Submitted:
24 April 2024
Posted:
24 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
2.1. Intrinsic Disorder Status of Members of Human Synuclein Family
2.2. Effect of Familial Point Mutations on the Intrinsic Disorder Propensity of α-Synuclein
2.3. Intrinsic Disorder Potential of α-, β-, and γ-Synucleins from other Species
2.4. Functional Disorder Analysis of Human Proteins Engaged in Interaction with Members of Synuclein Family
2.5. Functionality of Disorder in 11 most Disordered Proteins from the Joint α-β-γ Synuclein Interactome
2.5.1. MT3 (Metallothionein-3; UniProt ID: P25713; PPIDRPONDR® VSL2 = 100.0%; ADS PONDR® VSL2 = 0.9952)
2.5.2. CHMP2B (Charged Multivesicular Body Protein 2b; UniProt ID: Q9UQN3; PPIDRPONDR® VSL2 = 100.0%; ADS PONDR® VSL2 = 0.8144)
2.5.3. NRGN (Neurogranin, UniProt ID: Q92686; PPIDRPONDR® VSL2 = 100.0%; ADS PONDR® VSL2 = 0.8643)
2.5.4. CPLX1 (Complexin-1; UniProt ID: O14810; PPIDRPONDR® VSL2 = 100.0%; ADS PONDR® VSL2 = 0.8819)
2.5.5. CPLX2 (Complexin-2; UniProt ID: Q6PUV4; PPIDRPONDR® VSL2 = 100.0%; ADS PONDR® VSL2 = 0.9135)
2.5.6. NUCKS1 (Nuclear Ubiquitous Casein and Cyclin-Dependent Kinase Substrate 1; UniProt ID: Q9H1E3; PPIDRPONDR® VSL2 = 100.0%; ADS PONDR® VSL2 = 0.9879)
2.5.7. MBP (Myelin Basic Protein; UniProt ID: P02686; PPIDRPONDR® VSL2 = 100.0%; ADS PONDR® VSL2 = 0.8706)
2.5.8. CAST (Calpastatin; UniProt ID: P20810; PPIDRPONDR® VSL2 = 100.0%; ADS PONDR® VSL2 = 0.9547)
2.5.9. MAPT (Microtubule-Associated Protein tau; UniProt ID: P10636; PPIDRPONDR® VSL2 = 99.1%; ADS PONDR® VSL2 = 0.8612)
2.5.10. HEMGN (Hemogen; UniProt ID: Q9BXL5; PPIDRPONDR® VSL2 = 98.3%; ADS PONDR® VSL2 = 0.8304)
2.5.11. H1.2 (Histone H1.2; UniProt ID: P16403; PPIDRPONDR® VSL2 = 97.7%; ADS PONDR® VSL2 = 0.8947)
3. Materials and Methods
3.1. Overview
3.2. Sequence and Structure-Based Analysis
3.3. Disorder-Based Analysis of the Interactomes of Human Synucleins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Uversky, V.N. Looking at the recent advances in understanding alpha-synuclein and its aggregation through the proteoform prism. F1000Res 2017, 6, 525. [Google Scholar] [CrossRef] [PubMed]
- Stefanis, L. alpha-Synuclein in Parkinson’s disease. Cold Spring Harb Perspect Med 2012, 2, a009399. [Google Scholar] [CrossRef]
- Weinreb, P.H.; Zhen, W.; Poon, A.W.; Conway, K.A.; Lansbury, P.T., Jr. NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry 1996, 35, 13709–13715. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Fink, A.L. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J Biol Chem 2001, 276, 10737–10744. [Google Scholar] [CrossRef] [PubMed]
- Eliezer, D.; Kutluay, E.; Bussell, R., Jr.; Browne, G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol 2001, 307, 1061–1073. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Souillac, P.; Millett, I.S.; Doniach, S.; Jakes, R.; Goedert, M.; Fink, A.L. Biophysical properties of the synucleins and their propensities to fibrillate: Inhibition of alpha-synuclein assembly by beta- and gamma-synucleins. J Biol Chem 2002, 277, 11970–11978. [Google Scholar] [CrossRef]
- Uversky, V.N. A protein-chameleon: Conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J Biomol Struct Dyn 2003, 21, 211–234. [Google Scholar] [CrossRef]
- Sung, Y.H.; Eliezer, D. Secondary structure and dynamics of micelle bound beta- and gamma-synuclein. Protein Sci 2006, 15, 1162–1174. [Google Scholar] [CrossRef]
- Sung, Y.H.; Eliezer, D. Residual structure, backbone dynamics, and interactions within the synuclein family. J Mol Biol 2007, 372, 689–707. [Google Scholar] [CrossRef]
- Binolfi, A.; Theillet, F.X.; Selenko, P. Bacterial in-cell NMR of human alpha-synuclein: A disordered monomer by nature? Biochem Soc Trans 2012, 40, 950–954. [Google Scholar] [CrossRef]
- Limatola, A.; Eichmann, C.; Jacob, R.S.; Ben-Nissan, G.; Sharon, M.; Binolfi, A.; Selenko, P. Time-Resolved NMR Analysis of Proteolytic alpha-Synuclein Processing in vitro and in cellulo. Proteomics 2018, 18, e1800056. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.; Schneider, R.; Cantrelle, F.X.; Huvent, I.; Lippens, G. Studying Intrinsically Disordered Proteins under True In Vivo Conditions by Combined Cross-Polarization and Carbonyl-Detection NMR Spectroscopy. Angew Chem Int Ed Engl 2016, 55, 7418–7422. [Google Scholar] [CrossRef] [PubMed]
- Sciolino, N.; Burz, D.S.; Shekhtman, A. In-Cell NMR Spectroscopy of Intrinsically Disordered Proteins. Proteomics 2019, 19, e1800055. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.E.; Zhou, L.Z.; Pielak, G.J. Hydrogen exchange of disordered proteins in Escherichia coli. Protein Sci 2015, 24, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Theillet, F.X.; Binolfi, A.; Bekei, B.; Martorana, A.; Rose, H.M.; Stuiver, M.; Verzini, S.; Lorenz, D.; van Rossum, M.; Goldfarb, D.; et al. Structural disorder of monomeric alpha-synuclein persists in mammalian cells. Nature 2016, 530, 45–50. [Google Scholar] [CrossRef]
- Waudby, C.A.; Camilloni, C.; Fitzpatrick, A.W.; Cabrita, L.D.; Dobson, C.M.; Vendruscolo, M.; Christodoulou, J. In-cell NMR characterization of the secondary structure populations of a disordered conformation of alpha-synuclein within E. coli cells. PLoS ONE 2013, 8, e72286. [Google Scholar] [CrossRef] [PubMed]
- Galvin, J.E.; Lee, V.M.; Trojanowski, J.Q. Synucleinopathies: Clinical and pathological implications. Arch Neurol 2001, 58, 186–190. [Google Scholar] [CrossRef]
- Goedert, M. Filamentous nerve cell inclusions in neurodegenerative diseases: Tauopathies and alpha-synucleinopathies. Philos Trans R Soc Lond B Biol Sci 1999, 354, 1101–1118. [Google Scholar] [CrossRef]
- Goedert, M. Alpha-synuclein and neurodegenerative diseases. Nat Rev Neurosci 2001, 2, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M. Parkinson’s disease and other alpha-synucleinopathies. Clin Chem Lab Med 2001, 39, 308–312. [Google Scholar] [CrossRef]
- Goedert, M.; Falcon, B.; Clavaguera, F.; Tolnay, M. Prion-like mechanisms in the pathogenesis of tauopathies and synucleinopathies. Curr Neurol Neurosci Rep 2014, 14, 495. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Jakes, R.; Spillantini, M.G. The Synucleinopathies: Twenty Years On. J Parkinsons Dis 2017, 7, S51–S69. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Goedert, M. The alpha-synucleinopathies: Parkinson’s disease, dementia with Lewy bodies, and multiple system atrophy. Ann N Y Acad Sci 2000, 920, 16–27. [Google Scholar] [CrossRef] [PubMed]
- McKeith, I.G.; Dickson, D.W.; Lowe, J.; Emre, M.; O’Brien, J.T.; Feldman, H.; Cummings, J.; Duda, J.E.; Lippa, C.; Perry, E.K.; et al. Diagnosis and management of dementia with Lewy bodies: Third report of the DLB Consortium. Neurology 2005, 65, 1863–1872. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, K.; Yoshimoto, M.; Tsuji, S.; Takahashi, H. Alpha-synuclein immunoreactivity in glial cytoplasmic inclusions in multiple system atrophy. Neurosci Lett 1998, 249, 180–182. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Crowther, R.A.; Jakes, R.; Cairns, N.J.; Lantos, P.L.; Goedert, M. Filamentous alpha-synuclein inclusions link multiple system atrophy with Parkinson’s disease and dementia with Lewy bodies. Neurosci Lett 1998, 251, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Gai, W.P.; Power, J.H.; Blumbergs, P.C.; Blessing, W.W. Multiple-system atrophy: A new alpha-synuclein disease? Lancet 1998, 352, 547–548. [Google Scholar] [CrossRef]
- Trojanowski, J.Q.; Goedert, M.; Iwatsubo, T.; Lee, V.M. Fatal attractions: Abnormal protein aggregation and neuron death in Parkinson’s disease and Lewy body dementia. Cell Death Differ 1998, 5, 832–837. [Google Scholar] [CrossRef]
- Takeda, A.; Mallory, M.; Sundsmo, M.; Honer, W.; Hansen, L.; Masliah, E. Abnormal accumulation of NACP/alpha-synuclein in neurodegenerative disorders. Am J Pathol 1998, 152, 367–372. [Google Scholar]
- Lucking, C.B.; Brice, A. Alpha-synuclein and Parkinson’s disease. Cell Mol Life Sci 2000, 57, 1894–1908. [Google Scholar] [CrossRef]
- Arawaka, S.; Saito, Y.; Murayama, S.; Mori, H. Lewy body in neurodegeneration with brain iron accumulation type 1 is immunoreactive for alpha-synuclein. Neurology 1998, 51, 887–889. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. Alpha-synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, K.; Matsumoto, K.; Takayama, K.; Yoshimoto, M.; Takahashi, H. NACP, a presynaptic protein, immunoreactivity in Lewy bodies in Parkinson’s disease. Neurosci Lett 1997, 239, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Burre, J.; Sharma, M.; Sudhof, T.C. Cell Biology and Pathophysiology of alpha-Synuclein. Cold Spring Harb Perspect Med 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G. Synucleinopathies and tauopathies. In Basic Neurochemistry; Elsevier: 2012; pp. 829–843.
- Surguchov, A.; Surguchev, A. Synucleins: New Data on Misfolding, Aggregation and Role in Diseases. Biomedicines 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Crowther, R.A.; Jakes, R.; Hasegawa, M.; Goedert, M. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with lewy bodies. Proc Natl Acad Sci U S A 1998, 95, 6469–6473. [Google Scholar] [CrossRef] [PubMed]
- Trojanowski, J.Q.; Lee, V.M. Parkinson’s disease and related alpha-synucleinopathies are brain amyloidoses. Ann N Y Acad Sci 2003, 991, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Lundvig, D.; Lindersson, E.; Jensen, P.H. Pathogenic effects of alpha-synuclein aggregation. Brain Res Mol Brain Res 2005, 134, 3–17. [Google Scholar] [PubMed]
- Kosaka, K. Lewy bodies in cerebral cortex, report of three cases. Acta Neuropathol (Berl) 1978, 42, 127–134. [Google Scholar] [CrossRef]
- Kosaka, K.; Mehraein, P. Dementia-Parkinsonism syndrome with numerous Lewy bodies and senile plaques in cerebral cortex. Arch Psychiatr Nervenkr 1979, 226, 241–250. [Google Scholar] [CrossRef]
- Seidel, K.; Mahlke, J.; Siswanto, S.; Kruger, R.; Heinsen, H.; Auburger, G.; Bouzrou, M.; Grinberg, L.T.; Wicht, H.; Korf, H.W.; et al. The brainstem pathologies of Parkinson’s disease and dementia with Lewy bodies. Brain Pathol 2015, 25, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; Bagic, A. Olfactory pathogenesis of idiopathic Parkinson disease revisited. Mov Disord 2008, 23, 1076–1084. [Google Scholar] [CrossRef] [PubMed]
- Visanji, N.P.; Brooks, P.L.; Hazrati, L.N.; Lang, A.E. The prion hypothesis in Parkinson’s disease: Braak to the future. Acta Neuropathol Commun 2013, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Melki, R. Role of Different Alpha-Synuclein Strains in Synucleinopathies, Similarities with other Neurodegenerative Diseases. J Parkinsons Dis 2015, 5, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Peelaerts, W.; Bousset, L.; Van der Perren, A.; Moskalyuk, A.; Pulizzi, R.; Giugliano, M.; Van den Haute, C.; Melki, R.; Baekelandt, V. alpha-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature 2015, 522, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Tofaris, G.K.; Spillantini, M.G. Physiological and pathological properties of alpha-synuclein. Cell Mol Life Sci 2007, 64, 2194–2201. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Uversky, V.N.; Fink, A.L. Effect of familial Parkinson’s disease point mutations A30P and A53T on the structural properties, aggregation, and fibrillation of human alpha-synuclein. Biochemistry 2001, 40, 11604–11613. [Google Scholar] [CrossRef] [PubMed]
- Conway, K.A.; Harper, J.D.; Lansbury, P.T. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med 1998, 4, 1318–1320. [Google Scholar] [CrossRef] [PubMed]
- Conway, K.A.; Harper, J.D.; Lansbury, P.T., Jr. Fibrils formed in vitro from alpha-synuclein and two mutant forms linked to Parkinson’s disease are typical amyloid. Biochemistry 2000, 39, 2552–2563. [Google Scholar] [CrossRef]
- Conway, K.A.; Lee, S.J.; Rochet, J.C.; Ding, T.T.; Williamson, R.E.; Lansbury, P.T., Jr. Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson’s disease: Implications for pathogenesis and therapy. Proc Natl Acad Sci U S A 2000, 97, 571–576. [Google Scholar] [CrossRef]
- Lashuel, H.A.; Hartley, D.; Petre, B.M.; Walz, T.; Lansbury, P.T., Jr. Neurodegenerative disease: Amyloid pores from pathogenic mutations. Nature 2002, 418, 291. [Google Scholar] [CrossRef] [PubMed]
- Lashuel, H.A.; Petre, B.M.; Wall, J.; Simon, M.; Nowak, R.J.; Walz, T.; Lansbury, P.T., Jr. Alpha-synuclein, especially the Parkinson’s disease-associated mutants, forms pore-like annular and tubular protofibrils. J Mol Biol 2002, 322, 1089–1102. [Google Scholar] [CrossRef] [PubMed]
- Proukakis, C.; Dudzik, C.G.; Brier, T.; MacKay, D.S.; Cooper, J.M.; Millhauser, G.L.; Houlden, H.; Schapira, A.H. A novel alpha-synuclein missense mutation in Parkinson disease. Neurology 2013, 80, 1062–1064. [Google Scholar] [CrossRef]
- Appel-Cresswell, S.; Vilarino-Guell, C.; Encarnacion, M.; Sherman, H.; Yu, I.; Shah, B.; Weir, D.; Thompson, C.; Szu-Tu, C.; Trinh, J.; et al. Alpha-synuclein p.H50Q, a novel pathogenic mutation for Parkinson’s disease. Mov Disord 2013, 28, 811–813. [Google Scholar] [CrossRef]
- Khalaf, O.; Fauvet, B.; Oueslati, A.; Dikiy, I.; Mahul-Mellier, A.L.; Ruggeri, F.S.; Mbefo, M.K.; Vercruysse, F.; Dietler, G.; Lee, S.J.; et al. The H50Q mutation enhances alpha-synuclein aggregation, secretion, and toxicity. J Biol Chem 2014, 289, 21856–21876. [Google Scholar] [CrossRef]
- Dev, K.K.; Hofele, K.; Barbieri, S.; Buchman, V.L.; van der Putten, H. Part II: Alpha-synuclein and its molecular pathophysiological role in neurodegenerative disease. Neuropharmacology 2003, 45, 14–44. [Google Scholar] [CrossRef] [PubMed]
- da Costa, C.A.; Ancolio, K.; Checler, F. Wild-type but not Parkinson’s disease-related ala-53 --> Thr mutant alpha -synuclein protects neuronal cells from apoptotic stimuli. J Biol Chem 2000, 275, 24065–24069. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Fink, A.L. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinson’s disease and heavy metal exposure. J Biol Chem 2001, 276, 44284–44296. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Uversky, V.N. Metalloproteomics and metal toxicology of alpha-synuclein. Metallomics 2010, 2, 378–392. [Google Scholar] [CrossRef]
- Ahmad, A.; Burns, C.S.; Fink, A.L.; Uversky, V.N. Peculiarities of copper binding to alpha-synuclein. J Biomol Struct Dyn 2012, 29, 825–842. [Google Scholar] [CrossRef]
- Carboni, E.; Lingor, P. Insights on the interaction of alpha-synuclein and metals in the pathophysiology of Parkinson’s disease. Metallomics 2015, 7, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Bower, K.; Fink, A.L. Synergistic effects of pesticides and metals on the fibrillation of alpha-synuclein: Implications for Parkinson’s disease. Neurotoxicology 2002, 23, 527–536. [Google Scholar] [CrossRef]
- Uversky, V.N.; Li, J.; Fink, A.L. Pesticides directly accelerate the rate of alpha-synuclein fibril formation: A possible factor in Parkinson’s disease. FEBS Lett 2001, 500, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Maturana, M.G.; Pinheiro, A.S.; de Souza, T.L.; Follmer, C. Unveiling the role of the pesticides paraquat and rotenone on alpha-synuclein fibrillation in vitro. Neurotoxicology 2015, 46, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Ottolini, D.; Cali, T.; Szabo, I.; Brini, M. Alpha-synuclein at the intracellular and the extracellular side: Functional and dysfunctional implications. Biol Chem 2017, 398, 77–100. [Google Scholar] [CrossRef] [PubMed]
- Emanuele, M.; Chieregatti, E. Mechanisms of alpha-synuclein action on neurotransmission: Cell-autonomous and non-cell autonomous role. Biomolecules 2015, 5, 865–892. [Google Scholar] [CrossRef]
- Uversky, V.N. Alpha-synuclein misfolding and neurodegenerative diseases. Curr Protein Pept Sci 2008, 9, 507–540. [Google Scholar] [CrossRef]
- Payton, J.E.; Perrin, R.J.; Clayton, D.F.; George, J.M. Protein-protein interactions of alpha-synuclein in brain homogenates and transfected cells. Brain Res Mol Brain Res 2001, 95, 138–145. [Google Scholar] [CrossRef]
- Jin, J.; Li, G.J.; Davis, J.; Zhu, D.; Wang, Y.; Pan, C.; Zhang, J. Identification of novel proteins interacting with both a-synuclein and DJ-1. Mol Cell Proteomics 2006. [Google Scholar]
- Iwai, A.; Masliah, E.; Yoshimoto, M.; Ge, N.; Flanagan, L.; de Silva, H.A.; Kittel, A.; Saitoh, T. The precursor protein of non-A beta component of Alzheimer’s disease amyloid is a presynaptic protein of the central nervous system. Neuron 1995, 14, 467–475. [Google Scholar] [CrossRef]
- Hayashi, J.; Carver, J.A. beta-Synuclein: An Enigmatic Protein with Diverse Functionality. Biomolecules 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Liu, Y.E.; Jia, T.; Wang, M.; Liu, J.; Xiao, G.; Joseph, B.K.; Rosen, C.; Shi, Y.E. Identification of a breast cancer-specific gene, BCSG1, by direct differential cDNA sequencing. Cancer Res 1997, 57, 759–764. [Google Scholar] [PubMed]
- Dunker, A.K.; Obradovic, Z.; Romero, P.; Garner, E.C.; Brown, C.J. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform 2000, 11, 161–171. [Google Scholar] [PubMed]
- Uversky, V.N. The mysterious unfoldome: Structureless, underappreciated, yet vital part of any given proteome. J Biomed Biotechnol 2010, 2010, 568068. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 2004, 337, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Xue, B.; Dunker, A.K.; Uversky, V.N. Orderly order in protein intrinsic disorder distribution: Disorder in thirty five hundred proteomes from viruses and the three domains of life. J Biomol Struct Dyn In press. 2012. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Garner, E.; Guilliot, S.; Romero, P.; Albrecht, K.; Hart, J.; Obradovic, Z.; Kissinger, C.; Villafranca, J.E. Protein disorder and the evolution of molecular recognition: Theory, predictions and observations. Pac Symp Biocomput 1998, 473–484. [Google Scholar]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J Mol Biol 1999, 293, 321–331. [Google Scholar] [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J Mol Graph Model 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem Sci 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Daughdrill, G.W.; Pielak, G.J.; Uversky, V.N.; Cortese, M.S.; Dunker, A.K. Natively disordered proteins. In Handbook of Protein Folding; Buchner, J., Kiefhaber, T., Eds.; Wiley-VCH, Verlag GmbH & Co. KGaA: Weinheim, Germany, 2005; pp. 271–353. [Google Scholar]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim Biophys Acta 2010, 1804, 1231–1264. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Obradovic, Z. The protein trinity--linking function and disorder. Nat Biotechnol 2001, 19, 805–806. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci 2002, 11, 739–756. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta 2013, 1834, 932–951. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsic disorder-based protein interactions and their modulators. Curr Pharm Des 2013, 19, 4191–4213. [Google Scholar] [CrossRef]
