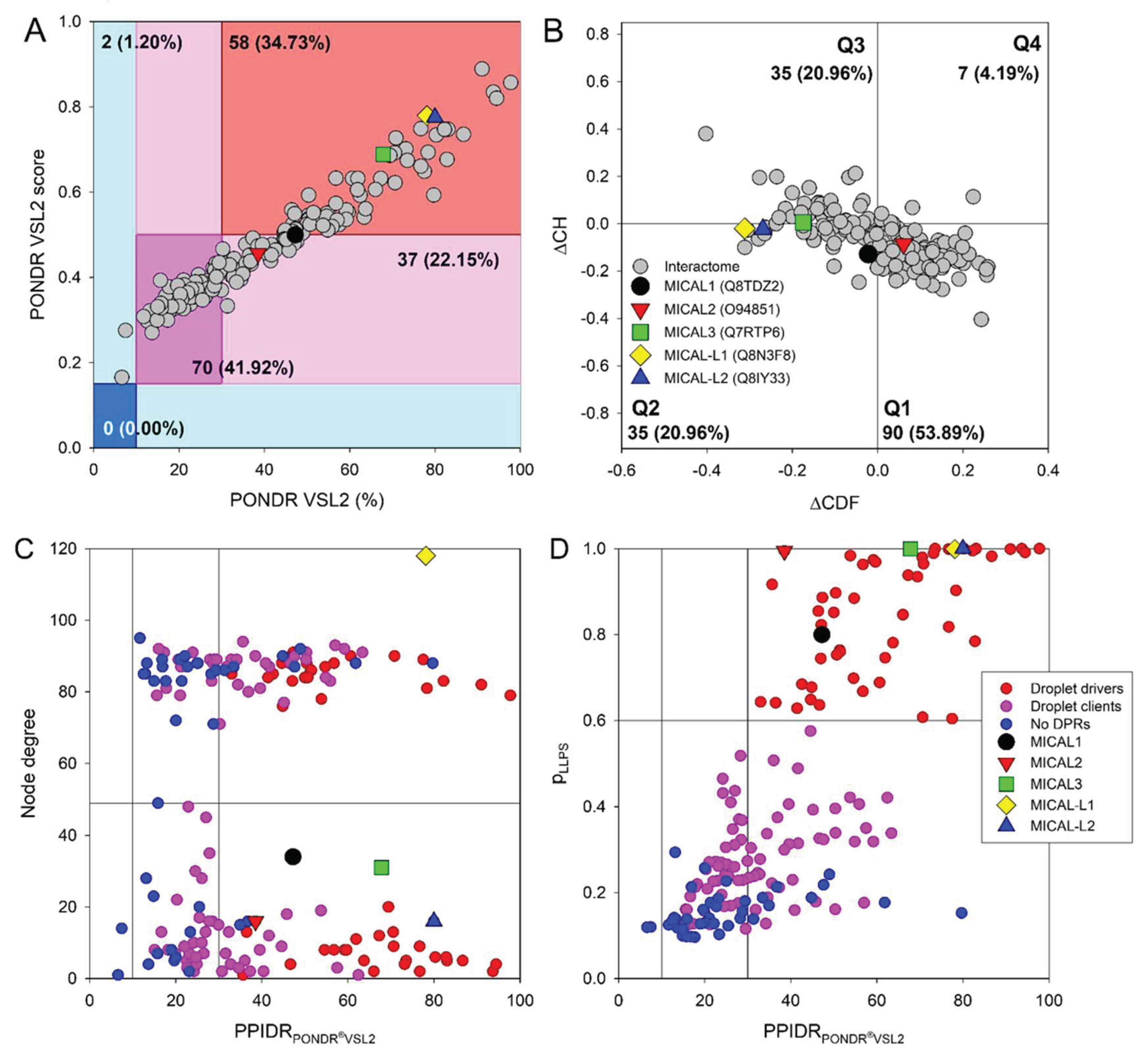
2.2. Global Analysis of the Interactome of Human MICAL Proteins
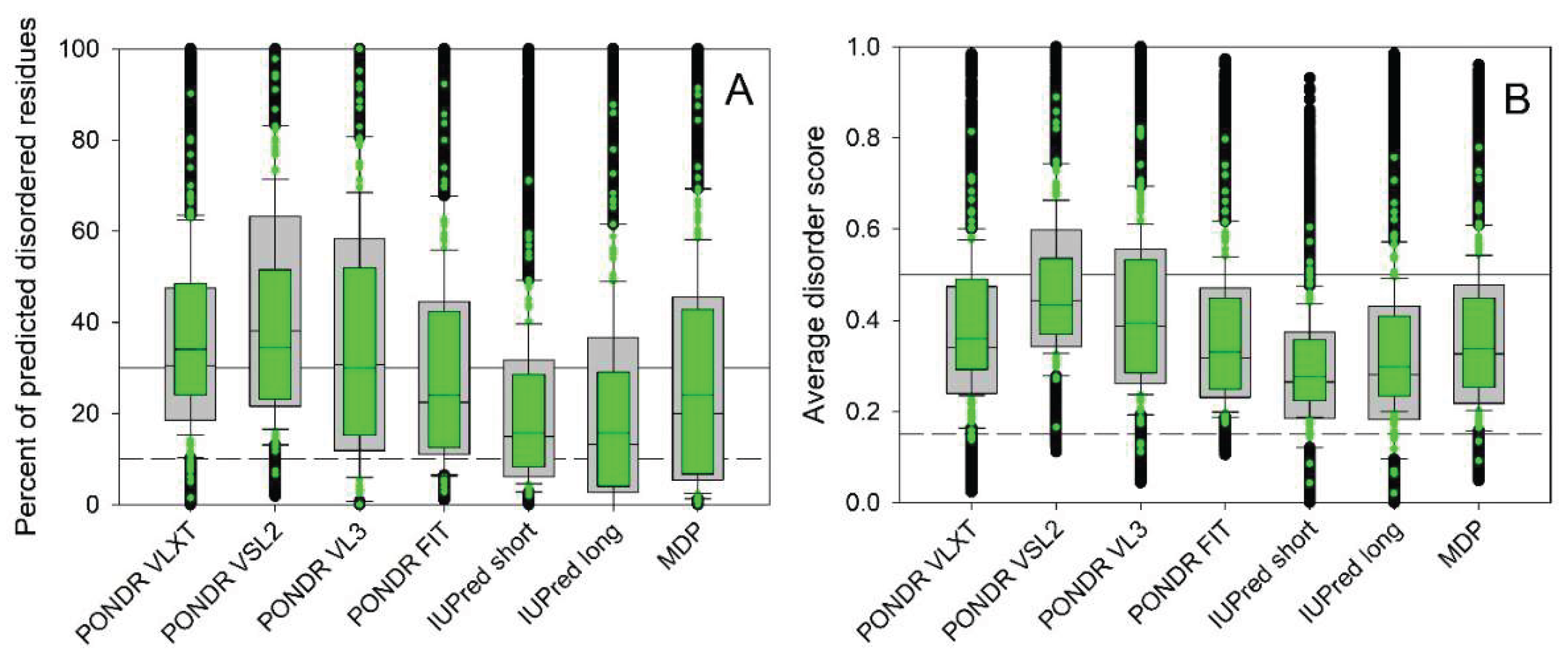
2.3. Global Analysis of the Prevalence of Intrinsic Disorder in MICAL Interactome
2.4. Functional Disorder Analysis of 5 Most Disordered MICAL Interactors
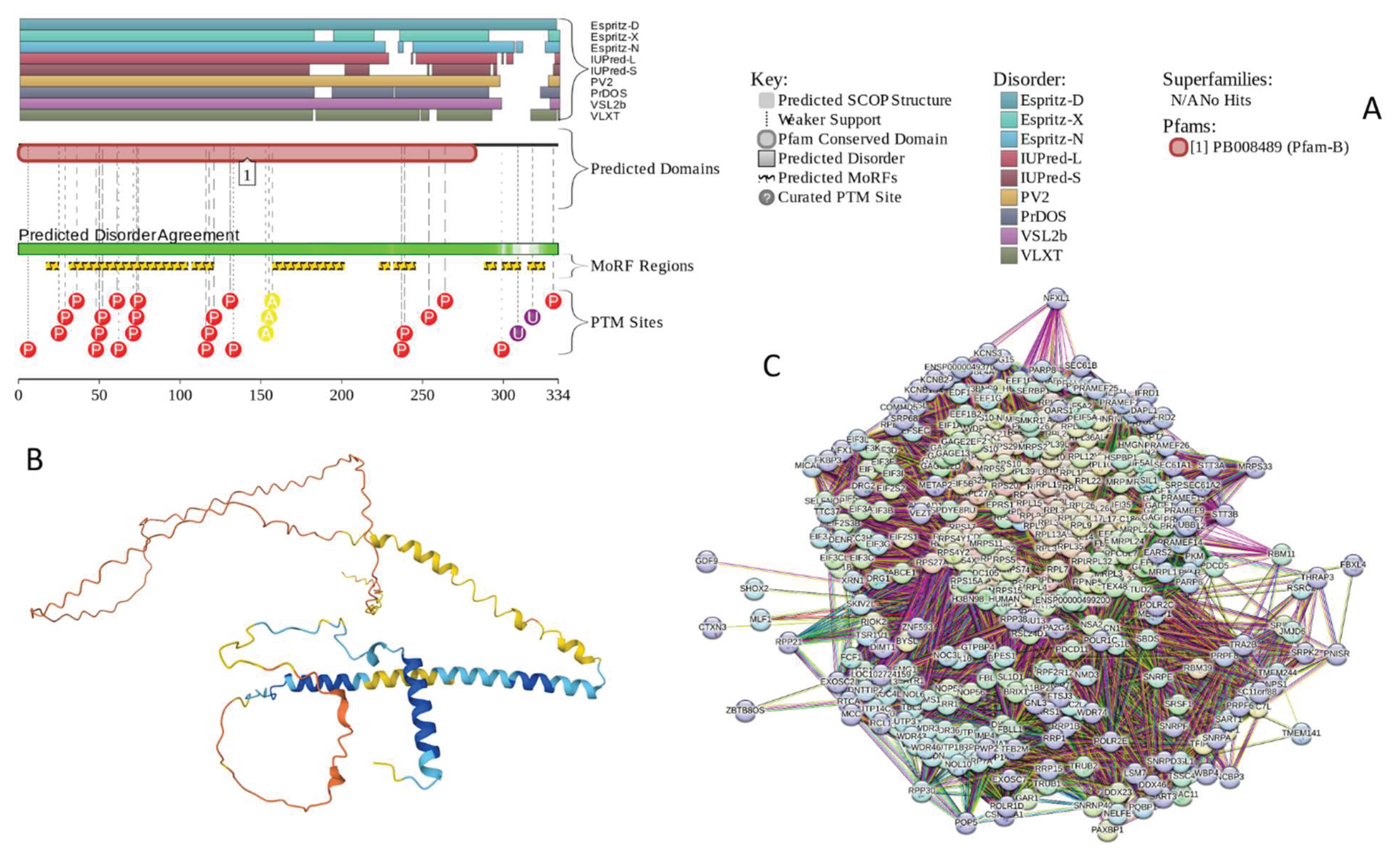
2.4.1. Protein Phosphatase 1 Regulatory Subunit 26 (PPP1R26; UniProt ID: Q5T8A7)
2.4.2. Serine/Arginine-Related Protein 53 (SRrp53; UniProt ID: Q96IZ7)
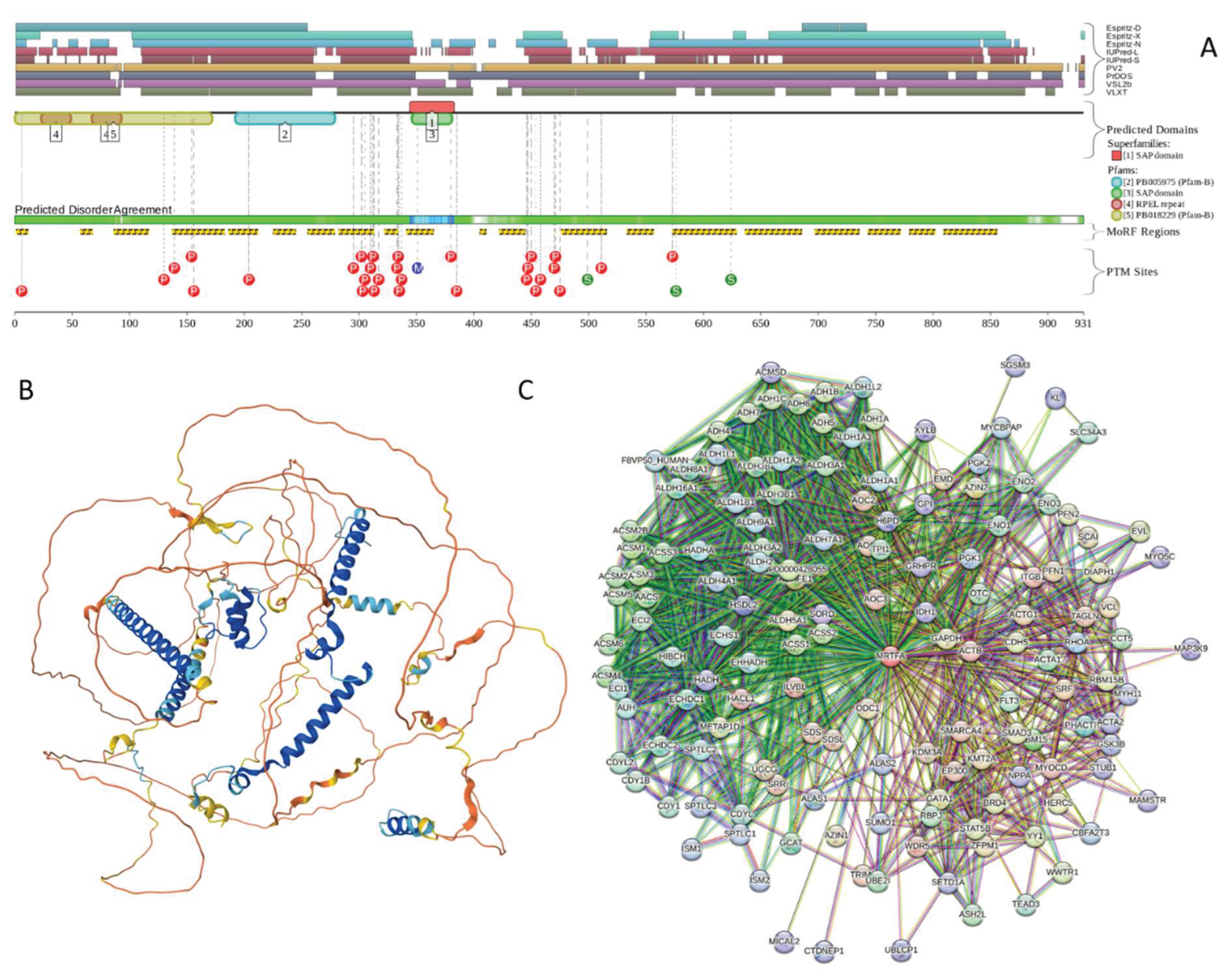
2.4.3. Myocardin-Related Transcription Factor A (MRTFA; UniProt ID: Q969V6)
2.4.4. ELKS/Rab6-Interacting/CAST Family Member 1 (ERC1; UniProt ID: Q8IUD2)
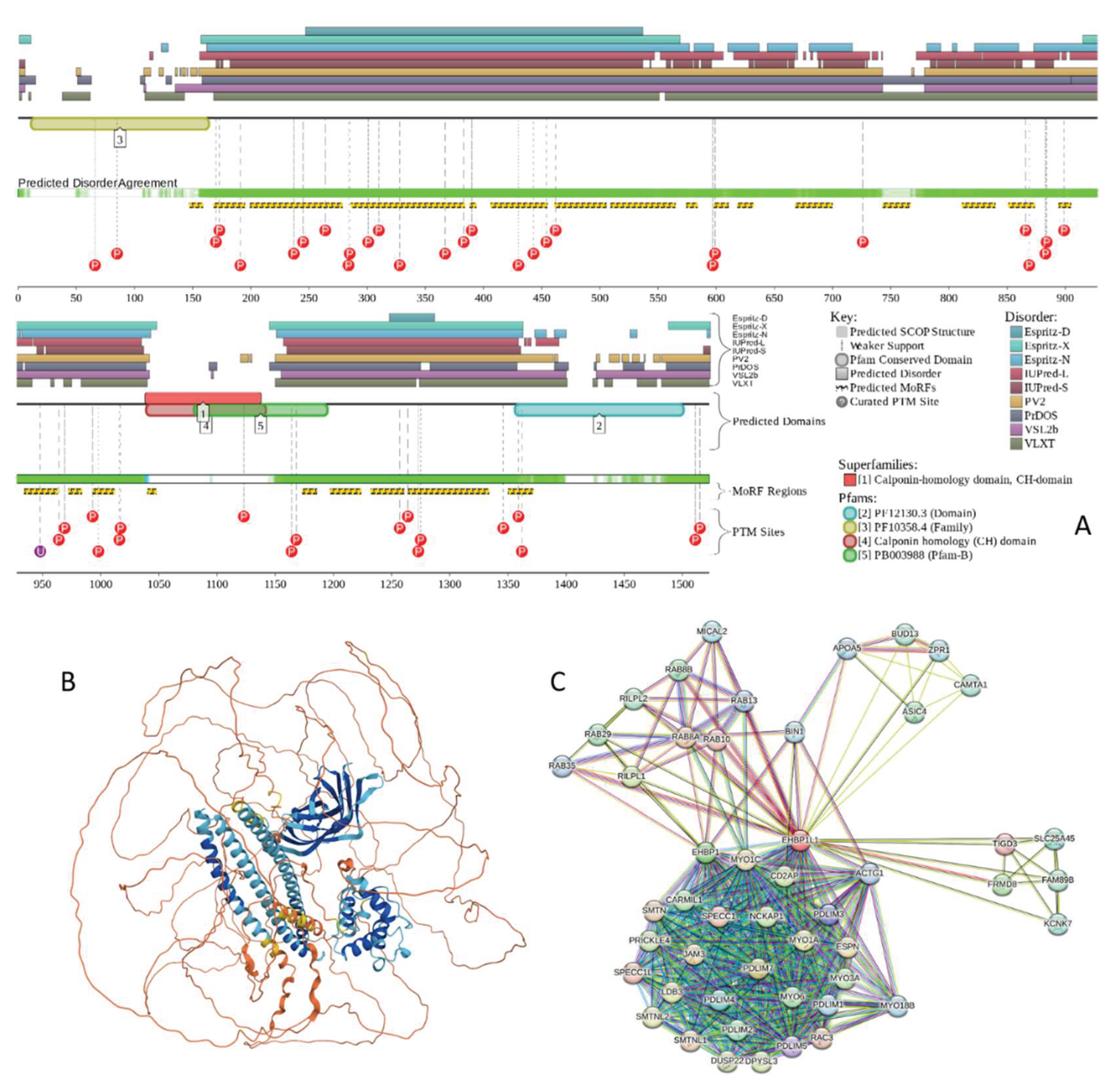
2.4.5. EH Domain-Binding Protein 1-Like Protein 1 (EHBP1L1; UniProt ID: Q8N3D4)
3. Materials and Methods
3.1. Protein Datasets
3.2. Bioinformatics Tools Utilized in This Study
3.2.1. STRING
3.2.2. RIDAO
3.2.3. IUPred
3.2.4. InterPro
3.2.5. FuzDrop
3.2.6. AlphaFold
3.2.7. D2P2
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, H.; Yesilyurt, H.G.; Yoon, J.; Terman, J.R. The MICALs are a family of F-actin dismantling oxidoreductases conserved from Drosophila to humans. Scientific reports 2018, 8, 937. [Google Scholar] [CrossRef] [PubMed]
- Vitali, T.; Maffioli, E.; Tedeschi, G.; Vanoni, M.A. Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and modulation by its CH, LIM and C-terminal domains. Arch Biochem Biophys 2016, 593, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Zucchini, D.; Caprini, G.; Pasterkamp, R.J.; Tedeschi, G.; Vanoni, M.A. Kinetic and spectroscopic characterization of the putative monooxygenase domain of human MICAL-1. Arch Biochem Biophys 2011, 515, 1–13. [Google Scholar] [CrossRef]
- Alto, L.T.; Terman, J.R. MICALs. Current Biology 2018, 28, R538–R541. [Google Scholar] [CrossRef]
- Giridharan, S.S.; Caplan, S. MICAL-family proteins: Complex regulators of the actin cytoskeleton. Antioxid Redox Signal 2014, 20, 2059–2073. [Google Scholar] [CrossRef]
- Rajan, S.; Terman, J.R.; Reisler, E. MICAL-mediated oxidation of actin and its effects on cytoskeletal and cellular dynamics. Frontiers in Cell and Developmental Biology 2023, 11, 1124202. [Google Scholar] [CrossRef]
- Grintsevich, E.E.; Ge, P.; Sawaya, M.R.; Yesilyurt, H.G.; Terman, J.R.; Zhou, Z.H.; Reisler, E. Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat Commun 2017, 8, 2183. [Google Scholar] [CrossRef]
- Manta, B.; Gladyshev, V.N. Regulated methionine oxidation by monooxygenases. Free Radic Biol Med 2017, 109, 141–155. [Google Scholar] [CrossRef]
- Fremont, S.; Romet-Lemonne, G.; Houdusse, A.; Echard, A. Emerging roles of MICAL family proteins - from actin oxidation to membrane trafficking during cytokinesis. J Cell Sci 2017, 130, 1509–1517. [Google Scholar] [CrossRef]