- Uversky, V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J 2015, 282, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. p53 Proteoforms and Intrinsic Disorder: An Illustration of the Protein Structure-Function Continuum Concept. Int J Mol Sci 2016, 17, 1874. [Google Scholar] [CrossRef]
- Uversky, V.N. Protein intrinsic disorder and structure-function continuum. Prog Mol Biol Transl Sci 2019, 166, 1–17. [Google Scholar] [CrossRef]
- Fonin, A.V.; Darling, A.L.; Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. Multi-functionality of proteins involved in GPCR and G protein signaling: Making sense of structure-function continuum with intrinsic disorder-based proteoforms. Cell Mol Life Sci 2019, 76, 4461–4492. [Google Scholar] [CrossRef]
- Uversky, V.N. p53 Proteoforms and Intrinsic Disorder: An Illustration of the Protein Structure-Function Continuum Concept. Int J Mol Sci 2016, 17. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Brown, C.J.; Lawson, J.D.; Obradovic, Z.; Dunker, A.K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 2002, 323, 573–584. [Google Scholar] [CrossRef]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets: The roles of intrinsic disorder in protein interaction networks. FEBS Journal 2005, 272, 5129–5148. [Google Scholar] [CrossRef]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Showing your ID: Intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit 2005, 18, 343–384. [Google Scholar] [CrossRef] [PubMed]
- Radivojac, P.; Iakoucheva, L.M.; Oldfield, C.J.; Obradovic, Z.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder and functional proteomics. Biophys J 2007, 92, 1439–1456. [Google Scholar] [CrossRef] [PubMed]
- Vucetic, S.; Xie, H.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J Proteome Res 2007, 6, 1899–1916. [Google Scholar] [CrossRef]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N.; Obradovic, Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res 2007, 6, 1882–1898. [Google Scholar] [CrossRef]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res 2007, 6, 1917–1932. [Google Scholar] [CrossRef] [PubMed]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing protein intrinsic disorder. Chem Rev 2014, 114, 6561–6588. [Google Scholar] [CrossRef]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem Rev 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Uversky, V.N. Multitude of binding modes attainable by intrinsically disordered proteins: A portrait gallery of disorder-based complexes. Chem Soc Rev 2011, 40, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Pejaver, V.; Hsu, W.L.; Xin, F.; Dunker, A.K.; Uversky, V.N.; Radivojac, P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci 2014, 23, 1077–1093. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Uversky, V.N. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv Colloid Interface Sci 2017, 239, 97–114. [Google Scholar] [CrossRef]
- Uversky, V.N. Recent Developments in the Field of Intrinsically Disordered Proteins: Intrinsic Disorder–Based Emergence in Cellular Biology in Light of the Physiological and Pathological Liquid–Liquid Phase Transitions. Annual Review of Biophysics 2021, 50, 135–156. [Google Scholar] [CrossRef] [PubMed]
- Mohan, A.; Sullivan, W.J., Jr.; Radivojac, P.; Dunker, A.K.; Uversky, V.N. Intrinsic disorder in pathogenic and non-pathogenic microbes: Discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol Biosyst 2008, 4, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Mok, K.H.; Muhandiram, R.; Park, K.H.; Suk, J.E.; Kim, D.H.; Chang, J.; Sung, Y.C.; Choi, K.Y.; Han, K.H. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem 2000, 275, 29426–29432. [Google Scholar] [CrossRef] [PubMed]
- Adkins, J.N.; Lumb, K.J. Intrinsic structural disorder and sequence features of the cell cycle inhibitor p57Kip2. Proteins 2002, 46, 1–7. [Google Scholar] [CrossRef]
- Chang, B.S.; Minn, A.J.; Muchmore, S.W.; Fesik, S.W.; Thompson, C.B. Identification of a novel regulatory domain in Bcl-X(L) and Bcl-2. EMBO J 1997, 16, 968–977. [Google Scholar] [CrossRef]
- Campbell, K.M.; Terrell, A.R.; Laybourn, P.J.; Lumb, K.J. Intrinsic structural disorder of the C-terminal activation domain from the bZIP transcription factor Fos. Biochemistry 2000, 39, 2708–2713. [Google Scholar] [CrossRef]
- Sunde, M.; McGrath, K.C.; Young, L.; Matthews, J.M.; Chua, E.L.; Mackay, J.P.; Death, A.K. TC-1 is a novel tumorigenic and natively disordered protein associated with thyroid cancer. Cancer Res 2004, 64, 2766–2773. [Google Scholar] [CrossRef] [PubMed]
- Glenner, G.G.; Wong, C.W. Alzheimer’s disease and Down’s syndrome: Sharing of a unique cerebrovascular amyloid fibril protein. Biochem Biophys Res Commun 1984, 122, 1131–1135. [Google Scholar] [CrossRef] [PubMed]
- Masters, C.L.; Multhaup, G.; Simms, G.; Pottgiesser, J.; Martins, R.N.; Beyreuther, K. Neuronal origin of a cerebral amyloid: Neurofibrillary tangles of Alzheimer’s disease contain the same protein as the amyloid of plaque cores and blood vessels. EMBO J 1985, 4, 2757–2763. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.M.; Balin, B.J.; Otvos, L., Jr.; Trojanowski, J.Q. A68: A major subunit of paired helical filaments and derivatized forms of normal Tau. Science 1991, 251, 675–678. [Google Scholar] [CrossRef]
- Ueda, K.; Fukushima, H.; Masliah, E.; Xia, Y.; Iwai, A.; Yoshimoto, M.; Otero, D.A.; Kondo, J.; Ihara, Y.; Saitoh, T. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci U S A 1993, 90, 11282–11286. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, K.E.; Dalton, A.J.; McLachlan, C.; Wen, G.Y.; Wisniewski, H.M. Alzheimer’s disease in Down’s syndrome: Clinicopathologic studies. Neurology 1985, 35, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B. Shattuck lecture--neurodegenerative diseases and prions. N Engl J Med 2001, 344, 1516–1526. [Google Scholar] [CrossRef]
- Zoghbi, H.Y.; Orr, H.T. Polyglutamine diseases: Protein cleavage and aggregation. Curr Opin Neurobiol 1999, 9, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; LeGall, T.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry 2006, 45, 10448–10460. [Google Scholar] [CrossRef]
- Uversky, V.N. Amyloidogenesis of natively unfolded proteins. Curr Alzheimer Res 2008, 5, 260–287. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Midic, U.; Xie, H.; Xue, B.; Vucetic, S.; Iakoucheva, L.M.; Obradovic, Z.; Dunker, A.K. Unfoldomics of human diseases: Linking protein intrinsic disorder with diseases. BMC Genomics 2009, 10 Suppl 1, S7. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci 2009, 14, 5188–5238. [Google Scholar] [CrossRef] [PubMed]
- Midic, U.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Protein disorder in the human diseasome: Unfoldomics of human genetic diseases. PLoS Computational Biology In press. 2008. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Fuxreiter, M.; Oldfield, C.J.; Simon, I.; Dunker, A.K.; Uversky, V.N. Close encounters of the third kind: Disordered domains and the interactions of proteins. Bioessays 2009, 31, 328–335. [Google Scholar] [CrossRef] [PubMed]
- George, J.M.; Jin, H.; Woods, W.S.; Clayton, D.F. Characterization of a novel protein regulated during the critical period for song learning in the zebra finch. Neuron 1995, 15, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Tanner, C.M. Is the cause of Parkinson’s disease environmental or hereditary? Evidence from twin studies. Adv Neurol 2003, 91, 133–142. [Google Scholar] [PubMed]
- Farrer, M.J. Genetics of Parkinson disease: Paradigm shifts and future prospects. Nat Rev Genet 2006, 7, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Olanow, C.W.; Tatton, W.G. Etiology and pathogenesis of Parkinson’s disease. Annu Rev Neurosci 1999, 22, 123–144. [Google Scholar] [CrossRef]
- Moghal, S.; Rajput, A.H.; D’Arcy, C.; Rajput, R. Prevalence of movement disorders in elderly community residents. Neuroepidemiology 1994, 13, 175–178. [Google Scholar] [CrossRef]
- Fahn, S. Description of Parkinson’s disease as a clinical syndrome. Ann N Y Acad Sci 2003, 991, 1–14. [Google Scholar] [CrossRef]
- Mizuno, Y.; Hattori, N.; Kitada, T.; Matsumine, H.; Mori, H.; Shimura, H.; Kubo, S.; Kobayashi, H.; Asakawa, S.; Minoshima, S.; et al. Familial Parkinson’s disease. Alpha-synuclein and parkin. Adv Neurol 2001, 86, 13–21. [Google Scholar]
- Van Den Eeden, S.K.; Tanner, C.M.; Bernstein, A.L.; Fross, R.D.; Leimpeter, A.; Bloch, D.A.; Nelson, L.M. Incidence of Parkinson’s disease: Variation by age, gender, and race/ethnicity. Am J Epidemiol 2003, 157, 1015–1022. [Google Scholar] [CrossRef]
- Forno, L.S. Neuropathology of Parkinson’s disease. J Neuropathol Exp Neurol 1996, 55, 259–272. [Google Scholar] [CrossRef]
- Zarranz, J.J.; Alegre, J.; Gomez-Esteban, J.C.; Lezcano, E.; Ros, R.; Ampuero, I.; Vidal, L.; Hoenicka, J.; Rodriguez, O.; Atares, B.; et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol 2004, 55, 164–173. [Google Scholar] [CrossRef]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R.; et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef]
- Kruger, R.; Kuhn, W.; Muller, T.; Woitalla, D.; Graeber, M.; Kosel, S.; Przuntek, H.; Epplen, J.T.; Schols, L.; Riess, O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet 1998, 18, 106–108. [Google Scholar] [CrossRef]
- Singleton, A.; Gwinn-Hardy, K.; Sharabi, Y.; Li, S.T.; Holmes, C.; Dendi, R.; Hardy, J.; Crawley, A.; Goldstein, D.S. Association between cardiac denervation and parkinsonism caused by alpha-synuclein gene triplication. Brain 2004, 127, 768–772. [Google Scholar] [CrossRef]
- Singleton, A.B.; Farrer, M.; Johnson, J.; Singleton, A.; Hague, S.; Kachergus, J.; Hulihan, M.; Peuralinna, T.; Dutra, A.; Nussbaum, R.; et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science 2003, 302, 841. [Google Scholar] [CrossRef] [PubMed]
- Farrer, M.; Kachergus, J.; Forno, L.; Lincoln, S.; Wang, D.S.; Hulihan, M.; Maraganore, D.; Gwinn-Hardy, K.; Wszolek, Z.; Dickson, D.; et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann Neurol 2004, 55, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Morar, A.S.; Olteanu, A.; Young, G.B.; Pielak, G.J. Solvent-induced collapse of alpha-synuclein and acid-denatured cytochrome c. Protein Sci 2001, 10, 2195–2199. [Google Scholar] [CrossRef]
- Bussell, R., Jr.; Eliezer, D. Residual structure and dynamics in Parkinson’s disease-associated mutants of alpha-synuclein. J Biol Chem 2001, 276, 45996–46003. [Google Scholar] [CrossRef]
- Dedmon, M.M.; Lindorff-Larsen, K.; Christodoulou, J.; Vendruscolo, M.; Dobson, C.M. Mapping long-range interactions in alpha-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J Am Chem Soc 2005, 127, 476–477. [Google Scholar] [CrossRef]
- Bertoncini, C.W.; Jung, Y.S.; Fernandez, C.O.; Hoyer, W.; Griesinger, C.; Jovin, T.M.; Zweckstetter, M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc Natl Acad Sci U S A 2005, 102, 1430–1435. [Google Scholar] [CrossRef]
- Hardenberg, M.; Horvath, A.; Ambrus, V.; Fuxreiter, M.; Vendruscolo, M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc Natl Acad Sci U S A 2020, 117, 33254–33262. [Google Scholar] [CrossRef]
- Vendruscolo, M.; Fuxreiter, M. Sequence Determinants of the Aggregation of Proteins Within Condensates Generated by Liquid-liquid Phase Separation. J Mol Biol 2022, 434, 167201. [Google Scholar] [CrossRef]
- Hardenberg, M.C.; Sinnige, T.; Casford, S.; Dada, S.T.; Poudel, C.; Robinson, E.A.; Fuxreiter, M.; Kaminksi, C.F.; Kaminski Schierle, G.S.; Nollen, E.A.A.; et al. Observation of an alpha-synuclein liquid droplet state and its maturation into Lewy body-like assemblies. J Mol Cell Biol 2021, 13, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Mo, X.; Wang, J.; Ye, X.; Yu, H.; Liu, Y. alpha-Synuclein phase separation and amyloid aggregation are modulated by C-terminal truncations. FEBS Lett 2022, 596, 1388–1400. [Google Scholar] [CrossRef]
- Huang, S.; Xu, B.; Liu, Y. Calcium promotes alpha-synuclein liquid-liquid phase separation to accelerate amyloid aggregation. Biochem Biophys Res Commun 2022, 603, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Singh, N.; Kumar, R.; Patel, K.; Pandey, S.; Datta, D.; Mahato, J.; Panigrahi, R.; Navalkar, A.; Mehra, S.; et al. alpha-Synuclein aggregation nucleates through liquid-liquid phase separation. Nat Chem 2020, 12, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Sawner, A.S.; Ray, S.; Yadav, P.; Mukherjee, S.; Panigrahi, R.; Poudyal, M.; Patel, K.; Ghosh, D.; Kummerant, E.; Kumar, A.; et al. Modulating alpha-Synuclein Liquid-Liquid Phase Separation. Biochemistry 2021, 60, 3676–3696. [Google Scholar] [CrossRef]
- Vendruscolo, M.; Fuxreiter, M. Protein condensation diseases: Therapeutic opportunities. Nat Commun 2022, 13, 5550. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Chen, X.; Rizo, J.; Jahn, R.; Sudhof, T.C. A broken alpha -helix in folded alpha -Synuclein. J Biol Chem 2003, 278, 15313–15318. [Google Scholar] [CrossRef] [PubMed]
- Davidson, W.S.; Jonas, A.; Clayton, D.F.; George, J.M. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem 1998, 273, 9443–9449. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Fink, A.L. Lipid binding inhibits alpha-synuclein fibril formation. J Biol Chem 2003, 278, 16873–16877. [Google Scholar] [CrossRef] [PubMed]
- Munishkina, L.A.; Phelan, C.; Uversky, V.N.; Fink, A.L. Conformational behavior and aggregation of alpha-synuclein in organic solvents: Modeling the effects of membranes. Biochemistry 2003, 42, 2720–2730. [Google Scholar] [CrossRef] [PubMed]
- Rao, J.N.; Jao, C.C.; Hegde, B.G.; Langen, R.; Ulmer, T.S. A combinatorial NMR and EPR approach for evaluating the structural ensemble of partially folded proteins. J Am Chem Soc 2010, 132, 8657–8668. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, T.S.; Bax, A.; Cole, N.B.; Nussbaum, R.L. Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem 2005, 280, 9595–9603. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.A.; Singh, V.K.; Jia, Z.; Forman-Kay, J.D. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Protein Sci 2006, 15, 2795–2804. [Google Scholar] [CrossRef] [PubMed]
- Kruger, R.; Kuhn, W.; Muller, T.; Woitalla, D.; Graeber, M.; Kosel, S.; Przuntek, H.; Epplen, J.T.; Schols, L.; Riess, O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet 1998, 18, 106–108. [Google Scholar] [CrossRef]
- Zarranz, J.J.; Alegre, J.; Gomez-Esteban, J.C.; Lezcano, E.; Ros, R.; Ampuero, I.; Vidal, L.; Hoenicka, J.; Rodriguez, O.; Atares, B.; et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol 2004, 55, 164–173. [Google Scholar] [CrossRef]
- Kiely, A.P.; Asi, Y.T.; Kara, E.; Limousin, P.; Ling, H.; Lewis, P.; Proukakis, C.; Quinn, N.; Lees, A.J.; Hardy, J.; et al. alpha-Synucleinopathy associated with G51D SNCA mutation: A link between Parkinson’s disease and multiple system atrophy? Acta Neuropathol 2013, 125, 753–769. [Google Scholar] [CrossRef]
- Lesage, S.; Anheim, M.; Letournel, F.; Bousset, L.; Honore, A.; Rozas, N.; Pieri, L.; Madiona, K.; Durr, A.; Melki, R.; et al. G51D alpha-synuclein mutation causes a novel parkinsonian-pyramidal syndrome. Ann Neurol 2013, 73, 459–471. [Google Scholar] [CrossRef]
- Pasanen, P.; Myllykangas, L.; Siitonen, M.; Raunio, A.; Kaakkola, S.; Lyytinen, J.; Tienari, P.J.; Poyhonen, M.; Paetau, A. Novel alpha-synuclein mutation A53E associated with atypical multiple system atrophy and Parkinson’s disease-type pathology. Neurobiol Aging 2014, 35, 2180–e2181. [Google Scholar] [CrossRef]
- Lemkau, L.R.; Comellas, G.; Kloepper, K.D.; Woods, W.S.; George, J.M.; Rienstra, C.M. Mutant protein A30P alpha-synuclein adopts wild-type fibril structure, despite slower fibrillation kinetics. J Biol Chem 2012, 287, 11526–11532. [Google Scholar] [CrossRef]
- Fredenburg, R.A.; Rospigliosi, C.; Meray, R.K.; Kessler, J.C.; Lashuel, H.A.; Eliezer, D.; Lansbury, P.T., Jr. The impact of the E46K mutation on the properties of alpha-synuclein in its monomeric and oligomeric states. Biochemistry 2007, 46, 7107–7118. [Google Scholar] [CrossRef]
- Pandey, N.; Schmidt, R.E.; Galvin, J.E. The alpha-synuclein mutation E46K promotes aggregation in cultured cells. Exp Neurol 2006, 197, 515–520. [Google Scholar] [CrossRef]
- Rutherford, N.J.; Moore, B.D.; Golde, T.E.; Giasson, B.I. Divergent effects of the H50Q and G51D SNCA mutations on the aggregation of alpha-synuclein. J Neurochem 2014, 131, 859–867. [Google Scholar] [CrossRef]
- Ghosh, D.; Sahay, S.; Ranjan, P.; Salot, S.; Mohite, G.M.; Singh, P.K.; Dwivedi, S.; Carvalho, E.; Banerjee, R.; Kumar, A.; et al. The newly discovered Parkinson’s disease associated Finnish mutation (A53E) attenuates alpha-synuclein aggregation and membrane binding. Biochemistry 2014, 53, 6419–6421. [Google Scholar] [CrossRef]
- Rutherford, N.J.; Giasson, B.I. The A53E alpha-synuclein pathological mutation demonstrates reduced aggregation propensity in vitro and in cell culture. Neurosci Lett 2015, 597, 43–48. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J Cell Biochem 2011, 112, 3256–3267. [Google Scholar] [CrossRef]
- Uversky, V.N. Analyzing IDPs in interactomes. In Intrinsically Disordered Proteins, Kragelund, B.B., Skriver, K., Eds.; Humana New York, NY, 2020; Volume Methods in Molecular Biology, pp. 895–945.