- McGarry, D.J.; Armstrong, G.; Castino, G.; Mason, S.; Clark, W.; Shaw, R.; McGarry, L.; Blyth, K.; Olson, M.F. MICAL1 regulates actin cytoskeleton organization, directional cell migration and the growth of human breast cancer cells as orthotopic xenograft tumours. Cancer Lett 2021, 519, 226–236. [Google Scholar] [CrossRef]
- Prifti, E.; Tsakiri, E.N.; Vourkou, E.; Stamatakis, G.; Samiotaki, M.; Skoulakis, E.M.C.; Papanikolopoulou, K. Mical modulates Tau toxicity via cysteine oxidation in vivo. Acta Neuropathol Commun 2022, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Beuchle, D.; Schwarz, H.; Langegger, M.; Koch, I.; Aberle, H. Drosophila MICAL regulates myofilament organization and synaptic structure. Mech Dev 2007, 124, 390–406. [Google Scholar] [CrossRef] [PubMed]
- Fischer, E. Einfluss der Configuration auf die Wirkung der Enzyme. Berichte der deutschen chemischen Gesellschaft 1894, 27, 2985–2993. [Google Scholar] [CrossRef]
- Lemieux, R.U.; Spohr, U. How Emil Fischer was led to the lock and key concept for enzyme specificity. Adv Carbohydr Chem Biochem 1994, 50, 1–20. [Google Scholar]
- Beadle, G.W.; Tatum, E.L. Genetic Control of Biochemical Reactions in Neurospora. Proc Natl Acad Sci U S A 1941, 27, 499–506. [Google Scholar] [CrossRef]
- Anfinsen, C.B. The formation and stabilization of protein structure. Biochem J 1972, 128, 737–749. [Google Scholar] [CrossRef]
- Anfinsen, C.B. Principles that govern the folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef]
- Anfinsen, C.B.; Haber, E.; Sela, M.; White, F.H., Jr. The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc Natl Acad Sci U S A 1961, 47, 1309–1314. [Google Scholar] [CrossRef]
- Dunker, A.K.; Babu, M.M.; Barbar, E.; Blackledge, M.; Bondos, S.E.; Dosztanyi, Z.; Dyson, H.J.; Forman-Kay, J.; Fuxreiter, M.; Gsponer, J.; et al. What's in a name? Why these proteins are intrinsically disordered: Why these proteins are intrinsically disordered. Intrinsically Disord Proteins 2013, 1, e24157. [Google Scholar] [CrossRef]
- Uversky, V.N.; Kulkarni, P. Intrinsically disordered proteins: Chronology of a discovery. Biophys Chem 2021, 279, 106694. [Google Scholar] [CrossRef]
- Ashraf, H.N.; Uversky, V.N. Intrinsic Disorder in the Host Proteins Entrapped in Rabies Virus Particles. Viruses 2024, 16, 916. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.M.; Verma, N.C.; Rao, C.; Uversky, V.N.; Nandi, C.K. Intrinsically disordered proteins of viruses: Involvement in the mechanism of cell regulation and pathogenesis. Prog Mol Biol Transl Sci 2020, 174, 1–78. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Paradoxes and wonders of intrinsic disorder: Stability of instability. Intrinsically Disord Proteins 2017, 5, e1327757. [Google Scholar] [CrossRef]
- Kc, S.; Nguyen, K.; Nicholson, V.; Walgren, A.; Trent, T.; Gollub, E.; Romero, S.; Holehouse, A.S.; Sukenik, S.; Boothby, T.C. Disordered proteins interact with the chemical environment to tune their protective function during drying. bioRxiv 2024. [Google Scholar] [CrossRef]
- Darling, A.L.; Uversky, V.N. Intrinsic Disorder and Posttranslational Modifications: The Darker Side of the Biological Dark Matter. Front Genet 2018, 9, 158. [Google Scholar] [CrossRef]
- Owen, I.; Shewmaker, F. The Role of Post-Translational Modifications in the Phase Transitions of Intrinsically Disordered Proteins. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef]
- McGarry, D.J.; Castino, G.; Lilla, S.; Carnet, A.; Kelly, L.; Micovic, K.; Zanivan, S.; Olson, M.F. MICAL1 activation by PAK1 mediates actin filament disassembly. Cell Rep 2022, 41, 111442. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, R.; Wang, Y.; Wang, Y.; Shuai, Y.; Ke, C.; Jin, R.; Wang, X.; Luo, J. Phosphorylation of MICAL2 by ARG promotes head and neck cancer tumorigenesis by regulating skeletal rearrangement. Oncogene 2022, 41, 334–346. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Brown, C.J.; Lawson, J.D.; Obradovic, Z.; Dunker, A.K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 2002, 323, 573–584. [Google Scholar] [CrossRef]
- Uversky, V.N. Targeting intrinsically disordered proteins in neurodegenerative and protein dysfunction diseases: another illustration of the D(2) concept. Expert Rev Proteomics 2010, 7, 543–564. [Google Scholar] [CrossRef]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys 2008, 37, 215–246. [Google Scholar] [CrossRef] [PubMed]
- Lucken-Ardjomande Hasler, S.; Vallis, Y.; Pasche, M.; McMahon, H.T. GRAF2, WDR44, and MICAL1 mediate Rab8/10/11-dependent export of E-cadherin, MMP14, and CFTR DeltaF508. J Cell Biol 2020, 219. [Google Scholar] [CrossRef] [PubMed]
- Alto, L.T.; Terman, J.R. Semaphorins and their Signaling Mechanisms. Methods Mol Biol 2017, 1493, 1–25. [Google Scholar] [CrossRef]
- Rai, A.; Oprisko, A.; Campos, J.; Fu, Y.; Friese, T.; Itzen, A.; Goody, R.S.; Gazdag, E.M.; Muller, M.P. bMERB domains are bivalent Rab8 family effectors evolved by gene duplication. Elife 2016, 5. [Google Scholar] [CrossRef]
- Horvath, M.; Schrofel, A.; Kowalska, K.; Sabo, J.; Vlasak, J.; Nourisanami, F.; Sobol, M.; Pinkas, D.; Knapp, K.; Koupilova, N.; et al. Structural basis of MICAL autoinhibition. Nat Commun 2024, 15, 9810. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Vendruscolo, M.; Fuxreiter, M. Sequence Determinants of the Aggregation of Proteins Within Condensates Generated by Liquid-liquid Phase Separation. J Mol Biol 2022, 434, 167201. [Google Scholar] [CrossRef]
- Horvath, A.; Miskei, M.; Ambrus, V.; Vendruscolo, M.; Fuxreiter, M. Sequence-based prediction of protein binding mode landscapes. PLoS Comput Biol 2020, 16, e1007864. [Google Scholar] [CrossRef]
- Miskei, M.; Horvath, A.; Vendruscolo, M.; Fuxreiter, M. Sequence-Based Prediction of Fuzzy Protein Interactions. J Mol Biol 2020, 432, 2289–2303. [Google Scholar] [CrossRef]