- Sun, X.; Xue, B.; Jones, W.T.; Rikkerink, E.; Dunker, A.K.; Uversky, V.N. A functionally required unfoldome from the plant kingdom: Intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol Biol 2011, 77, 205–223. [Google Scholar] [CrossRef]
- Xue, B.; Oldfield, C.J.; Van, Y.Y.; Dunker, A.K.; Uversky, V.N. Protein intrinsic disorder and induced pluripotent stem cells. Mol Biosyst 2012, 8, 134–150. [Google Scholar] [CrossRef]
- Huang, F.; Oldfield, C.; Meng, J.; Hsu, W.L.; Xue, B.; Uversky, V.N.; Romero, P.; Dunker, A.K. Subclassifying disordered proteins by the CH-CDF plot method. Pac Symp Biocomput 2012, 128–139. [Google Scholar]
- Vasak, M.; Hasler, D.W. Metallothioneins: New functional and structural insights. Curr Opin Chem Biol 2000, 4, 177–183. [Google Scholar] [CrossRef]
- Quaife, C.J.; Findley, S.D.; Erickson, J.C.; Froelick, G.J.; Kelly, E.J.; Zambrowicz, B.P.; Palmiter, R.D. Induction of a new metallothionein isoform (MT-IV) occurs during differentiation of stratified squamous epithelia. Biochemistry 1994, 33, 7250–7259. [Google Scholar] [CrossRef]
- Moffatt, P.; Denizeau, F. Metallothionein in physiological and physiopathological processes. Drug Metab Rev 1997, 29, 261–307. [Google Scholar] [CrossRef]
- Ding, Z.C.; Zheng, Q.; Cai, B.; Ni, F.Y.; Yu, W.H.; Teng, X.C.; Gao, Y.; Liu, F.; Chen, D.; Wang, Y.; et al. Study on structure-property-reactivity-function relationship of human neuronal growth inhibitory factor (hGIF). J Inorg Biochem 2008, 102, 1965–1972. [Google Scholar] [CrossRef]
- Bogumil, R.; Faller, P.; Binz, P.A.; Vasak, M.; Charnock, J.M.; Garner, C.D. Structural characterization of Cu(I) and Zn(II) sites in neuronal-growth-inhibitory factor by extended X-ray absorption fine structure (EXAFS). Eur J Biochem 1998, 255, 172–177. [Google Scholar] [CrossRef]
- Sewell, A.K.; Jensen, L.T.; Erickson, J.C.; Palmiter, R.D.; Winge, D.R. Bioactivity of metallothionein-3 correlates with its novel beta domain sequence rather than metal binding properties. Biochemistry 1995, 34, 4740–4747. [Google Scholar] [CrossRef]
- Hasler, D.W.; Jensen, L.T.; Zerbe, O.; Winge, D.R.; Vasak, M. Effect of the two conserved prolines of human growth inhibitory factor (metallothionein-3) on its biological activity and structure fluctuation: Comparison with a mutant protein. Biochemistry 2000, 39, 14567–14575. [Google Scholar] [CrossRef]
- Romero-Isart, N.; Jensen, L.T.; Zerbe, O.; Winge, D.R.; Vasak, M. Engineering of metallothionein-3 neuroinhibitory activity into the inactive isoform metallothionein-1. J Biol Chem 2002, 277, 37023–37028. [Google Scholar] [CrossRef]
- Uchida, Y.; Takio, K.; Titani, K.; Ihara, Y.; Tomonaga, M. The growth inhibitory factor that is deficient in the Alzheimer’s disease brain is a 68 amino acid metallothionein-like protein. Neuron 1991, 7, 337–347. [Google Scholar] [CrossRef]
- Vasak, M.; Meloni, G. Mammalian Metallothionein-3: New Functional and Structural Insights. Int J Mol Sci 2017, 18. [Google Scholar] [CrossRef]
- Jiang, Z.; Shen, B.; Xiang, J. Metal-dependent interactions of metallothionein-3 beta-domain with amyloid-beta peptide and related physiological implications. J Inorg Biochem 2019, 196, 110693. [Google Scholar] [CrossRef]
- Koh, J.Y.; Lee, S.J. Metallothionein-3 as a multifunctional player in the control of cellular processes and diseases. Mol Brain 2020, 13, 116. [Google Scholar] [CrossRef]
- Uchida, Y. Growth-inhibitory factor, metallothionein-like protein, and neurodegenerative diseases. Biol Signals 1994, 3, 211–215. [Google Scholar] [CrossRef]
- Howells, C.; West, A.K.; Chung, R.S. Neuronal growth-inhibitory factor (metallothionein-3): Evaluation of the biological function of growth-inhibitory factor in the injured and neurodegenerative brain. FEBS J 2010, 277, 2931–2939. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Q.; Cai, B.; Li, H.; Sze, K.H.; Huang, Z.X.; Wu, H.M.; Sun, H. Solution structure and dynamics of human metallothionein-3 (MT-3). FEBS Lett 2006, 580, 795–800. [Google Scholar] [CrossRef]
- Oz, G.; Zangger, K.; Armitage, I.M. Three-dimensional structure and dynamics of a brain specific growth inhibitory factor: Metallothionein-3. Biochemistry 2001, 40, 11433–11441. [Google Scholar] [CrossRef]
- Yuan, A.T.; Korkola, N.C.; Stillman, M.J. Apo-metallothionein-3 cooperatively forms tightly compact structures under physiological conditions. J Biol Chem 2023, 299, 102899. [Google Scholar] [CrossRef]
- Meszaros, B.; Erdos, G.; Dosztanyi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Pountney, D.L.; Dickson, T.C.; Power, J.H.; Vickers, J.C.; West, A.J.; Gai, W.P. Association of metallothionein-III with oligodendroglial cytoplasmic inclusions in multiple system atrophy. Neurotox Res 2011, 19, 115–122. [Google Scholar] [CrossRef]
- Ugbode, C.; West, R.J.H. Lessons learned from CHMP2B, implications for frontotemporal dementia and amyotrophic lateral sclerosis. Neurobiol Dis 2021, 147, 105144. [Google Scholar] [CrossRef]
- Rusten, T.E.; Stenmark, H. How do ESCRT proteins control autophagy? J Cell Sci 2009, 122, 2179–2183. [Google Scholar] [CrossRef]
- Bhutta, M.S.; McInerny, C.J.; Gould, G.W. ESCRT function in cytokinesis: Location, dynamics and regulation by mitotic kinases. Int J Mol Sci 2014, 15, 21723–21739. [Google Scholar] [CrossRef]
- Caballe, A.; Martin-Serrano, J. ESCRT machinery and cytokinesis: The road to daughter cell separation. Traffic 2011, 12, 1318–1326. [Google Scholar] [CrossRef]
- Radulovic, M.; Schink, K.O.; Wenzel, E.M.; Nahse, V.; Bongiovanni, A.; Lafont, F.; Stenmark, H. ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J 2018, 37. [Google Scholar] [CrossRef]
- Lata, S.; Schoehn, G.; Solomons, J.; Pires, R.; Gottlinger, H.G.; Weissenhorn, W. Structure and function of ESCRT-III. Biochem Soc Trans 2009, 37, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Krasniak, C.S.; Ahmad, S.T. The role of CHMP2B(Intron5) in autophagy and frontotemporal dementia. Brain Res 2016, 1649, 151–157. [Google Scholar] [CrossRef]
- Skibinski, G.; Parkinson, N.J.; Brown, J.M.; Chakrabarti, L.; Lloyd, S.L.; Hummerich, H.; Nielsen, J.E.; Hodges, J.R.; Spillantini, M.G.; Thusgaard, T.; et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet 2005, 37, 806–808. [Google Scholar] [CrossRef]
- Bugiani, O. The many ways to frontotemporal degeneration and beyond. Neurol Sci 2007, 28, 241–244. [Google Scholar] [CrossRef]
- Urwin, H.; Ghazi-Noori, S.; Collinge, J.; Isaacs, A. The role of CHMP2B in frontotemporal dementia. Biochem Soc Trans 2009, 37, 208–212. [Google Scholar] [CrossRef]
- Siuda, J.; Fujioka, S.; Wszolek, Z.K. Parkinsonian syndrome in familial frontotemporal dementia. Parkinsonism Relat Disord 2014, 20, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Babst, M.; Wendland, B.; Estepa, E.J.; Emr, S.D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J 1998, 17, 2982–2993. [Google Scholar] [CrossRef] [PubMed]
- Bodon, G.; Chassefeyre, R.; Pernet-Gallay, K.; Martinelli, N.; Effantin, G.; Hulsik, D.L.; Belly, A.; Goldberg, Y.; Chatellard-Causse, C.; Blot, B.; et al. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem 2011, 286, 40276–40286. [Google Scholar] [CrossRef] [PubMed]
- Stuchell-Brereton, M.D.; Skalicky, J.J.; Kieffer, C.; Karren, M.A.; Ghaffarian, S.; Sundquist, W.I. ESCRT-III recognition by VPS4 ATPases. Nature 2007, 449, 740–744. [Google Scholar] [CrossRef]
- Xiang, Y.; Xin, J.; Le, W.; Yang, Y. Neurogranin: A Potential Biomarker of Neurological and Mental Diseases. Front Aging Neurosci 2020, 12, 584743. [Google Scholar] [CrossRef]
- Represa, A.; Deloulme, J.C.; Sensenbrenner, M.; Ben-Ari, Y.; Baudier, J. Neurogranin: Immunocytochemical localization of a brain-specific protein kinase C substrate. J Neurosci 1990, 10, 3782–3792. [Google Scholar] [CrossRef]