- Gerhard, D.S.; Wagner, L.; Feingold, E.A.; Shenmen, C.M.; Grouse, L.H.; Schuler, G.; Klein, S.L.; Old, S.; Rasooly, R.; Good, P.; et al. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC). Genome Res 2004, 14, 2121–2127. [Google Scholar] [CrossRef]
- Hattori, A.; Okumura, K.; Nagase, T.; Kikuno, R.; Hirosawa, M.; Ohara, O. Characterization of long cDNA clones from human adult spleen. DNA Res 2000, 7, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Ota, T.; Suzuki, Y.; Nishikawa, T.; Otsuki, T.; Sugiyama, T.; Irie, R.; Wakamatsu, A.; Hayashi, K.; Sato, H.; Nagai, K.; et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet 2004, 36, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.R.; Zaidi, S.; Fang, Y.Y.; Uversky, V.N.; Radivojac, P.; Oldfield, C.J.; Cortese, M.S.; Sickmeier, M.; LeGall, T.; Obradovic, Z.; et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci U S A 2006, 103, 8390–8395. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Matsuura, Y.; Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res 2025, 53, D672–D677. [Google Scholar] [CrossRef]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res 2024, 52, D672–D678. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Lundquist, M.R.; Storaska, A.J.; Liu, T.C.; Larsen, S.D.; Evans, T.; Neubig, R.R.; Jaffrey, S.R. Redox modification of nuclear actin by MICAL-2 regulates SRF signaling. Cell 2014, 156, 563–576. [Google Scholar] [CrossRef]
- Feuerstein, R.; Wang, X.; Song, D.; Cooke, N.E.; Liebhaber, S.A. The LIM/double zinc-finger motif functions as a protein dimerization domain. Proc Natl Acad Sci U S A 1994, 91, 10655–10659. [Google Scholar] [CrossRef]
- Olejniczak, M.; Storz, G. ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers? Mol Microbiol 2017, 104, 905–915. [Google Scholar] [CrossRef]
- Grigoriev, I.; Yu, K.L.; Martinez-Sanchez, E.; Serra-Marques, A.; Smal, I.; Meijering, E.; Demmers, J.; Peranen, J.; Pasterkamp, R.J.; van der Sluijs, P.; et al. Rab6, Rab8, and MICAL3 cooperate in controlling docking and fusion of exocytotic carriers. Curr Biol 2011, 21, 967–974. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, F.; Yu, K.L.; Tas, R.; Grigoriev, I.; Remmelzwaal, S.; Serra-Marques, A.; Kapitein, L.C.; Heck, A.J.; Akhmanova, A. MICAL3 Flavoprotein Monooxygenase Forms a Complex with Centralspindlin and Regulates Cytokinesis. J Biol Chem 2016, 291, 20617–20629. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Raicar, G.; Tsunoda, T.; Patil, A.; Sharma, A. OPAL: prediction of MoRF regions in intrinsically disordered protein sequences. Bioinformatics 2018, 34, 1850–1858. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Fukuda, M. Rab35 establishes the EHD1-association site by coordinating two distinct effectors during neurite outgrowth. J Cell Sci 2013, 126, 2424–2435. [Google Scholar] [CrossRef]
- Kobayashi, H.; Etoh, K.; Ohbayashi, N.; Fukuda, M. Rab35 promotes the recruitment of Rab8, Rab13 and Rab36 to recycling endosomes through MICAL-L1 during neurite outgrowth. Biol Open 2014, 3, 803–814. [Google Scholar] [CrossRef]
- Sikora, R.; Bun, P.; Danglot, L.; Alqabandi, M.; Bassereau, P.; Niedergang, F.; Galli, T.; Zahraoui, A. MICAL-L1 is required for cargo protein delivery to the cell surface. Biol Open 2021, 10. [Google Scholar] [CrossRef]
- Giridharan, S.S.; Cai, B.; Vitale, N.; Naslavsky, N.; Caplan, S. Cooperation of MICAL-L1, syndapin2, and phosphatidic acid in tubular recycling endosome biogenesis. Mol Biol Cell 2013, 24, 1776–1790. [Google Scholar] [CrossRef]
- Rahajeng, J.; Giridharan, S.S.; Naslavsky, N.; Caplan, S. Collapsin response mediator protein-2 (Crmp2) regulates trafficking by linking endocytic regulatory proteins to dynein motors. J Biol Chem 2010, 285, 31918–31922. [Google Scholar] [CrossRef]
- Sharma, M.; Giridharan, S.S.; Rahajeng, J.; Naslavsky, N.; Caplan, S. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol Biol Cell 2009, 20, 5181–5194. [Google Scholar] [CrossRef]
- Xie, S.; Farmer, T.; Naslavsky, N.; Caplan, S. MICAL-L1 coordinates ciliogenesis by recruiting EHD1 to the primary cilium. J Cell Sci 2019, 132. [Google Scholar] [CrossRef]
- Compeer, E.B.; Boes, M. MICAL-L1-related and unrelated mechanisms underlying elongated tubular endosomal network (ETEN) in human dendritic cells. Commun Integr Biol 2014, 7, e994969. [Google Scholar] [CrossRef]
- Abou-Zeid, N.; Pandjaitan, R.; Sengmanivong, L.; David, V.; Le Pavec, G.; Salamero, J.; Zahraoui, A. MICAL-like1 mediates epidermal growth factor receptor endocytosis. Mol Biol Cell 2011, 22, 3431–3441. [Google Scholar] [CrossRef] [PubMed]
- Nahm, M.; Park, S.; Lee, J.; Lee, S. MICAL-like Regulates Fasciclin II Membrane Cycling and Synaptic Development. Mol Cells 2016, 39, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Rahajeng, J.; Giridharan, S.S.; Cai, B.; Naslavsky, N.; Caplan, S. MICAL-L1 is a tubular endosomal membrane hub that connects Rab35 and Arf6 with Rab8a. Traffic 2012, 13, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Jovic, M.; Kieken, F.; Naslavsky, N.; Sorgen, P.L.; Caplan, S. Eps15 homology domain 1-associated tubules contain phosphatidylinositol-4-phosphate and phosphatidylinositol-(4,5)-bisphosphate and are required for efficient recycling. Mol Biol Cell 2009, 20, 2731–2743. [Google Scholar] [CrossRef]
- Braun, A.; Pinyol, R.; Dahlhaus, R.; Koch, D.; Fonarev, P.; Grant, B.D.; Kessels, M.M.; Qualmann, B. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol Biol Cell 2005, 16, 3642–3658. [Google Scholar] [CrossRef]
- Etoh, K.; Fukuda, M. Rab10 regulates tubular endosome formation through KIF13A and KIF13B motors. J Cell Sci 2019, 132. [Google Scholar] [CrossRef]
- Sakane, A.; Honda, K.; Sasaki, T. Rab13 regulates neurite outgrowth in PC12 cells through its effector protein, JRAB/MICAL-L2. Mol Cell Biol 2010, 30, 1077–1087. [Google Scholar] [CrossRef]