- Chen, S.J.; Klann, E.; Gower, M.C.; Powell, C.M.; Sessoms, J.S.; Sweatt, J.D. Studies with synthetic peptide substrates derived from the neuronal protein neurogranin reveal structural determinants of potency and selectivity for protein kinase C. Biochemistry 1993, 32, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Baudier, J.; Deloulme, J.C.; Van Dorsselaer, A.; Black, D.; Matthes, H.W. Purification and characterization of a brain-specific protein kinase C substrate, neurogranin (p17). Identification of a consensus amino acid sequence between neurogranin and neuromodulin (GAP43) that corresponds to the protein kinase C phosphorylation site and the calmodulin-binding domain. J Biol Chem 1991, 266, 229–237. [Google Scholar] [PubMed]
- Gerendasy, D.D.; Herron, S.R.; Watson, J.B.; Sutcliffe, J.G. Mutational and biophysical studies suggest RC3/neurogranin regulates calmodulin availability. J Biol Chem 1994, 269, 22420–22426. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, B.; Morley, P.; Whitfield, J. Ca2+-calmodulin and protein kinase Cs: A hypothetical synthesis of their conflicting convergences on shared substrate domains. Trends Neurosci 1999, 22, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Gerendasy, D. Homeostatic tuning of Ca2+ signal transduction by members of the calpacitin protein family. J Neurosci Res 1999, 58, 107–119. [Google Scholar] [CrossRef]
- Fyfe, I. Alzheimer disease: Neurogranin in the CSF signals early Alzheimer disease and predicts disease progression. Nat Rev Neurol 2015, 11, 609. [Google Scholar] [CrossRef]
- Hellwig, K.; Kvartsberg, H.; Portelius, E.; Andreasson, U.; Oberstein, T.J.; Lewczuk, P.; Blennow, K.; Kornhuber, J.; Maler, J.M.; Zetterberg, H.; et al. Neurogranin and YKL-40: Independent markers of synaptic degeneration and neuroinflammation in Alzheimer’s disease. Alzheimers Res Ther 2015, 7, 74. [Google Scholar] [CrossRef]
- Kester, M.I.; Teunissen, C.E.; Crimmins, D.L.; Herries, E.M.; Ladenson, J.H.; Scheltens, P.; van der Flier, W.M.; Morris, J.C.; Holtzman, D.M.; Fagan, A.M. Neurogranin as a Cerebrospinal Fluid Biomarker for Synaptic Loss in Symptomatic Alzheimer Disease. JAMA Neurol 2015, 72, 1275–1280. [Google Scholar] [CrossRef]
- Tarawneh, R.; D’Angelo, G.; Crimmins, D.; Herries, E.; Griest, T.; Fagan, A.M.; Zipfel, G.J.; Ladenson, J.H.; Morris, J.C.; Holtzman, D.M. Diagnostic and Prognostic Utility of the Synaptic Marker Neurogranin in Alzheimer Disease. JAMA Neurol 2016, 73, 561–571. [Google Scholar] [CrossRef]
- Portelius, E.; Zetterberg, H.; Skillback, T.; Tornqvist, U.; Andreasson, U.; Trojanowski, J.Q.; Weiner, M.W.; Shaw, L.M.; Mattsson, N.; Blennow, K.; et al. Cerebrospinal fluid neurogranin: Relation to cognition and neurodegeneration in Alzheimer’s disease. Brain 2015, 138, 3373–3385. [Google Scholar] [CrossRef]
- Kvartsberg, H.; Duits, F.H.; Ingelsson, M.; Andreasen, N.; Ohrfelt, A.; Andersson, K.; Brinkmalm, G.; Lannfelt, L.; Minthon, L.; Hansson, O.; et al. Cerebrospinal fluid levels of the synaptic protein neurogranin correlates with cognitive decline in prodromal Alzheimer’s disease. Alzheimers Dement 2015, 11, 1180–1190. [Google Scholar] [CrossRef] [PubMed]
- Portelius, E.; Olsson, B.; Hoglund, K.; Cullen, N.C.; Kvartsberg, H.; Andreasson, U.; Zetterberg, H.; Sandelius, A.; Shaw, L.M.; Lee, V.M.Y.; et al. Cerebrospinal fluid neurogranin concentration in neurodegeneration: Relation to clinical phenotypes and neuropathology. Acta Neuropathol 2018, 136, 363–376. [Google Scholar] [CrossRef]
- Blennow, K.; Diaz-Lucena, D.; Zetterberg, H.; Villar-Pique, A.; Karch, A.; Vidal, E.; Hermann, P.; Schmitz, M.; Ferrer Abizanda, I.; Zerr, I.; et al. CSF neurogranin as a neuronal damage marker in CJD: A comparative study with AD. J Neurol Neurosurg Psychiatry 2019, 90, 846–853. [Google Scholar] [CrossRef] [PubMed]
- Hodges, A.; Strand, A.D.; Aragaki, A.K.; Kuhn, A.; Sengstag, T.; Hughes, G.; Elliston, L.A.; Hartog, C.; Goldstein, D.R.; Thu, D.; et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet 2006, 15, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Runne, H.; Kuhn, A.; Wild, E.J.; Pratyaksha, W.; Kristiansen, M.; Isaacs, J.D.; Regulier, E.; Delorenzi, M.; Tabrizi, S.J.; Luthi-Carter, R. Analysis of potential transcriptomic biomarkers for Huntington’s disease in peripheral blood. Proc Natl Acad Sci U S A 2007, 104, 14424–14429. [Google Scholar] [CrossRef] [PubMed]
- Lista, S.; Santos-Lozano, A.; Emanuele, E.; Mercuri, N.B.; Gabelle, A.; Lopez-Ortiz, S.; Martin-Hernandez, J.; Maisto, N.; Imbimbo, C.; Caraci, F.; et al. Monitoring synaptic pathology in Alzheimer’s disease through fluid and PET imaging biomarkers: A comprehensive review and future perspectives. Mol Psychiatry 2024. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Lu, Q.; Kang, H.; Suridjan, I.; Kollmorgen, G.; Wild, N.; Deming, Y.; Van Hulle, C.A.; Anderson, R.M.; Zetterberg, H.; et al. CSF metabolites associated with biomarkers of Alzheimer’s disease pathology. Front Aging Neurosci 2023, 15, 1214932. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Gobom, J.; Sjodin, S.; Brinkmalm, G.; Ashton, N.J.; Svensson, J.; Johansson, P.; Portelius, E.; Zetterberg, H.; Blennow, K.; et al. Cerebrospinal fluid biomarker panel for synaptic dysfunction in Alzheimer’s disease. Alzheimers Dement (Amst) 2021, 13, e12179. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, T.; Blandino, V.; Maniscalco, L.; Matranga, D.; Graziano, F.; Guajana, F.; Agnello, L.; Lo Sasso, B.; Gambino, C.M.; Giglio, R.V.; et al. Biomarkers Related to Synaptic Dysfunction to Discriminate Alzheimer’s Disease from Other Neurological Disorders. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.K.; Chien, W.J.; Arunkumar, A.I. Conformation of a protein kinase C substrate NG(28-43), and its analog in aqueous and sodium dodecyl sulfate micelle solutions. Biophys J 1997, 72, 554–566. [Google Scholar] [CrossRef]
- Ran, X.; Miao, H.H.; Sheu, F.S.; Yang, D. Structural and dynamic characterization of a neuron-specific protein kinase C substrate, neurogranin. Biochemistry 2003, 42, 5143–5150. [Google Scholar] [CrossRef]
- Ishizuka, T.; Saisu, H.; Odani, S.; Abe, T. Synaphin: A protein associated with the docking/fusion complex in presynaptic terminals. Biochem Biophys Res Commun 1995, 213, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- McMahon, H.T.; Missler, M.; Li, C.; Sudhof, T.C. Complexins: Cytosolic proteins that regulate SNAP receptor function. Cell 1995, 83, 111–119. [Google Scholar] [CrossRef]
- Takahashi, S.; Yamamoto, H.; Matsuda, Z.; Ogawa, M.; Yagyu, K.; Taniguchi, T.; Miyata, T.; Kaba, H.; Higuchi, T.; Okutani, F.; et al. Identification of two highly homologous presynaptic proteins distinctly localized at the dendritic and somatic synapses. FEBS Lett 1995, 368, 455–460. [Google Scholar] [CrossRef]
- Ono, S.; Baux, G.; Sekiguchi, M.; Fossier, P.; Morel, N.F.; Nihonmatsu, I.; Hirata, K.; Awaji, T.; Takahashi, S.; Takahashi, M. Regulatory roles of complexins in neurotransmitter release from mature presynaptic nerve terminals. Eur J Neurosci 1998, 10, 2143–2152. [Google Scholar] [CrossRef]
- Yamada, M.; Saisu, H.; Ishizuka, T.; Takahashi, H.; Abe, T. Immunohistochemical distribution of the two isoforms of synaphin/complexin involved in neurotransmitter release: Localization at the distinct central nervous system regions and synaptic types. Neuroscience 1999, 93, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Itakura, M.; Misawa, H.; Sekiguchi, M.; Takahashi, S.; Takahashi, M. Transfection analysis of functional roles of complexin I and II in the exocytosis of two different types of secretory vesicles. Biochem Biophys Res Commun 1999, 265, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Reim, K.; Mansour, M.; Varoqueaux, F.; McMahon, H.T.; Sudhof, T.C.; Brose, N.; Rosenmund, C. Complexins regulate a late step in Ca2+-dependent neurotransmitter release. Cell 2001, 104, 71–81. [Google Scholar] [CrossRef]