- Sakane, A.; Alamir Mahmoud Abdallah, A.; Nakano, K.; Honda, K.; Kitamura, T.; Imoto, I.; Matsushita, N.; Sasaki, T. Junctional Rab13-binding protein (JRAB) regulates cell spreading via filamins. Genes Cells 2013, 18, 810–822. [Google Scholar] [CrossRef]
- Nishimura, N.; Sasaki, T. Regulation of epithelial cell adhesion and repulsion: role of endocytic recycling. J Med Invest 2008, 55, 9–16. [Google Scholar] [CrossRef]
- Sakane, A.; Yoshizawa, S.; Nishimura, M.; Tsuchiya, Y.; Matsushita, N.; Miyake, K.; Horikawa, K.; Imoto, I.; Mizuguchi, C.; Saito, H.; et al. Conformational plasticity of JRAB/MICAL-L2 provides "law and order" in collective cell migration. Mol Biol Cell 2016, 27, 3095–3108. [Google Scholar] [CrossRef]
- Sakane, A.; Yano, T.A.; Uchihashi, T.; Horikawa, K.; Hara, Y.; Imoto, I.; Kurisu, S.; Yamada, H.; Takei, K.; Sasaki, T. JRAB/MICAL-L2 undergoes liquid-liquid phase separation to form tubular recycling endosomes. Commun Biol 2021, 4, 551. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, R.; Nishimura, N.; Nakatsuji, H.; Arase, S.; Sasaki, T. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol Biol Cell 2008, 19, 971–983. [Google Scholar] [CrossRef]
- Min, P.; Zhao, S.; Liu, L.; Zhang, Y.; Ma, Y.; Zhao, X.; Wang, Y.; Song, Y.; Zhu, C.; Jiang, H.; et al. MICAL-L2 potentiates Cdc42-dependent EGFR stability and promotes gastric cancer cell migration. J Cell Mol Med 2019, 23, 4475–4488. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.Y.; Zhang, W.M.; Yang, X.M.; Cui, L.; Li, J.; Zhang, Y.L.; Wang, Y.H.; Ao, J.P.; Ma, M.Z.; Lu, H.; et al. Silencing of MICAL-L2 suppresses malignancy of ovarian cancer by inducing mesenchymal-epithelial transition. Cancer Lett 2015, 363, 71–82. [Google Scholar] [CrossRef]
- Ioannou, M.S.; Bell, E.S.; Girard, M.; Chaineau, M.; Hamlin, J.N.; Daubaras, M.; Monast, A.; Park, M.; Hodgson, L.; McPherson, P.S. DENND2B activates Rab13 at the leading edge of migrating cells and promotes metastatic behavior. J Cell Biol 2015, 208, 629–648. [Google Scholar] [CrossRef]
- Min, P.; Zhang, L.; Wang, Y.; Qi, C.; Song, Y.; Bibi, M.; Zhang, Y.; Ma, Y.; Zhao, X.; Yu, M.; et al. MICAL-L2 Is Essential for c-Myc Deubiquitination and Stability in Non-small Cell Lung Cancer Cells. Front Cell Dev Biol 2020, 8, 575903. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, F.; Xia, T.; Wang, Q.; Zhang, Y.; Du, J. High MICAL-L2 expression and its role in the prognosis of colon adenocarcinoma. BMC Cancer 2022, 22, 487. [Google Scholar] [CrossRef]
- Castresana, J.; Saraste, M. Does Vav bind to F-actin through a CH domain? FEBS Lett 1995, 374, 149–151. [Google Scholar] [CrossRef]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J Cell Biochem 2011, 112, 3256–3267. [Google Scholar] [CrossRef]
- Uversky, V.N. Analyzing IDPs in interactomes. In Intrinsically Disordered Proteins, Kragelund, B.B., Skriver, K., Eds.; Humana New York, NY, 2020; Volume Methods in Molecular Biology, pp. 895-945.
- Mohammed, A.S.; Uversky, V.N. Intrinsic Disorder as a Natural Preservative: High Levels of Intrinsic Disorder in Proteins Found in the 2600-Year-Old Human Brain. Biology (Basel) 2022, 11. [Google Scholar] [CrossRef]
- Sun, X.; Xue, B.; Jones, W.T.; Rikkerink, E.; Dunker, A.K.; Uversky, V.N. A functionally required unfoldome from the plant kingdom: intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol Biol 2011, 77, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Oldfield, C.J.; Van, Y.Y.; Dunker, A.K.; Uversky, V.N. Protein intrinsic disorder and induced pluripotent stem cells. Mol Biosyst 2012, 8, 134–150. [Google Scholar] [CrossRef] [PubMed]
- Mohan, A.; Sullivan, W.J., Jr.; Radivojac, P.; Dunker, A.K.; Uversky, V.N. Intrinsic disorder in pathogenic and non-pathogenic microbes: discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol Biosyst 2008, 4, 328–340. [Google Scholar] [CrossRef]
- Huang, F.; Oldfield, C.; Meng, J.; Hsu, W.L.; Xue, B.; Uversky, V.N.; Romero, P.; Dunker, A.K. Subclassifying disordered proteins by the CH-CDF plot method. Pac Symp Biocomput 2012, 128–139. [Google Scholar]
- Hendrickx, A.; Beullens, M.; Ceulemans, H.; Den Abt, T.; Van Eynde, A.; Nicolaescu, E.; Lesage, B.; Bollen, M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol 2009, 16, 365–371. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, J.; Lu, W.; Li, Y.; Du, X.; Ning, T.; Lu, G.; Ke, Y. KIAA0649, a 1A6/DRIM-interacting protein with the oncogenic potential. Biochem Biophys Res Commun 2005, 334, 884–890. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, J.; Jin, H.; Sheng, S. Whole transcriptome sequencing identifies tumor-specific mutations in human oral squamous cell carcinoma. BMC Med Genomics 2013, 6, 28. [Google Scholar] [CrossRef]
- Yang, Y.; Ren, P.; Liu, X.; Sun, X.; Zhang, C.; Du, X.; Xing, B. PPP1R26 drives hepatocellular carcinoma progression by controlling glycolysis and epithelial-mesenchymal transition. J Exp Clin Cancer Res 2022, 41, 101. [Google Scholar] [CrossRef]
- Perez, Y.; Menascu, S.; Cohen, I.; Kadir, R.; Basha, O.; Shorer, Z.; Romi, H.; Meiri, G.; Rabinski, T.; Ofir, R.; et al. RSRC1 mutation affects intellect and behaviour through aberrant splicing and transcription, downregulating IGFBP3. Brain 2018, 141, 961–970. [Google Scholar] [CrossRef]
- Cazalla, D.; Newton, K.; Caceres, J.F. A novel SR-related protein is required for the second step of Pre-mRNA splicing. Mol Cell Biol 2005, 25, 2969–2980. [Google Scholar] [CrossRef]
- Chen, L.; Li, W.; Qiu, W.; Ren, W.; Li, Q.; Han, B.; Zhou, L.; Cheng, L.; Zhang, H.; Ye, Q. RSRC1 SUMOylation enhances SUMOylation and inhibits transcriptional activity of estrogen receptor beta. FEBS Lett 2015, 589, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- Record, J.; Malinova, D.; Zenner, H.L.; Plagnol, V.; Nowak, K.; Syed, F.; Bouma, G.; Curtis, J.; Gilmour, K.; Cale, C.; et al. Immunodeficiency and severe susceptibility to bacterial infection associated with a loss-of-function homozygous mutation of MKL1. Blood 2015, 126, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Brandt, D.T.; Baarlink, C.; Kitzing, T.M.; Kremmer, E.; Ivaska, J.; Nollau, P.; Grosse, R. SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of beta1-integrin. Nat Cell Biol 2009, 11, 557–568. [Google Scholar] [CrossRef]
- Osmanagic-Myers, S.; Kiss, A.; Manakanatas, C.; Hamza, O.; Sedlmayer, F.; Szabo, P.L.; Fischer, I.; Fichtinger, P.; Podesser, B.K.; Eriksson, M.; et al. Endothelial progerin expression causes cardiovascular pathology through an impaired mechanoresponse. J Clin Invest 2019, 129, 531–545. [Google Scholar] [CrossRef]
- Scharenberg, M.A.; Chiquet-Ehrismann, R.; Asparuhova, M.B. Megakaryoblastic leukemia protein-1 (MKL1): Increasing evidence for an involvement in cancer progression and metastasis. Int J Biochem Cell Biol 2010, 42, 1911–1914. [Google Scholar] [CrossRef]
- Mouilleron, S.; Langer, C.A.; Guettler, S.; McDonald, N.Q.; Treisman, R. Structure of a pentavalent G-actin*MRTF-A complex reveals how G-actin controls nucleocytoplasmic shuttling of a transcriptional coactivator. Sci Signal 2011, 4, ra40. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E.V. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci 2000, 25, 112–114. [Google Scholar] [CrossRef]
- Patwardhan, A.; Bardin, S.; Miserey-Lenkei, S.; Larue, L.; Goud, B.; Raposo, G.; Delevoye, C. Routing of the RAB6 secretory pathway towards the lysosome related organelle of melanocytes. Nat Commun 2017, 8, 15835. [Google Scholar] [CrossRef]
- Ducut Sigala, J.L.; Bottero, V.; Young, D.B.; Shevchenko, A.; Mercurio, F.; Verma, I.M. Activation of transcription factor NF-kappaB requires ELKS, an IkappaB kinase regulatory subunit. Science 2004, 304, 1963–1967. [Google Scholar] [CrossRef]
- Israel, A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb Perspect Biol 2010, 2, a000158. [Google Scholar] [CrossRef]
- Nakata, T.; Kitamura, Y.; Shimizu, K.; Tanaka, S.; Fujimori, M.; Yokoyama, S.; Ito, K.; Emi, M. Fusion of a novel gene, ELKS, to RET due to translocation t(10;12)(q11;p13) in a papillary thyroid carcinoma. Genes Chromosomes Cancer 1999, 25, 97–103. [Google Scholar] [CrossRef]