- Krishnakumar, S.S.; Radoff, D.T.; Kummel, D.; Giraudo, C.G.; Li, F.; Khandan, L.; Baguley, S.W.; Coleman, J.; Reinisch, K.M.; Pincet, F.; et al. A conformational switch in complexin is required for synaptotagmin to trigger synaptic fusion. Nat Struct Mol Biol 2011, 18, 934–940. [Google Scholar] [CrossRef]
- Lottermoser, J.A.; Dittman, J.S. Complexin Membrane Interactions: Implications for Synapse Evolution and Function. J Mol Biol 2023, 435, 167774. [Google Scholar] [CrossRef]
- Gispert, S.; Kurz, A.; Brehm, N.; Rau, K.; Walter, M.; Riess, O.; Auburger, G. Complexin-1 and Foxp1 Expression Changes Are Novel Brain Effects of Alpha-Synuclein Pathology. Mol Neurobiol 2015, 52, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Reim, K.; Chen, X.; Chao, H.T.; Deng, H.; Rizo, J.; Brose, N.; Rosenmund, C. Distinct domains of complexin I differentially regulate neurotransmitter release. Nat Struct Mol Biol 2007, 14, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Hobson, R.J.; Liu, Q.; Watanabe, S.; Jorgensen, E.M. Complexin maintains vesicles in the primed state in C. elegans. Curr Biol 2011, 21, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Hu, Z.; Fenz, K.M.; Fernandez, J.; Dittman, J.S. Complexin has opposite effects on two modes of synaptic vesicle fusion. Curr Biol 2011, 21, 97–105. [Google Scholar] [CrossRef]
- Lai, Y.; Choi, U.B.; Zhang, Y.; Zhao, M.; Pfuetzner, R.A.; Wang, A.L.; Diao, J.; Brunger, A.T. N-terminal domain of complexin independently activates calcium-triggered fusion. Proc Natl Acad Sci U S A 2016, 113, E4698–E4707. [Google Scholar] [CrossRef] [PubMed]
- Zdanowicz, R.; Kreutzberger, A.; Liang, B.; Kiessling, V.; Tamm, L.K.; Cafiso, D.S. Complexin Binding to Membranes and Acceptor t-SNAREs Explains Its Clamping Effect on Fusion. Biophys J 2017, 113, 1235–1250. [Google Scholar] [CrossRef] [PubMed]
- Bera, M.; Ramakrishnan, S.; Coleman, J.; Krishnakumar, S.S.; Rothman, J.E. Molecular determinants of complexin clamping and activation function. Elife 2022, 11. [Google Scholar] [CrossRef]
- Pabst, S.; Hazzard, J.W.; Antonin, W.; Sudhof, T.C.; Jahn, R.; Rizo, J.; Fasshauer, D. Selective interaction of complexin with the neuronal SNARE complex. Determination of the binding regions. J Biol Chem 2000, 275, 19808–19818. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tomchick, D.R.; Kovrigin, E.; Arac, D.; Machius, M.; Sudhof, T.C.; Rizo, J. Three-dimensional structure of the complexin/SNARE complex. Neuron 2002, 33, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Bowen, M.E.; Weninger, K.; Ernst, J.; Chu, S.; Brunger, A.T. Single-molecule studies of synaptotagmin and complexin binding to the SNARE complex. Biophys J 2005, 89, 690–702. [Google Scholar] [CrossRef]
- Malsam, J.; Seiler, F.; Schollmeier, Y.; Rusu, P.; Krause, J.M.; Sollner, T.H. The carboxy-terminal domain of complexin I stimulates liposome fusion. Proc Natl Acad Sci U S A 2009, 106, 2001–2006. [Google Scholar] [CrossRef] [PubMed]
- Kaeser-Woo, Y.J.; Yang, X.; Sudhof, T.C. C-terminal complexin sequence is selectively required for clamping and priming but not for Ca2+ triggering of synaptic exocytosis. J Neurosci 2012, 32, 2877–2885. [Google Scholar] [CrossRef] [PubMed]
- Wragg, R.T.; Snead, D.; Dong, Y.; Ramlall, T.F.; Menon, I.; Bai, J.; Eliezer, D.; Dittman, J.S. Synaptic vesicles position complexin to block spontaneous fusion. Neuron 2013, 77, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Snead, D.; Wragg, R.T.; Dittman, J.S.; Eliezer, D. Membrane curvature sensing by the C-terminal domain of complexin. Nat Commun 2014, 5, 4955. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Lai, Y.; Li, X.; Wang, M.; Leitz, J.; Hu, Y.; Zhang, Y.; Choi, U.B.; Cipriano, D.; Pfuetzner, R.A.; et al. C-terminal domain of mammalian complexin-1 localizes to highly curved membranes. Proc Natl Acad Sci U S A 2016, 113, E7590–E7599. [Google Scholar] [CrossRef] [PubMed]
- Courtney, K.C.; Wu, L.; Mandal, T.; Swift, M.; Zhang, Z.; Alaghemandi, M.; Wu, Z.; Bradberry, M.M.; Deo, C.; Lavis, L.D.; et al. The complexin C-terminal amphipathic helix stabilizes the fusion pore open state by sculpting membranes. Nat Struct Mol Biol 2022, 29, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Trimbuch, T.; Rosenmund, C. Should I stop or should I go? The role of complexin in neurotransmitter release. Nat Rev Neurosci 2016, 17, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Falkowski, M.A.; Thomas, D.D.; Groblewski, G.E. Complexin 2 modulates vesicle-associated membrane protein (VAMP) 2-regulated zymogen granule exocytosis in pancreatic acini. J Biol Chem 2010, 285, 35558–35566. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, S.; Nakanishi, M.; Hirashima, N. Complexin II facilitates exocytotic release in mast cells by enhancing Ca2+ sensitivity of the fusion process. J Cell Sci 2005, 118, 2239–2246. [Google Scholar] [CrossRef]
- Tsuru, E.; Oryu, K.; Sawada, K.; Nishihara, M.; Tsuda, M. Complexin 2 regulates secretion of immunoglobulin in antibody-secreting cells. Immun Inflamm Dis 2019, 7, 318–325. [Google Scholar] [CrossRef]
- Tsai, P.S.; Brewis, I.A.; van Maaren, J.; Gadella, B.M. Involvement of complexin 2 in docking, locking and unlocking of different SNARE complexes during sperm capacitation and induced acrosomal exocytosis. PLoS ONE 2012, 7, e32603. [Google Scholar] [CrossRef] [PubMed]
- DiProspero, N.A.; Chen, E.Y.; Charles, V.; Plomann, M.; Kordower, J.H.; Tagle, D.A. Early changes in Huntington’s disease patient brains involve alterations in cytoskeletal and synaptic elements. J Neurocytol 2004, 33, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Parplys, A.C.; Zhao, W.; Sharma, N.; Groesser, T.; Liang, F.; Maranon, D.G.; Leung, S.G.; Grundt, K.; Dray, E.; Idate, R.; et al. NUCKS1 is a novel RAD51AP1 paralog important for homologous recombination and genome stability. Nucleic Acids Res 2015, 43, 9817–9834. [Google Scholar] [CrossRef] [PubMed]
- Symonowicz, K.; Dus-Szachniewicz, K.; Wozniak, M.; Murawski, M.; Kolodziej, P.; Osiecka, B.; Jurczyszyn, K.; Ziolkowski, P. Immunohistochemical study of nuclear ubiquitous casein and cyclin-dependent kinase substrate 1 in invasive breast carcinoma of no special type. Exp Ther Med 2014, 8, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Drosos, Y.; Kouloukoussa, M.; Ostvold, A.C.; Grundt, K.; Goutas, N.; Vlachodimitropoulos, D.; Havaki, S.; Kollia, P.; Kittas, C.; Marinos, E.; et al. NUCKS overexpression in breast cancer. Cancer Cell Int 2009, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Cheong, J.Y.; Kim, Y.B.; Woo, J.H.; Kim, D.K.; Yeo, M.; Yang, S.J.; Yang, K.S.; Soon, S.K.; Wang, H.J.; Kim, B.W.; et al. Identification of NUCKS1 as a putative oncogene and immunodiagnostic marker of hepatocellular carcinoma. Gene 2016, 584, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, X.; Li, X.; Bao, H.; Li, G.; Li, N.; Li, H.; Dou, J. NUCKS1 Acts as a Promising Novel Biomarker for the Prognosis of Patients with Hepatocellular Carcinoma. Cancer Biother Radiopharm 2023, 38, 720–725. [Google Scholar] [CrossRef]
- Shi, C.; Qin, L.; Gao, H.; Gu, L.; Yang, C.; Liu, H.; Liu, T. NUCKS nuclear elevated expression indicates progression and prognosis of ovarian cancer. Tumour Biol 2017, 39, 1010428317714631. [Google Scholar] [CrossRef]
- Huang, Y.K.; Kang, W.M.; Ma, Z.Q.; Liu, Y.Q.; Zhou, L.; Yu, J.C. NUCKS1 promotes gastric cancer cell aggressiveness by upregulating IGF-1R and subsequently activating the PI3K/Akt/mTOR signaling pathway. Carcinogenesis 2019, 40, 370–379. [Google Scholar] [CrossRef]
- Gu, L.; Xia, B.; Zhong, L.; Ma, Y.; Liu, L.; Yang, L.; Lou, G. NUCKS1 overexpression is a novel biomarker for recurrence-free survival in cervical squamous cell carcinoma. Tumour Biol 2014, 35, 7831–7836. [Google Scholar] [CrossRef]
- Zhu, L.L.; Shi, J.J.; Guo, Y.D.; Yang, C.; Wang, R.L.; Li, S.S.; Gan, D.X.; Ma, P.X.; Li, J.Q.; Su, H.C. NUCKS1 promotes the progression of colorectal cancer via activating PI3K/AKT/mTOR signaling pathway. Neoplasma 2023, 70, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Ji, R.; He, H.; Li, N.; Han, C.; Han, J.; Li, X.; Zhang, L.; Wang, Y.; Zhao, W. NUCKS1, a LINC00629-upregulated gene, facilitated osteosarcoma progression and metastasis by elevating asparagine synthesis. Cell Death Dis 2023, 14, 489. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Xu, J.; Zhao, R.; Qi, Y.; Ji, Y.; Ma, K. Upregulation of NUCKS1 in Lung Adenocarcinoma is Associated with a Poor Prognosis. Cancer Invest 2021, 39, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Choi, B.S.; Kim, S.S.; Roh, T.Y.; Park, J.; Yoon, C.H. NUCKS1, a novel Tat coactivator, plays a crucial role in HIV-1 replication by increasing Tat-mediated viral transcription on the HIV-1 LTR promoter. Retrovirology 2014, 11, 67. [Google Scholar] [CrossRef]
- Qiu, B.; Han, W.; Tergaonkar, V. NUCKS: A potential biomarker in cancer and metabolic disease. Clin Sci (Lond) 2015, 128, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cheng, L.; Lu, Z.J.; Sun, X.Y.; Li, J.Y.; Peng, R. Association of three candidate genetic variants in RAB7L1/NUCKS1, MCCC1 and STK39 with sporadic Parkinson’s disease in Han Chinese. J Neural Transm (Vienna) 2016, 123, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Seth, P.K. Functional association between NUCKS1 gene and Parkinson disease: A potential susceptibility biomarker. Bioinformation 2019, 15, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xia, C.; Sun, W.; Qin, X.; Qiu, Y.; Zhu, Z. Genetic Polymorphism of NUCKS1 Is Associated With the Susceptibility of Adolescent Idiopathic Scoliosis. Spine (Phila Pa 1976) 2017, 42, 1629–1634. [Google Scholar] [CrossRef] [PubMed]
- Ostvold, A.C.; Holtlund, J.; Laland, S.G. A novel, highly phosphorylated protein, of the high-mobility group type, present in a variety of proliferating and non-proliferating mammalian cells. Eur J Biochem 1985, 153, 469–475. [Google Scholar] [CrossRef]
- Maelandsmo, G.M.; Ostvold, A.C.; Laland, S.G. Phosphorylation of the high-mobility-group-like protein P1 by casein kinase-2. Eur J Biochem 1989, 184, 529–534. [Google Scholar] [CrossRef]
- Ostvold, A.C.; Norum, J.H.; Mathiesen, S.; Wanvik, B.; Sefland, I.; Grundt, K. Molecular cloning of a mammalian nuclear phosphoprotein NUCKS, which serves as a substrate for Cdk1 in vivo. Eur J Biochem 2001, 268, 2430–2440. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, E.J.; Scherer, S.S. On the molecular architecture of myelinated fibers. Histochem Cell Biol 2000, 113, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Rosenbluth, J.; Mierzwa, A.; Shroff, S. Molecular architecture of myelinated nerve fibers: Leaky paranodal junctions and paranodal dysmyelination. Neuroscientist 2013, 19, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Tzimourakas, A.; Giasemi, S.; Mouratidou, M.; Karagogeos, D. Structure-function analysis of protein complexes involved in the molecular architecture of juxtaparanodal regions of myelinated fibers. Biotechnol J 2007, 2, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Baumann, N.; Pham-Dinh, D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev 2001, 81, 871–927. [Google Scholar] [CrossRef] [PubMed]
- Harauz, G.; Ishiyama, N.; Hill, C.M.; Bates, I.R.; Libich, D.S.; Fares, C. Myelin basic protein-diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron 2004, 35, 503–542. [Google Scholar] [CrossRef] [PubMed]
- Kramer, E.M.; Schardt, A.; Nave, K.A. Membrane traffic in myelinating oligodendrocytes. Microsc Res Tech 2001, 52, 656–671. [Google Scholar] [CrossRef] [PubMed]
- Kon, T.; Tanji, K.; Mori, F.; Kimura, A.; Kakita, A.; Wakabayashi, K. Immunoreactivity of myelin-associated oligodendrocytic basic protein in Lewy bodies. Neuropathology 2019, 39, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.H.; Halliday, G.M.; Kim, W.S. Exploring myelin dysfunction in multiple system atrophy. Exp Neurobiol 2014, 23, 337–344. [Google Scholar] [CrossRef]
- Smith, R. The basic protein of CNS myelin: Its structure and ligand binding. J Neurochem 1992, 59, 1589–1608. [Google Scholar] [CrossRef]
- Hill, C.M.; Bates, I.R.; White, G.F.; Hallett, F.R.; Harauz, G. Effects of the osmolyte trimethylamine-N-oxide on conformation, self-association, and two-dimensional crystallization of myelin basic protein. J Struct Biol 2002, 139, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.M.; Haines, J.D.; Antler, C.E.; Bates, I.R.; Libich, D.S.; Harauz, G. Terminal deletion mutants of myelin basic protein: New insights into self-association and phospholipid interactions. Micron 2003, 34, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Sedzik, J.; Kirschner, D.A. Is myelin basic protein crystallizable? Neurochem Res 1992, 17, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Nixon, R.A.; Saito, K.I.; Grynspan, F.; Griffin, W.R.; Katayama, S.; Honda, T.; Mohan, P.S.; Shea, T.B.; Beermann, M. Calcium-activated neutral proteinase (calpain) system in aging and Alzheimer’s disease. Ann N Y Acad Sci 1994, 747, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, K.K. The calpain family and human disease. Trends Mol Med 2001, 7, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Hata, S.; Kawabata, Y.; Sorimachi, H. Structure, activation, and biology of calpain. Diabetes 2004, 53 (Suppl. 1), S12–S18. [Google Scholar] [CrossRef]
- Mellgren, R.L.; Rozanov, C.B. Calpain II-dependent solubilization of a nuclear protein kinase at micromolar calcium concentrations. Biochem Biophys Res Commun 1990, 168, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.K.; Dasgupta, S.; Banik, N.L.; Hogan, E.L. Regulation of the calcium-activated neutral proteinase (CANP) of bovine brain by myelin lipids. Biochim Biophys Acta 1990, 1038, 195–198. [Google Scholar] [CrossRef]
- Saido, T.C.; Shibata, M.; Takenawa, T.; Murofushi, H.; Suzuki, K. Positive regulation of mu-calpain action by polyphosphoinositides. J Biol Chem 1992, 267, 24585–24590. [Google Scholar] [CrossRef]
- Salamino, F.; De Tullio, R.; Mengotti, P.; Viotti, P.L.; Melloni, E.; Pontremoli, S. Site-directed activation of calpain is promoted by a membrane-associated natural activator protein. Biochem J 1993, 290 ( Pt 1) Pt 1, 191–197. [Google Scholar] [CrossRef]
- Suzuki, K.; Ohno, S. Calcium activated neutral protease--structure-function relationship and functional implications. Cell Struct Funct 1990, 15, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Murachi, T. Intracellular regulatory system involving calpain and calpastatin. Biochem Int 1989, 18, 263–294. [Google Scholar] [PubMed]
- Nixon, R.A. Calcium-activated neutral proteinases as regulators of cellular function. Implications for Alzheimer’s disease pathogenesis. Ann N Y Acad Sci 1989, 568, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Nixon, R.A.; Quackenbush, R.; Vitto, A. Multiple calcium-activated neutral proteinases (CANP) in mouse retinal ganglion cell neurons: Specificities for endogenous neuronal substrates and comparison to purified brain CANP. J Neurosci 1986, 6, 1252–1263. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Ma, H.; Takano, E.; Yang, H.Q.; Hatanaka, M.; Maki, M. Molecular diversity in amino-terminal domains of human calpastatin by exon skipping. J Biol Chem 1992, 267, 8437–8442. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Ishida-Takahashi, A.; Takahashi, C.; Takano, E.; Murachi, T.; Hatanaka, M. Phosphorylation and subcellular distribution of calpastatin in human hematopoietic system cells. J Biol Chem 1991, 266, 3968–3972. [Google Scholar] [CrossRef]