- Sala, K.; Corbetta, A.; Minici, C.; Tonoli, D.; Murray, D.H.; Cammarota, E.; Ribolla, L.; Ramella, M.; Fesce, R.; Mazza, D.; et al. The ERC1 scaffold protein implicated in cell motility drives the assembly of a liquid phase. Sci Rep 2019, 9, 13530. [Google Scholar] [CrossRef] [PubMed]
- Iwano, T.; Sobajima, T.; Takeda, S.; Harada, A.; Yoshimura, S.I. The Rab GTPase-binding protein EHBP1L1 and its interactors CD2AP/CIN85 negatively regulate the length of primary cilia via actin remodeling. J Biol Chem 2023, 299, 102985. [Google Scholar] [CrossRef] [PubMed]
- Nakajo, A.; Yoshimura, S.; Togawa, H.; Kunii, M.; Iwano, T.; Izumi, A.; Noguchi, Y.; Watanabe, A.; Goto, A.; Sato, T.; et al. EHBP1L1 coordinates Rab8 and Bin1 to regulate apical-directed transport in polarized epithelial cells. J Cell Biol 2016, 212, 297–306. [Google Scholar] [CrossRef]
- Wu, J.; Moriwaki, K.; Asuka, T.; Nakai, R.; Kanda, S.; Taniguchi, M.; Sugiyama, T.; Yoshimura, S.I.; Kunii, M.; Nagasawa, T.; et al. EHBP1L1, an apicobasal polarity regulator, is critical for nuclear polarization during enucleation of erythroblasts. Blood Adv 2023, 7, 3382–3394. [Google Scholar] [CrossRef]
- Kunii, M.; Harada, A. Molecular mechanisms of polarized transport to the apical plasma membrane. Front Cell Dev Biol 2024, 12, 1477173. [Google Scholar] [CrossRef]
- Pan, Y.; Shu, G.; Fu, L.; Huang, K.; Zhou, X.; Gui, C.; Liu, H.; Jin, X.; Chen, M.; Li, P.; et al. EHBP1L1 Drives Immune Evasion in Renal Cell Carcinoma through Binding and Stabilizing JAK1. Adv Sci (Weinh) 2023, 10, e2206792. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Dayhoff, G.W., 2nd; Uversky, V.N. Rapid prediction and analysis of protein intrinsic disorder. Protein Sci 2022, 31, e4496. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Oldfield, C.J.; Xue, B.; Hsu, W.L.; Meng, J.; Liu, X.; Shen, L.; Romero, P.; Uversky, V.N.; Dunker, A. Improving protein order-disorder classification using charge-hydropathy plots. BMC Bioinformatics 2014, 15 Suppl 17, S4. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol 2005, 347, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Meszaros, B.; Erdos, G.; Dosztanyi, Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Meszaros, B.; Simon, I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics 2009, 25, 2745–2746. [Google Scholar] [CrossRef]
- Meszaros, B.; Simon, I.; Dosztanyi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput Biol 2009, 5, e1000376. [Google Scholar] [CrossRef]
- Cheng, Y.; Oldfield, C.J.; Meng, J.; Romero, P.; Uversky, V.N.; Dunker, A.K. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry 2007, 46, 13468–13477. [Google Scholar] [CrossRef]
- Mohan, A.; Oldfield, C.J.; Radivojac, P.; Vacic, V.; Cortese, M.S.; Dunker, A.K.; Uversky, V.N. Analysis of molecular recognition features (MoRFs). J Mol Biol 2006, 362, 1043–1059. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Romero, P.; Uversky, V.N.; Dunker, A.K. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 2005, 44, 12454–12470. [Google Scholar] [CrossRef]
- Vacic, V.; Oldfield, C.J.; Mohan, A.; Radivojac, P.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res 2007, 6, 2351–2366. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Meng, J.; Yang, J.Y.; Yang, M.Q.; Uversky, V.N.; Dunker, A.K. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 2008, 9 Suppl 1, S1. [Google Scholar] [CrossRef]
- Hsu, W.L.; Oldfield, C.; Meng, J.; Huang, F.; Xue, B.; Uversky, V.N.; Romero, P.; Dunker, A.K. Intrinsic protein disorder and protein-protein interactions. Pac Symp Biocomput 2012, 116–127. [Google Scholar]
- Hsu, W.L.; Oldfield, C.J.; Xue, B.; Meng, J.; Huang, F.; Romero, P.; Uversky, V.N.; Dunker, A.K. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci 2013, 22, 258–273. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: the protein sequence classification resource in 2025. Nucleic Acids Res 2025, 53, D444–D456. [Google Scholar] [CrossRef]
- Hardenberg, M.; Horvath, A.; Ambrus, V.; Fuxreiter, M.; Vendruscolo, M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc Natl Acad Sci U S A 2020, 117, 33254–33262. [Google Scholar] [CrossRef]
- Hirose, T.; Ninomiya, K.; Nakagawa, S.; Yamazaki, T. A guide to membraneless organelles and their various roles in gene regulation. Nature Reviews Molecular Cell Biology 2023, 24, 288–304. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res 2024, 52, D368–D375. [Google Scholar] [CrossRef]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D(2)P(2): database of disordered protein predictions. Nucleic Acids Res 2013, 41, D508–516. [Google Scholar] [CrossRef]
- Li, X.; Romero, P.; Rani, M.; Dunker, A.K.; Obradovic, Z. Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform Ser Workshop Genome Inform 1999, 10, 30–40. [Google Scholar] [PubMed]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kinoshita, K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res 2007, 35, W460–464. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