- Nakamura, M.; Inomata, M.; Imajoh, S.; Suzuki, K.; Kawashima, S. Fragmentation of an endogenous inhibitor upon complex formation with high- and low-Ca2+-requiring forms of calcium-activated neutral proteases. Biochemistry 1989, 28, 449–455. [Google Scholar] [CrossRef]
- Emori, Y.; Kawasaki, H.; Imajoh, S.; Minami, Y.; Suzuki, K. All four repeating domains of the endogenous inhibitor for calcium-dependent protease independently retain inhibitory activity. Expression of the cDNA fragments in Escherichia coli. J Biol Chem 1988, 263, 2364–2370. [Google Scholar] [CrossRef]
- Wendt, A.; Thompson, V.F.; Goll, D.E. Interaction of calpastatin with calpain: A review. Biol Chem 2004, 385, 465–472. [Google Scholar] [CrossRef]
- Takano, E.; Ma, H.; Yang, H.Q.; Maki, M.; Hatanaka, M. Preference of calcium-dependent interactions between calmodulin-like domains of calpain and calpastatin subdomains. FEBS Lett 1995, 362, 93–97. [Google Scholar] [CrossRef]
- Tompa, P.; Mucsi, Z.; Orosz, G.; Friedrich, P. Calpastatin subdomains A and C are activators of calpain. J Biol Chem 2002, 277, 9022–9026. [Google Scholar] [CrossRef] [PubMed]
- Uemori, T.; Shimojo, T.; Asada, K.; Asano, T.; Kimizuka, F.; Kato, I.; Maki, M.; Hatanaka, M.; Murachi, T.; Hanzawa, H.; et al. Characterization of a functional domain of human calpastatin. Biochem Biophys Res Commun 1990, 166, 1485–1493. [Google Scholar] [CrossRef] [PubMed]
- Konno, T.; Tanaka, N.; Kataoka, M.; Takano, E.; Maki, M. A circular dichroism study of preferential hydration and alcohol effects on a denatured protein, pig calpastatin domain I. Biochim Biophys Acta 1997, 1342, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Kiss, R.; Kovacs, D.; Tompa, P.; Perczel, A. Local structural preferences of calpastatin, the intrinsically unstructured protein inhibitor of calpain. Biochemistry 2008, 47, 6936–6945. [Google Scholar] [CrossRef] [PubMed]
- Ballatore, C.; Brunden, K.R.; Huryn, D.M.; Trojanowski, J.Q.; Lee, V.M.; Smith, A.B., 3rd. Microtubule stabilizing agents as potential treatment for Alzheimer’s disease and related neurodegenerative tauopathies. J Med Chem 2012, 55, 8979–8996. [Google Scholar] [CrossRef] [PubMed]
- Drechsel, D.N.; Hyman, A.A.; Cobb, M.H.; Kirschner, M.W. Modulation of the dynamic instability of tubulin assembly by the microtubule-associated protein tau. Mol Biol Cell 1992, 3, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Zhang, B.; Lee, V.M.; Trojanowski, J.Q. Axonal transport defects: A common theme in neurodegenerative diseases. Acta Neuropathol 2005, 109, 5–13. [Google Scholar] [CrossRef]
- Kuret, J.; Congdon, E.E.; Li, G.; Yin, H.; Yu, X.; Zhong, Q. Evaluating triggers and enhancers of tau fibrillization. Microsc Res Tech 2005, 67, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Kuret, J.; Chirita, C.N.; Congdon, E.E.; Kannanayakal, T.; Li, G.; Necula, M.; Yin, H.; Zhong, Q. Pathways of tau fibrillization. Biochim Biophys Acta 2005, 1739, 167–178. [Google Scholar] [CrossRef]
- Lee, V.M.; Goedert, M.; Trojanowski, J.Q. Neurodegenerative tauopathies. Annu Rev Neurosci 2001, 24, 1121–1159. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Goedert, M.; Crowther, R.A.; Murrell, J.R.; Farlow, M.R.; Ghetti, B. Familial multiple system tauopathy with presenile dementia: A disease with abundant neuronal and glial tau filaments. Proc Natl Acad Sci U S A 1997, 94, 4113–4118. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.M.; Trojanowski, J.Q. Neurodegenerative tauopathies: Human disease and transgenic mouse models. Neuron 1999, 24, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Josephs, K.A.; Hodges, J.R.; Snowden, J.S.; Mackenzie, I.R.; Neumann, M.; Mann, D.M.; Dickson, D.W. Neuropathological background of phenotypical variability in frontotemporal dementia. Acta Neuropathol 2011, 122, 137–153. [Google Scholar] [CrossRef] [PubMed]
- Arendt, T.; Stieler, J.T.; Holzer, M. Tau and tauopathies. Brain Res Bull 2016, 126, 238–292. [Google Scholar] [CrossRef] [PubMed]
- Gotz, J.; Halliday, G.; Nisbet, R.M. Molecular Pathogenesis of the Tauopathies. Annu Rev Pathol 2019, 14, 239–261. [Google Scholar] [CrossRef] [PubMed]
- Kneynsberg, A.; Combs, B.; Christensen, K.; Morfini, G.; Kanaan, N.M. Axonal Degeneration in Tauopathies: Disease Relevance and Underlying Mechanisms. Front Neurosci 2017, 11, 572. [Google Scholar] [CrossRef] [PubMed]
- Uemura, N.; Uemura, M.T.; Luk, K.C.; Lee, V.M.; Trojanowski, J.Q. Cell-to-Cell Transmission of Tau and alpha-Synuclein. Trends Mol Med 2020, 26, 936–952. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, G.S.; Lee, V.M.Y.; Trojanowski, J.Q. Mechanisms of Cell-to-Cell Transmission of Pathological Tau: A Review. JAMA Neurol 2019, 76, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Drewes, G.; Trinczek, B.; Illenberger, S.; Biernat, J.; Schmitt-Ulms, G.; Meyer, H.E.; Mandelkow, E.M.; Mandelkow, E. Microtubule-associated protein/microtubule affinity-regulating kinase (p110mark). A novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J Biol Chem 1995, 270, 7679–7688. [Google Scholar] [CrossRef]
- Clark, L.N.; Poorkaj, P.; Wszolek, Z.; Geschwind, D.H.; Nasreddine, Z.S.; Miller, B.; Li, D.; Payami, H.; Awert, F.; Markopoulou, K.; et al. Pathogenic implications of mutations in the tau gene in pallido-ponto-nigral degeneration and related neurodegenerative disorders linked to chromosome 17. Proc Natl Acad Sci U S A 1998, 95, 13103–13107. [Google Scholar] [CrossRef]
- Narayanan, R.L.; Durr, U.H.; Bibow, S.; Biernat, J.; Mandelkow, E.; Zweckstetter, M. Automatic assignment of the intrinsically disordered protein Tau with 441-residues. J Am Chem Soc 2010, 132, 11906–11907. [Google Scholar] [CrossRef] [PubMed]
- Mukrasch, M.D.; Bibow, S.; Korukottu, J.; Jeganathan, S.; Biernat, J.; Griesinger, C.; Mandelkow, E.; Zweckstetter, M. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol 2009, 7, e34. [Google Scholar] [CrossRef] [PubMed]
- Ambadipudi, S.; Reddy, J.G.; Biernat, J.; Mandelkow, E.; Zweckstetter, M. Residue-specific identification of phase separation hot spots of Alzheimer’s-related protein tau. Chem Sci 2019, 10, 6503–6507. [Google Scholar] [CrossRef] [PubMed]
- Ukmar-Godec, T.; Wegmann, S.; Zweckstetter, M. Biomolecular condensation of the microtubule-associated protein tau. Semin Cell Dev Biol 2020, 99, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Rai, S.K.; Savastano, A.; Singh, P.; Mukhopadhyay, S.; Zweckstetter, M. Liquid-liquid phase separation of tau: From molecular biophysics to physiology and disease. Protein Sci 2021, 30, 1294–1314. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Zweckstetter, M. Phase separation of the microtubule-associated protein tau. Essays Biochem 2022, 66, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Li, C.; Meng, L.; Tian, Y.; He, M.; Yuan, X.; Zhang, G.; Zhang, Z.; Xiong, J.; Chen, G.; et al. Tau accelerates alpha-synuclein aggregation and spreading in Parkinson’s disease. Brain 2022, 145, 3454–3471. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.L.; Lee, E.B.; Xie, S.X.; Rennert, L.; Suh, E.; Bredenberg, C.; Caswell, C.; Van Deerlin, V.M.; Yan, N.; Yousef, A.; et al. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain 2018, 141, 2181–2193. [Google Scholar] [CrossRef] [PubMed]
- Twohig, D.; Nielsen, H.M. alpha-synuclein in the pathophysiology of Alzheimer’s disease. Mol Neurodegener 2019, 14, 23. [Google Scholar] [CrossRef]
- Jin, M.; Wang, S.; Gao, X.; Zou, Z.; Hirotsune, S.; Sun, L. Pathological and physiological functional cross-talks of alpha-synuclein and tau in the central nervous system. Neural Regen Res 2024, 19, 855–862. [Google Scholar] [CrossRef]
- Kayed, R.; Dettmer, U.; Lesne, S.E. Soluble endogenous oligomeric alpha-synuclein species in neurodegenerative diseases: Expression, spreading, and cross-talk. J Parkinsons Dis 2020, 10, 791–818. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zhang, S.; Ma, X.; Jia, C.; Liu, Z.; Huang, C.; Liu, C.; Li, D. Structural basis of the interplay between alpha-synuclein and Tau in regulating pathological amyloid aggregation. J Biol Chem 2020, 295, 7470–7480. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.; Sorrentino, Z.; Weinrich, M.; Giasson, B.I.; Chakrabarty, P. Differential cross-seeding properties of tau and alpha-synuclein in mouse models of tauopathy and synucleinopathy. Brain Commun 2020, 2, fcaa090. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Fu, Y.; Miyata, T.; Matsumoto, S.; Shinoda, T.; Itoh, K.; Harada, A.; Hirotsune, S.; Jin, M. Functional Cooperation of alpha-Synuclein and Tau Is Essential for Proper Corticogenesis. J Neurosci 2022, 42, 7031–7046. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Xu, W.X.; Wang, S.Y.; Zhan, Y.Q.; Jiang, Y.; Cai, W.M.; Yang, X.M. Isolation and Characterization of EDAG-1, A Novel Gene Related to Regulation in Hematopoietic System. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 2001, 33, 641–646. [Google Scholar]
- Yang, L.V.; Nicholson, R.H.; Kaplan, J.; Galy, A.; Li, L. Hemogen is a novel nuclear factor specifically expressed in mouse hematopoietic development and its human homologue EDAG maps to chromosome 9q22, a region containing breakpoints of hematological neoplasms. Mech Dev 2001, 104, 105–111. [Google Scholar] [CrossRef]
- Liu, C.C.; Chou, Y.L.; Ch’ang, L.Y. Down-regulation of human NDR gene in megakaryocytic differentiation of erythroleukemia K562 cells. J Biomed Sci 2004, 11, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Zhan, Y.Q.; Xu, C.W.; Xu, W.X.; Wang, S.Y.; Lv, J.; Zhou, Y.; Yue, P.B.; Chen, B.; Yang, X.M. EDAG regulates the proliferation and differentiation of hematopoietic cells and resists cell apoptosis through the activation of nuclear factor-kappa B. Cell Death Differ 2004, 11, 1299–1308. [Google Scholar] [CrossRef]
- An, L.L.; Li, G.; Wu, K.F.; Ma, X.T.; Zheng, G.G.; Qiu, L.G.; Song, Y.H. High expression of EDAG and its significance in AML. Leukemia 2005, 19, 1499–1502. [Google Scholar] [CrossRef]
- Yang, L.V.; Heng, H.H.; Wan, J.; Southwood, C.M.; Gow, A.; Li, L. Alternative promoters and polyadenylation regulate tissue-specific expression of Hemogen isoforms during hematopoiesis and spermatogenesis. Dev Dyn 2003, 228, 606–616. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, W.X.; Zhan, Y.Q.; Li, C.Y.; Xu, C.W.; Zheng, H.; Li, F.F.; Yang, X.M.; Wang, S.Y. [Expression of EDAG-1 gene in human leukemia and lymphoma cell lines]. Ai Zheng 2004, 23, 1238–1243. [Google Scholar]
- Chen, D.L.; Hu, Z.Q.; Zheng, X.F.; Wang, X.Y.; Xu, Y.Z.; Li, W.Q.; Fang, H.S.; Kan, L.; Wang, S.Y. EDAG-1 promotes proliferation and invasion of human thyroid cancer cells by activating MAPK/Erk and AKT signal pathways. Cancer Biol Ther 2016, 17, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Millan-Arino, L.; Izquierdo-Bouldstridge, A.; Jordan, A. Specificities and genomic distribution of somatic mammalian histone H1 subtypes. Biochim Biophys Acta 2016, 1859, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Fyodorov, D.V.; Zhou, B.R.; Skoultchi, A.I.; Bai, Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol 2018, 19, 192–206. [Google Scholar] [CrossRef]
- Salinas-Pena, M.; Rebollo, E.; Jordan, A. Imaging analysis of six human histone H1 variants reveals universal enrichment of H1.2, H1.3, and H1.5 at the nuclear periphery and nucleolar H1X presence. Elife 2024, 12. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Lailler, N.; Zhang, Y.; Kumar, A.; Uppal, K.; Liu, Z.; Lee, E.K.; Wu, H.; Medrzycki, M.; Pan, C.; et al. High-resolution mapping of h1 linker histone variants in embryonic stem cells. PLoS Genet 2013, 9, e1003417. [Google Scholar] [CrossRef]
- Izzo, A.; Kamieniarz-Gdula, K.; Ramirez, F.; Noureen, N.; Kind, J.; Manke, T.; van Steensel, B.; Schneider, R. The genomic landscape of the somatic linker histone subtypes H1.1 to H1.5 in human cells. Cell Rep 2013, 3, 2142–2154. [Google Scholar] [CrossRef]
- Millan-Arino, L.; Islam, A.B.; Izquierdo-Bouldstridge, A.; Mayor, R.; Terme, J.M.; Luque, N.; Sancho, M.; Lopez-Bigas, N.; Jordan, A. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res 2014, 42, 4474–4493. [Google Scholar] [CrossRef]
- Serna-Pujol, N.; Salinas-Pena, M.; Mugianesi, F.; Le Dily, F.; Marti-Renom, M.A.; Jordan, A. Coordinated changes in gene expression, H1 variant distribution and genome 3D conformation in response to H1 depletion. Nucleic Acids Res 2022, 50, 3892–3910. [Google Scholar] [CrossRef]
- Lai, S.; Jia, J.; Cao, X.; Zhou, P.K.; Gao, S. Molecular and Cellular Functions of the Linker Histone H1.2. Front Cell Dev Biol 2021, 9, 773195. [Google Scholar] [CrossRef]
- Roque, A.; Sortino, R.; Ventura, S.; Ponte, I.; Suau, P. Histone H1 Favors Folding and Parallel Fibrillar Aggregation of the 1-42 Amyloid-beta Peptide. Langmuir 2015, 31, 6782–6790. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Mizianty, M.J.; Xue, B.; Kurgan, L.; Uversky, V.N. More than just tails: Intrinsic disorder in histone proteins. Mol Biosyst 2012, 8, 1886–1901. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Kruger, S.; Mann, M. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol Cell Proteomics 2007, 6, 72–87. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhou, X.; Taghizadeh, K.; Dong, M.; Dedon, P.C. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc Natl Acad Sci U S A 2007, 104, 60–65. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Mann, M. Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res 2008, 36, 570–577. [Google Scholar] [CrossRef]
- Weiss, T.; Hergeth, S.; Zeissler, U.; Izzo, A.; Tropberger, P.; Zee, B.M.; Dundr, M.; Garcia, B.A.; Daujat, S.; Schneider, R. Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenetics Chromatin 2010, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Izzo, A.; Schneider, R. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim Biophys Acta 2016, 1859, 486–495. [Google Scholar] [CrossRef]
- Goers, J.; Manning-Bog, A.B.; McCormack, A.L.; Millett, I.S.; Doniach, S.; Di Monte, D.A.; Uversky, V.N.; Fink, A.L. Nuclear localization of alpha-synuclein and its interaction with histones. Biochemistry 2003, 42, 8465–8471. [Google Scholar] [CrossRef] [PubMed]
- UniProt_Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Dayhoff, G.W., 2nd; Uversky, V.N. Rapid prediction and analysis of protein intrinsic disorder. Protein Sci 2022, 31, e4496. [Google Scholar] [CrossRef] [PubMed]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D(2)P(2): Database of disordered protein predictions. Nucleic Acids Res 2013, 41, D508–D516. [Google Scholar] [CrossRef] [PubMed]
- Hatos, A.; Tosatto, S.C.E.; Vendruscolo, M.; Fuxreiter, M. FuzDrop on AlphaFold: Visualizing the sequence-dependent propensity of liquid-liquid phase separation and aggregation of proteins. Nucleic Acids Res 2022, 50, W337–W344. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
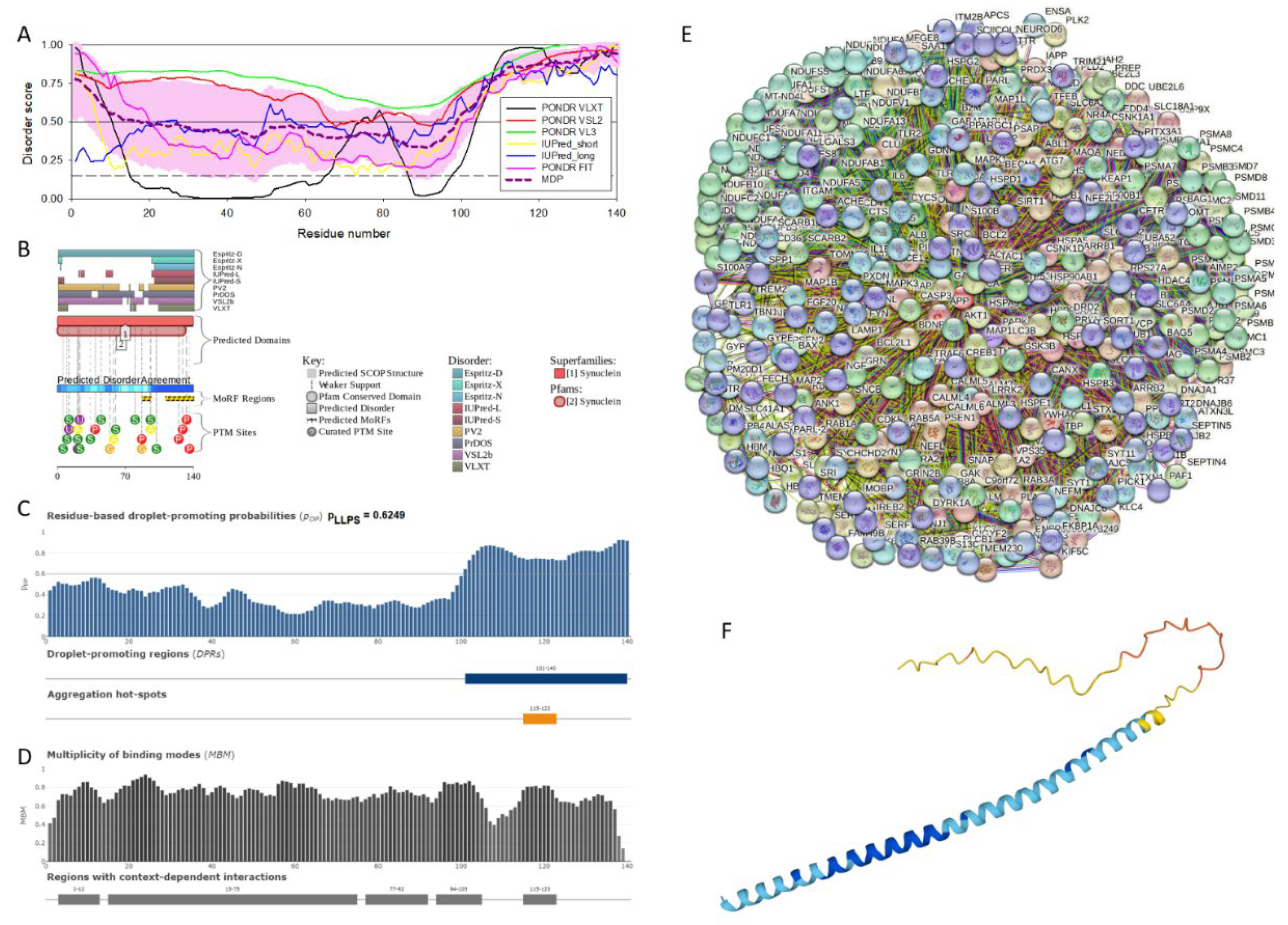
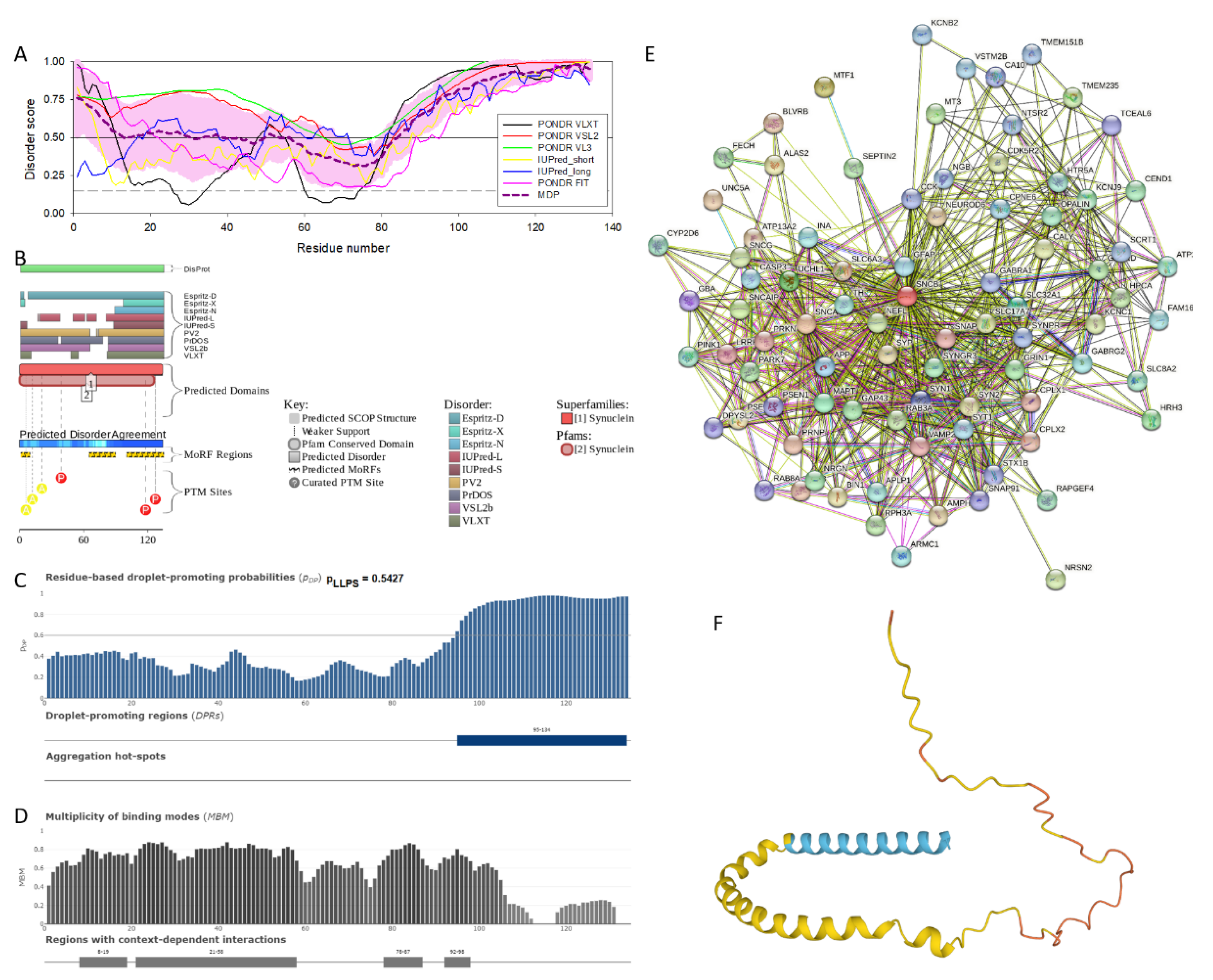
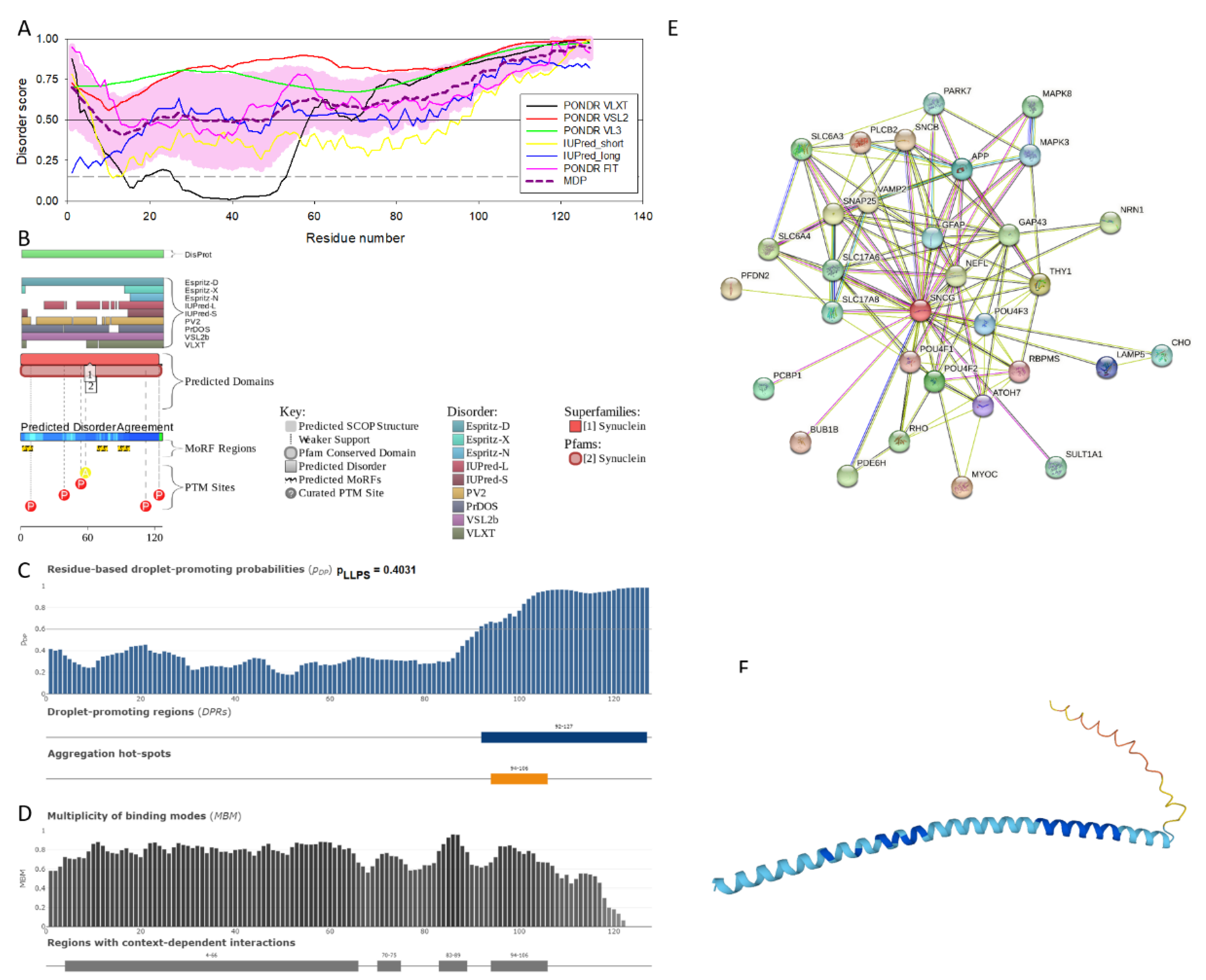
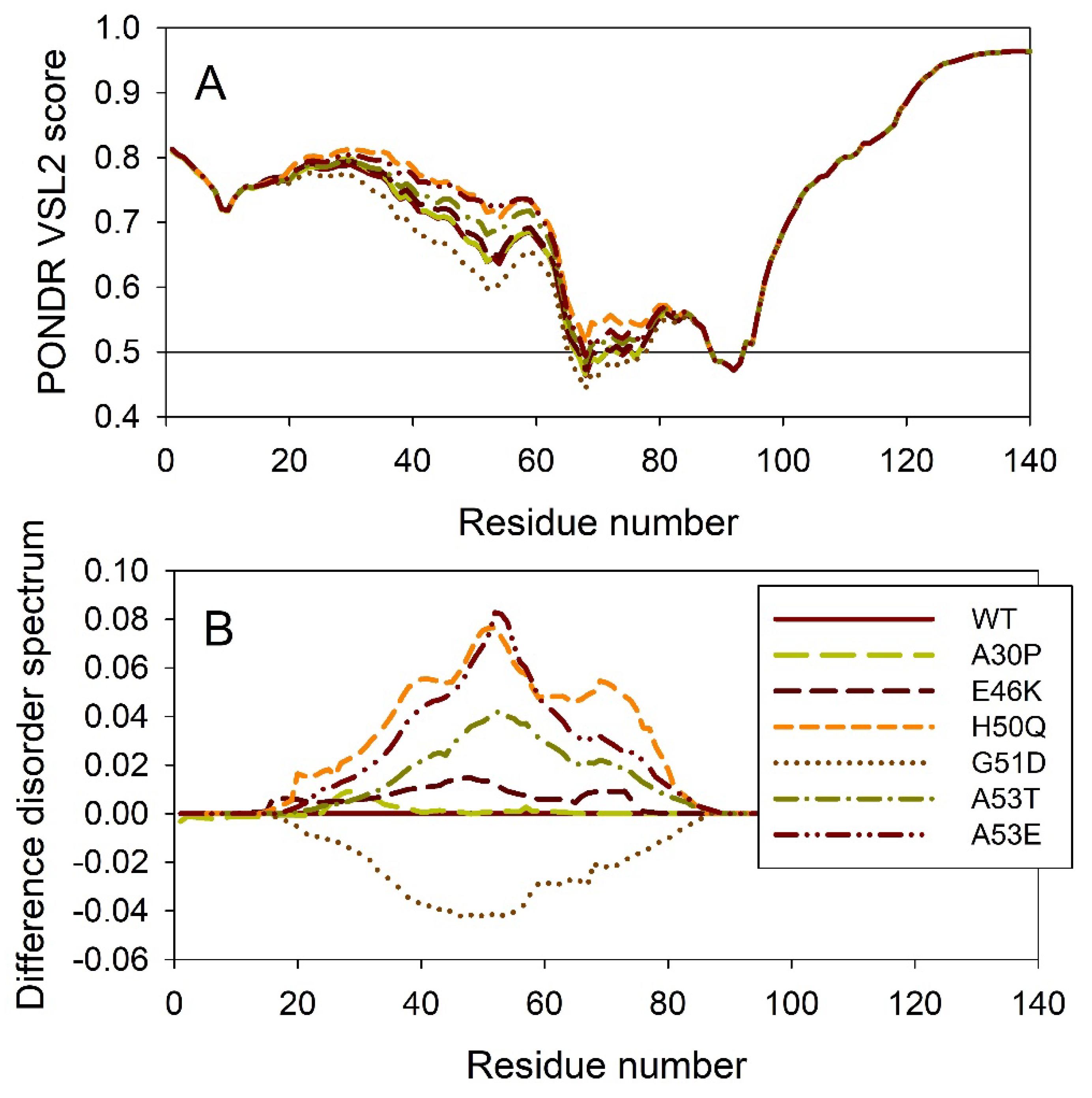
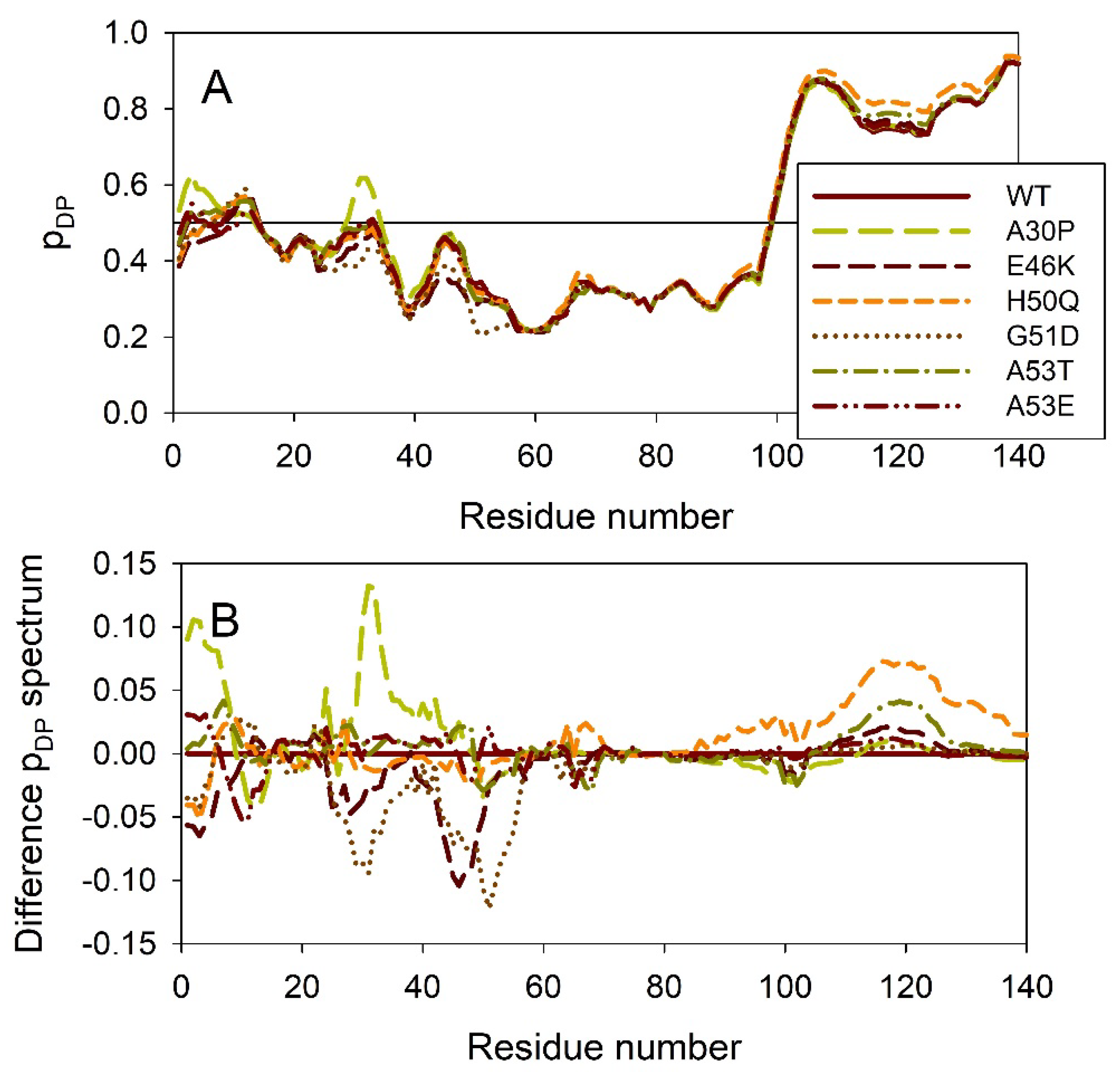
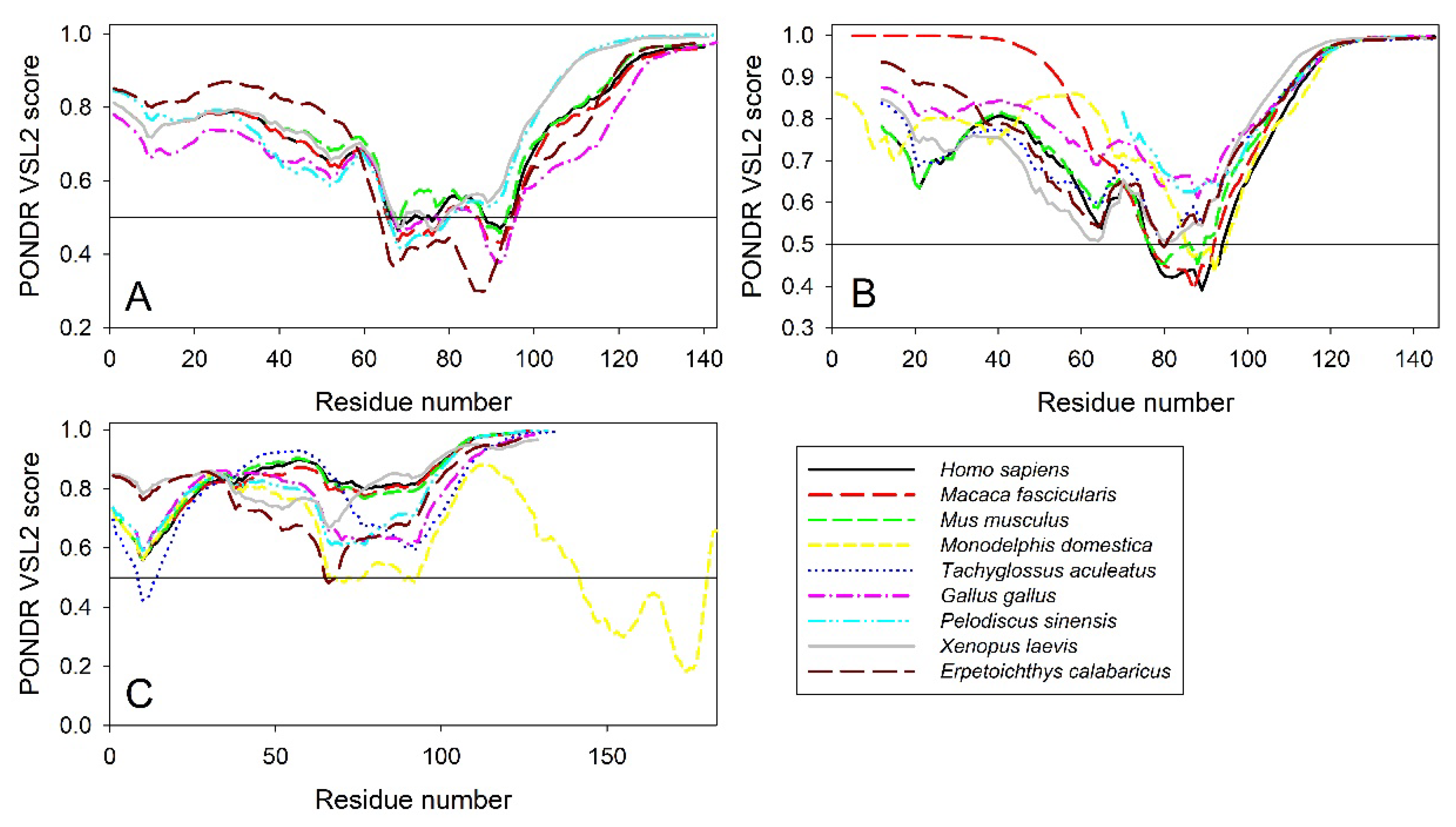

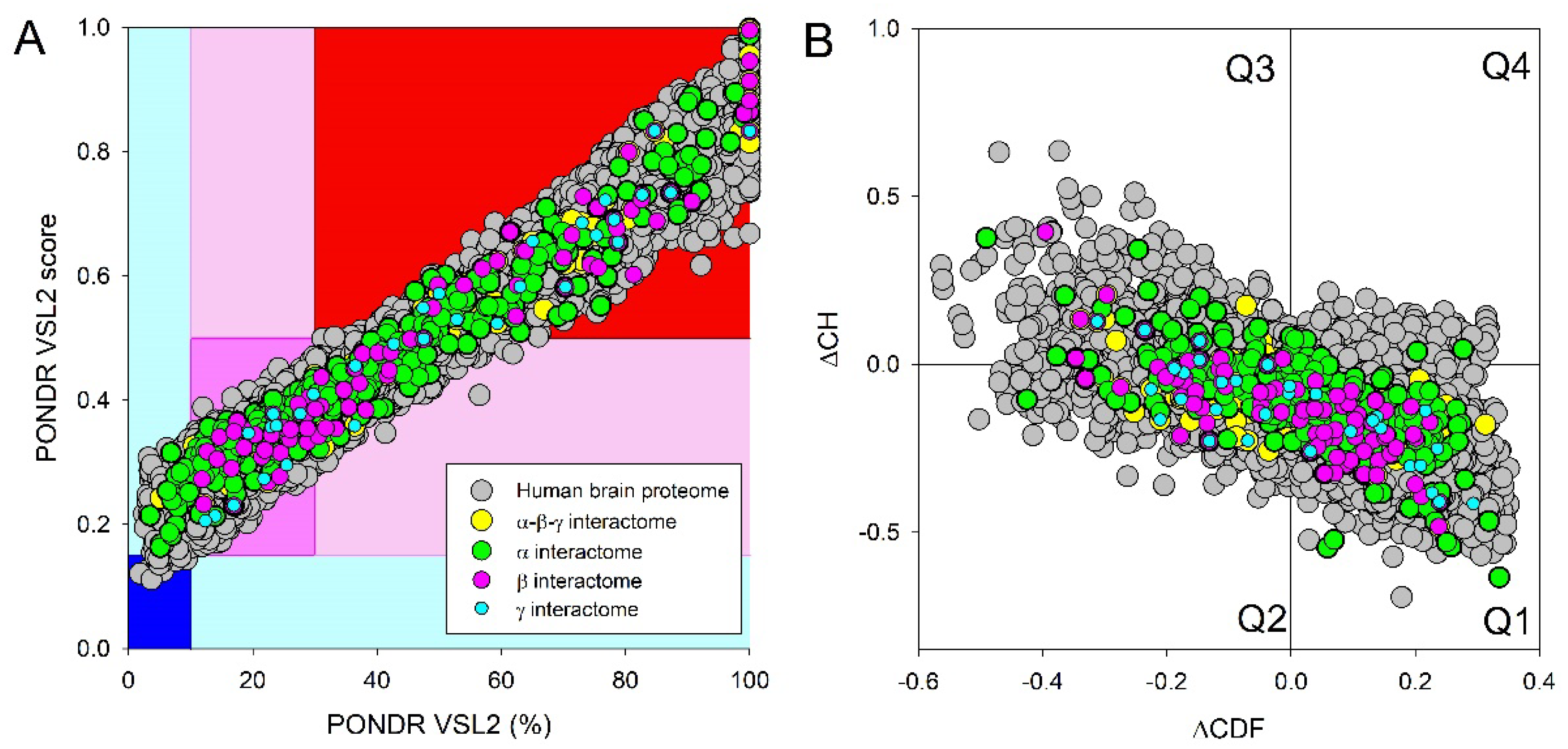
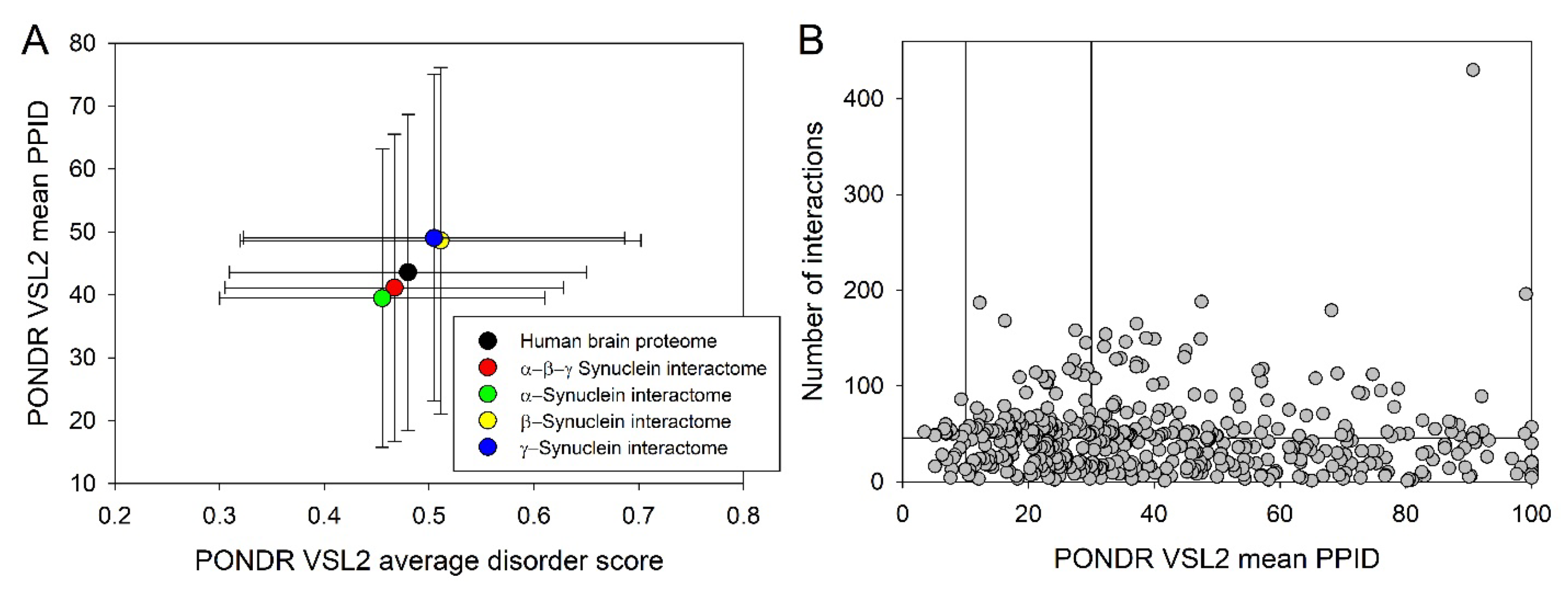
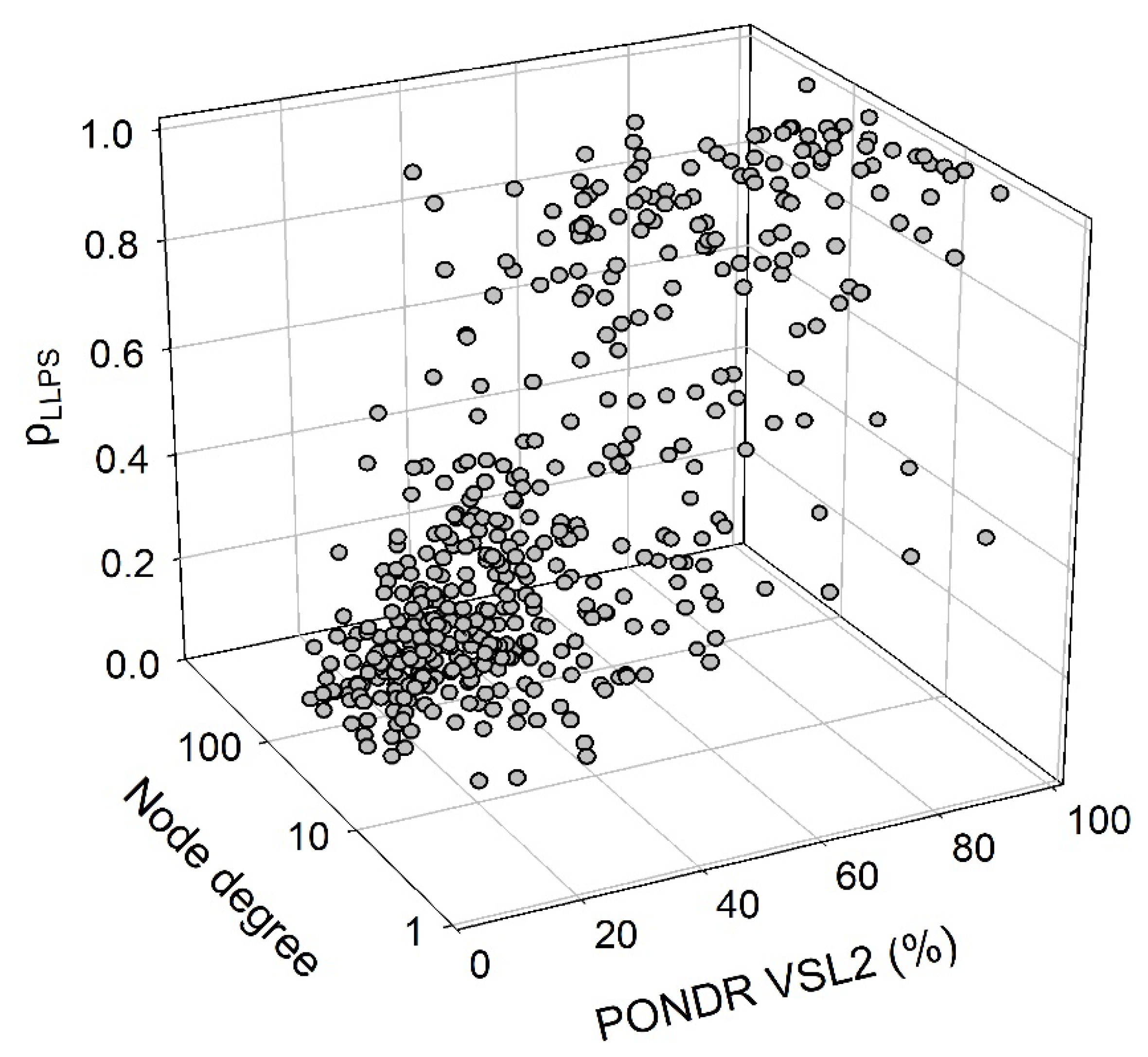
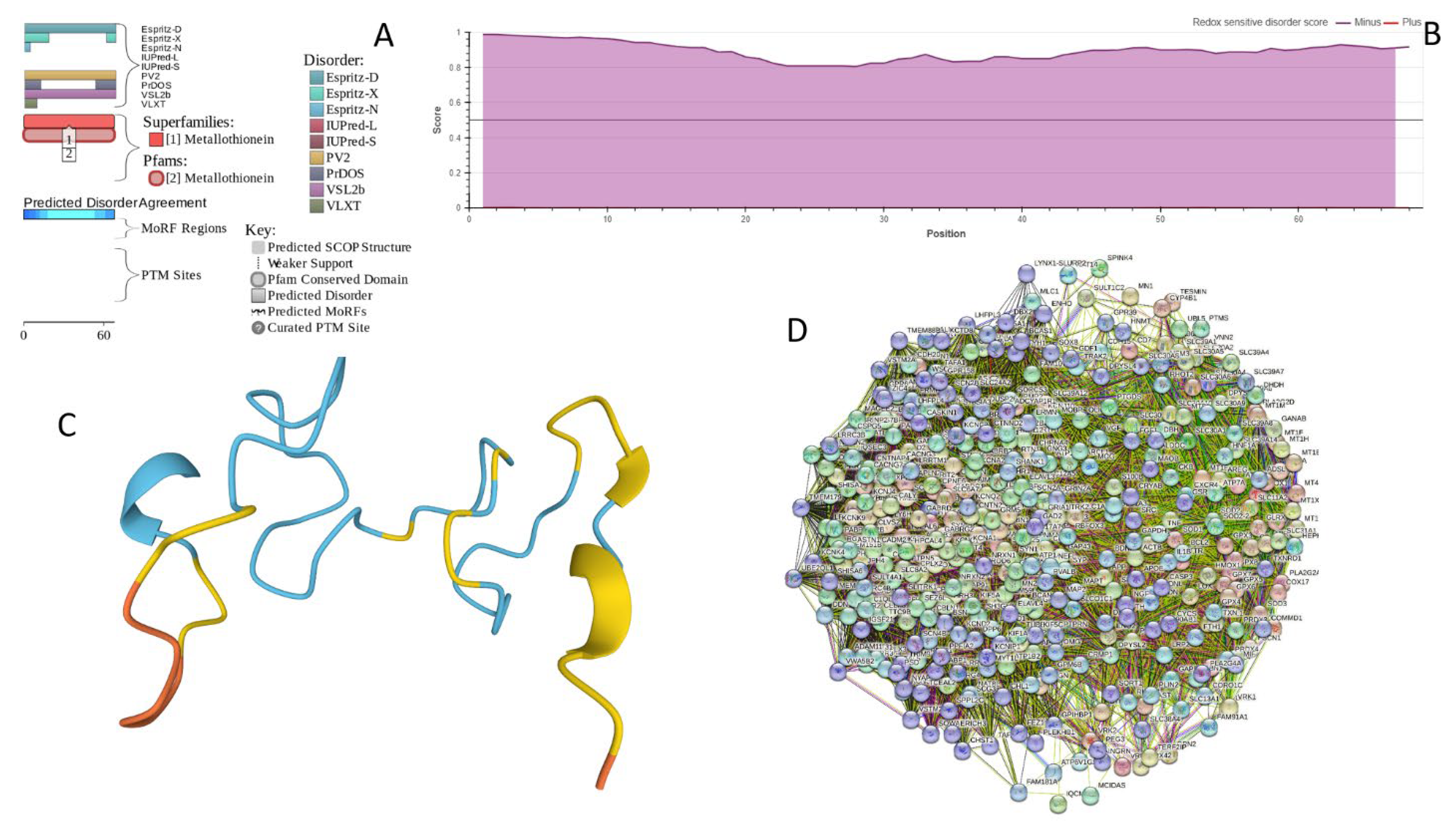
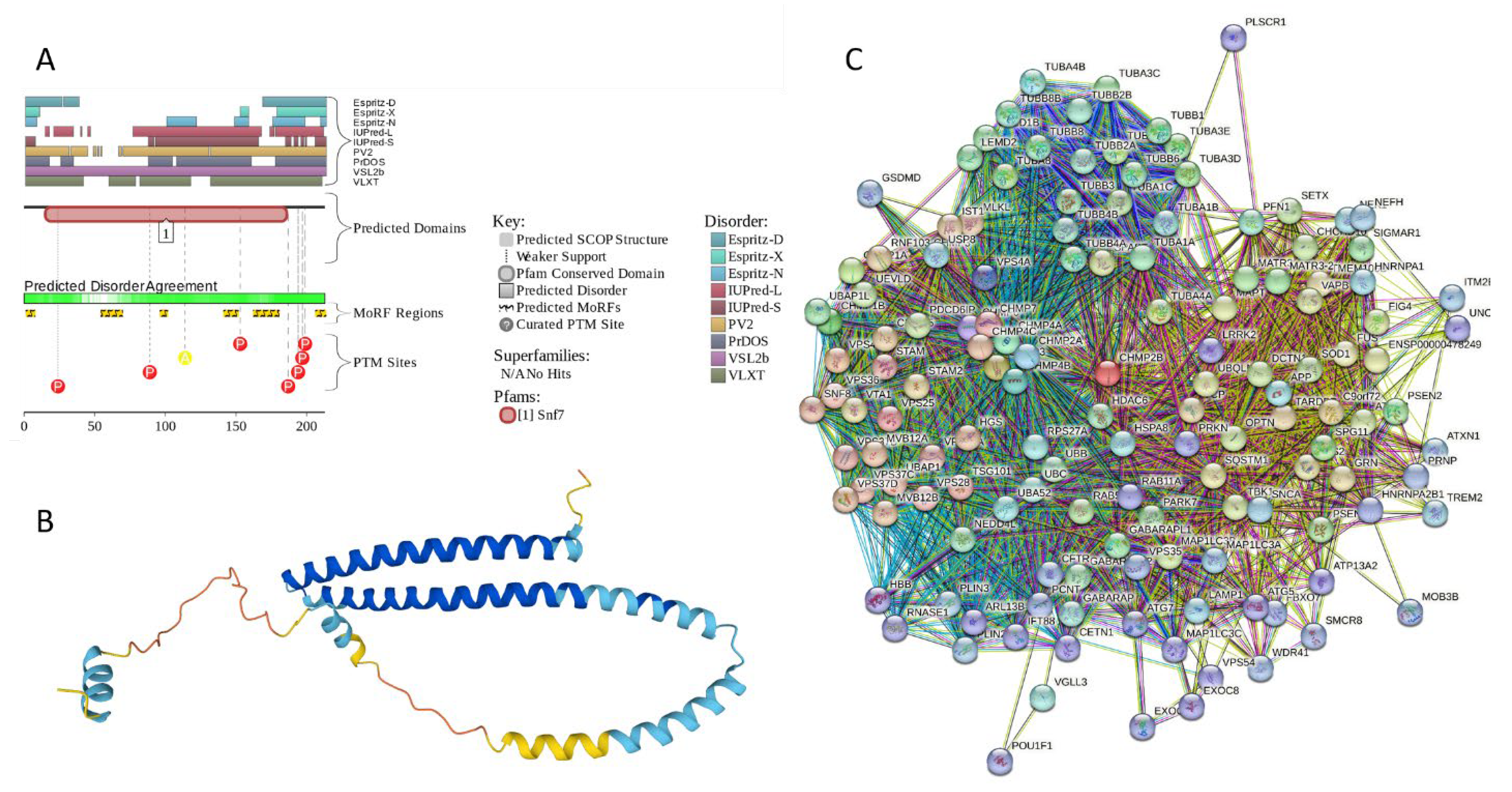
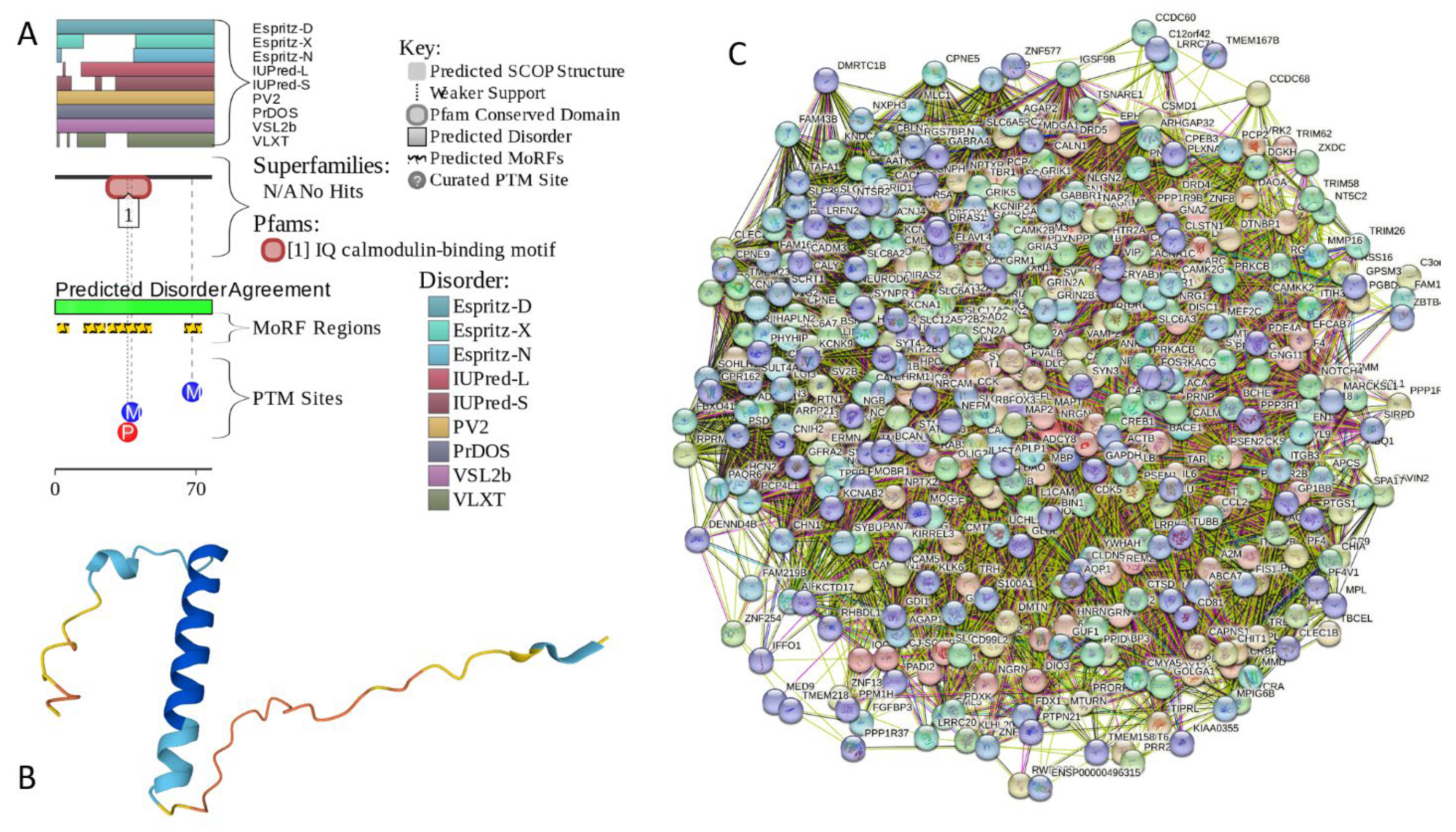
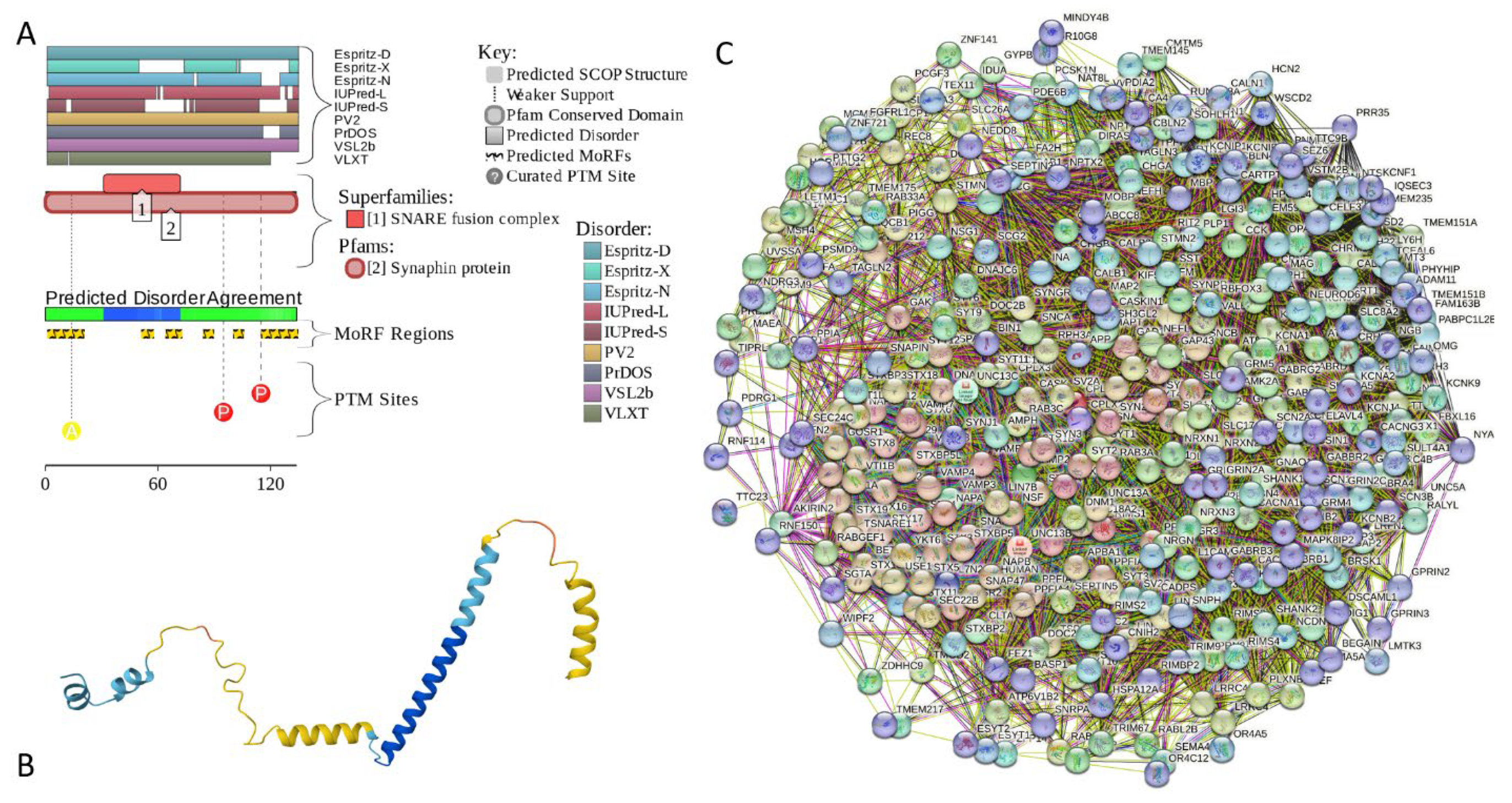
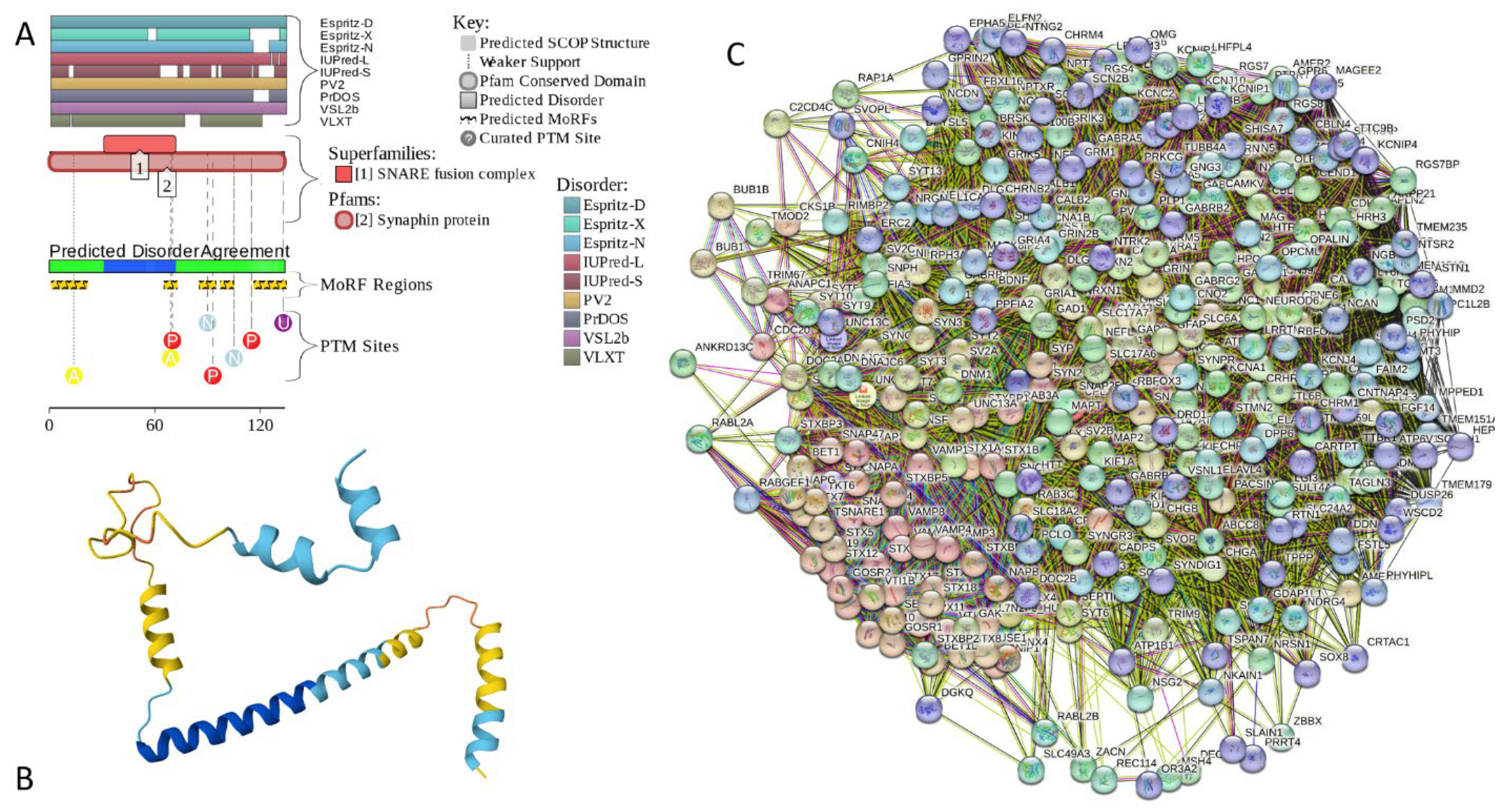
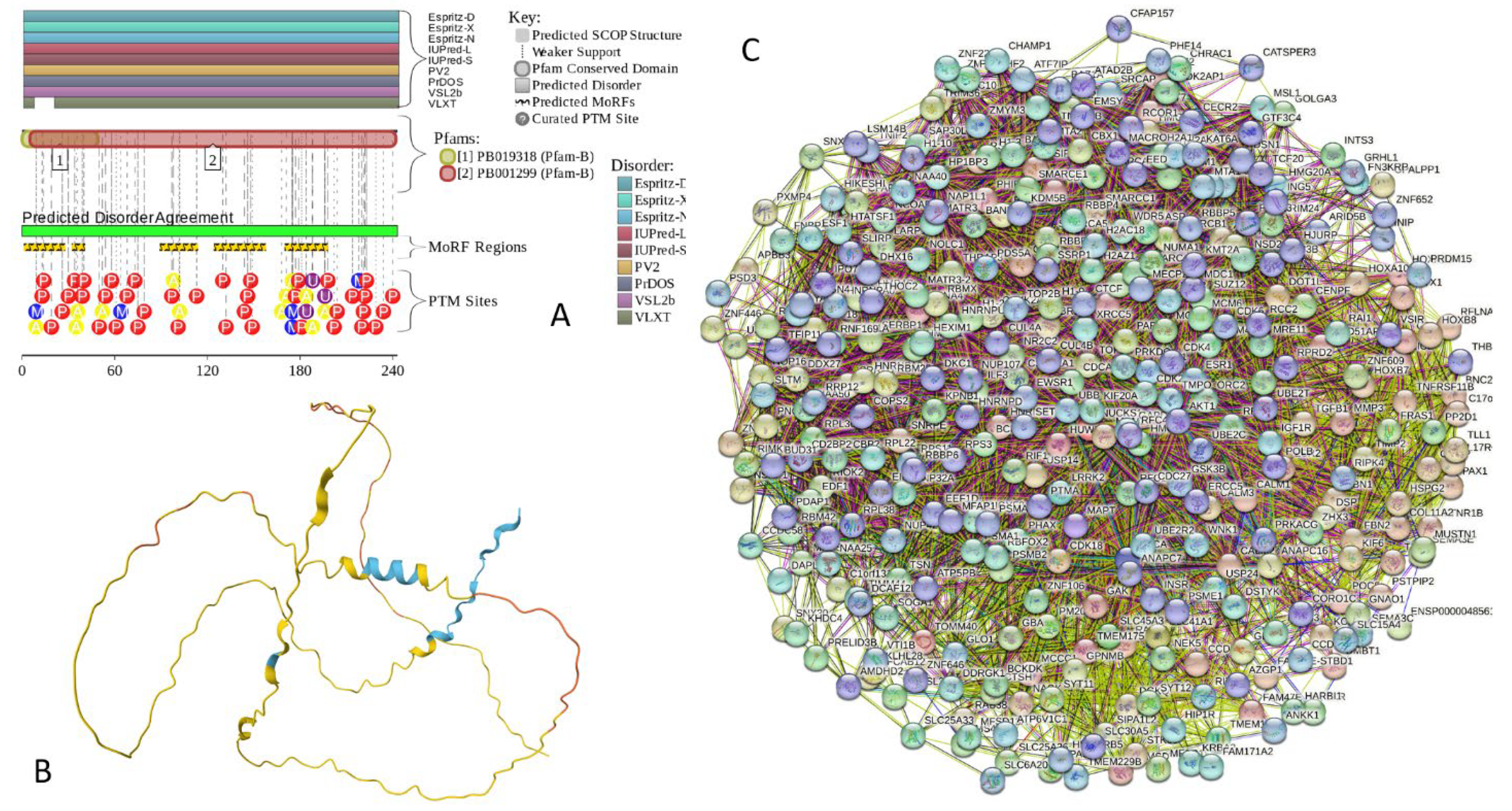
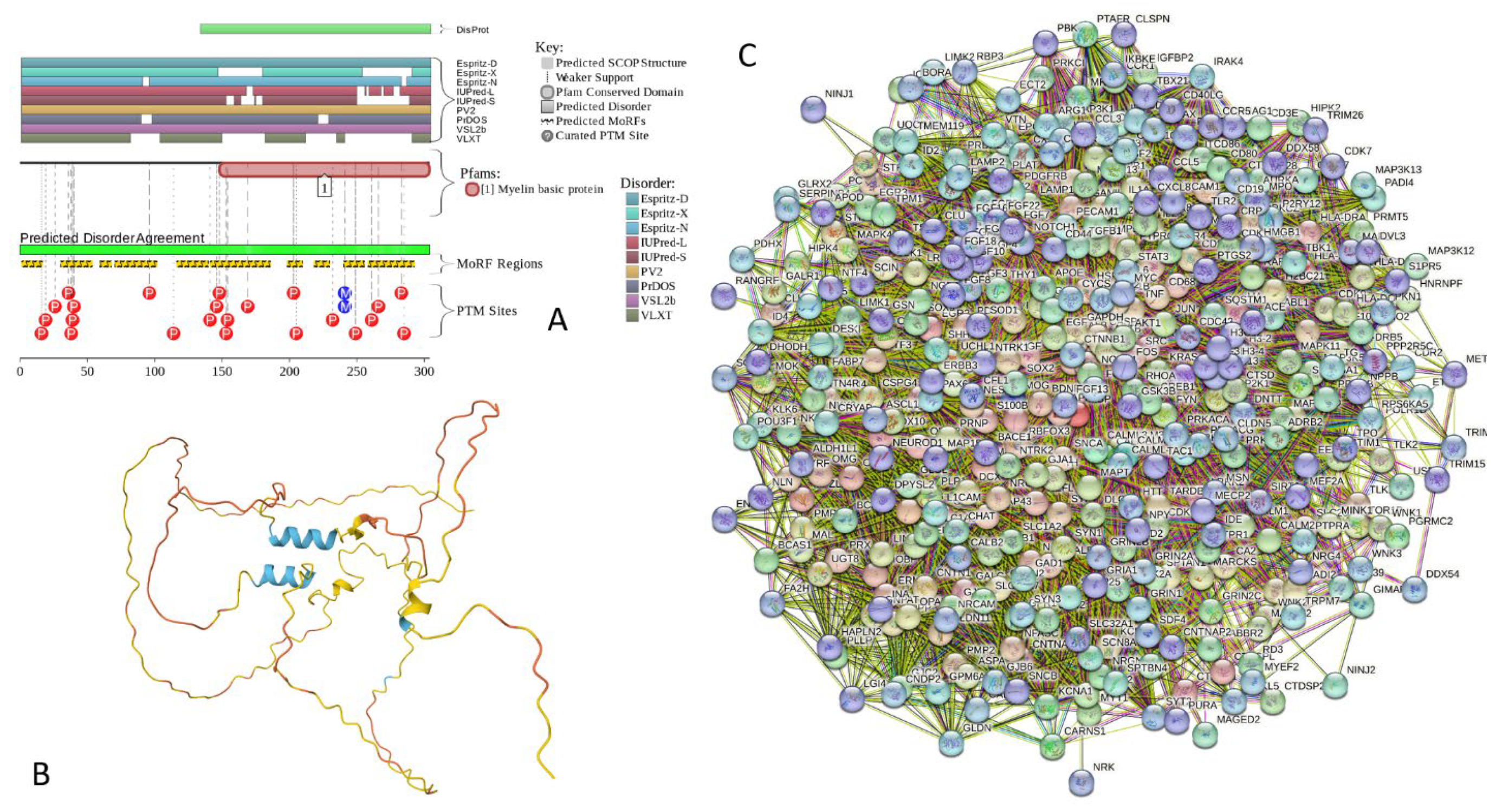
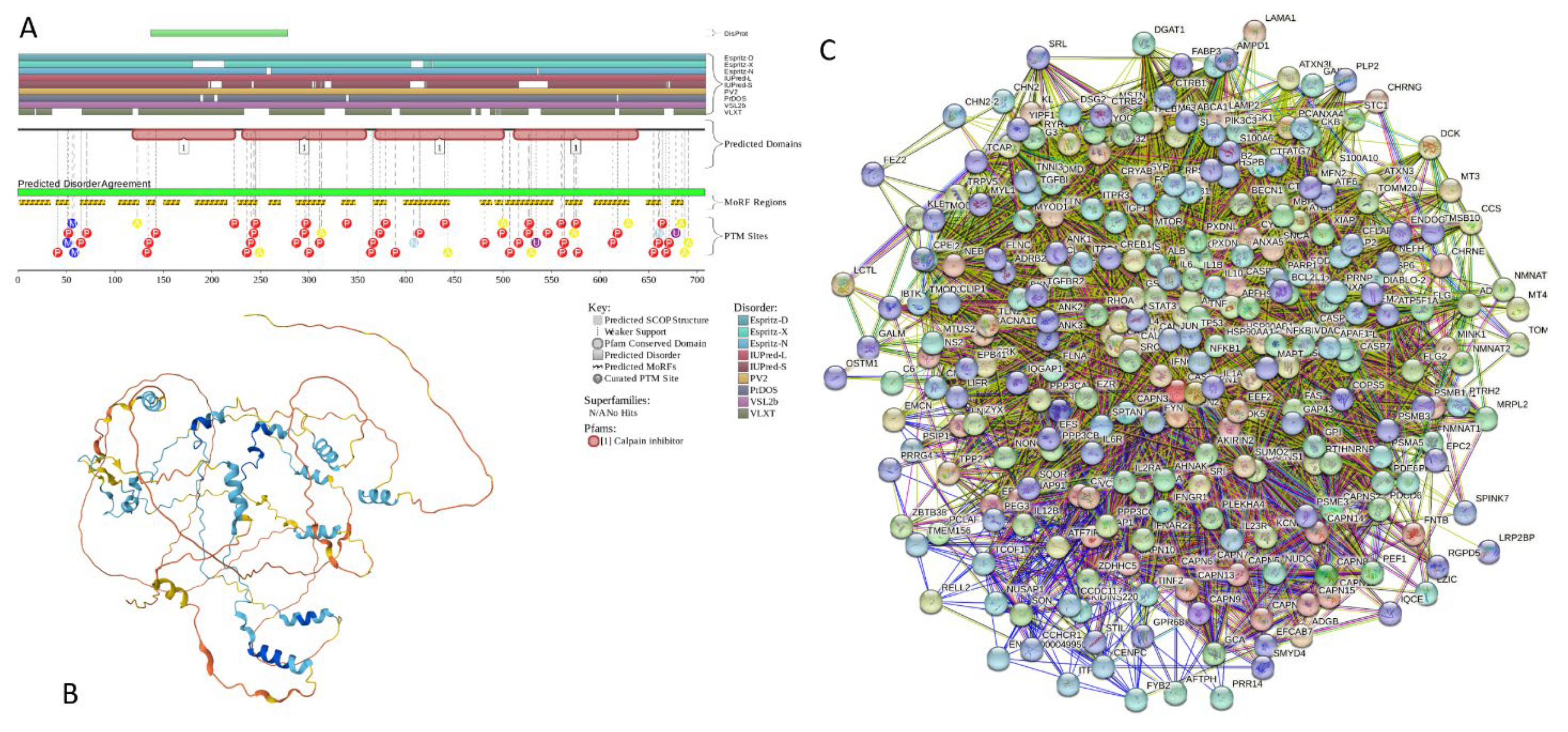
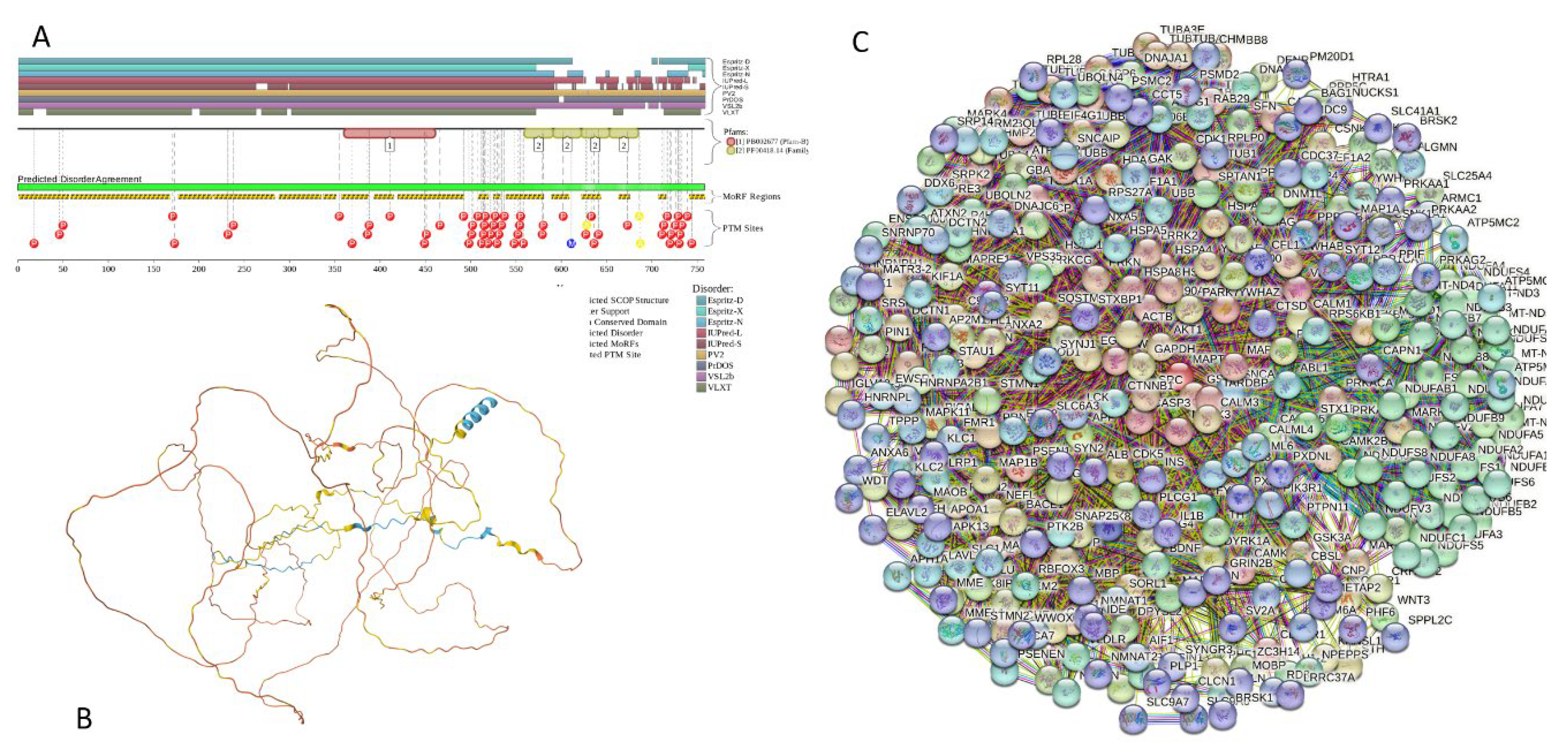
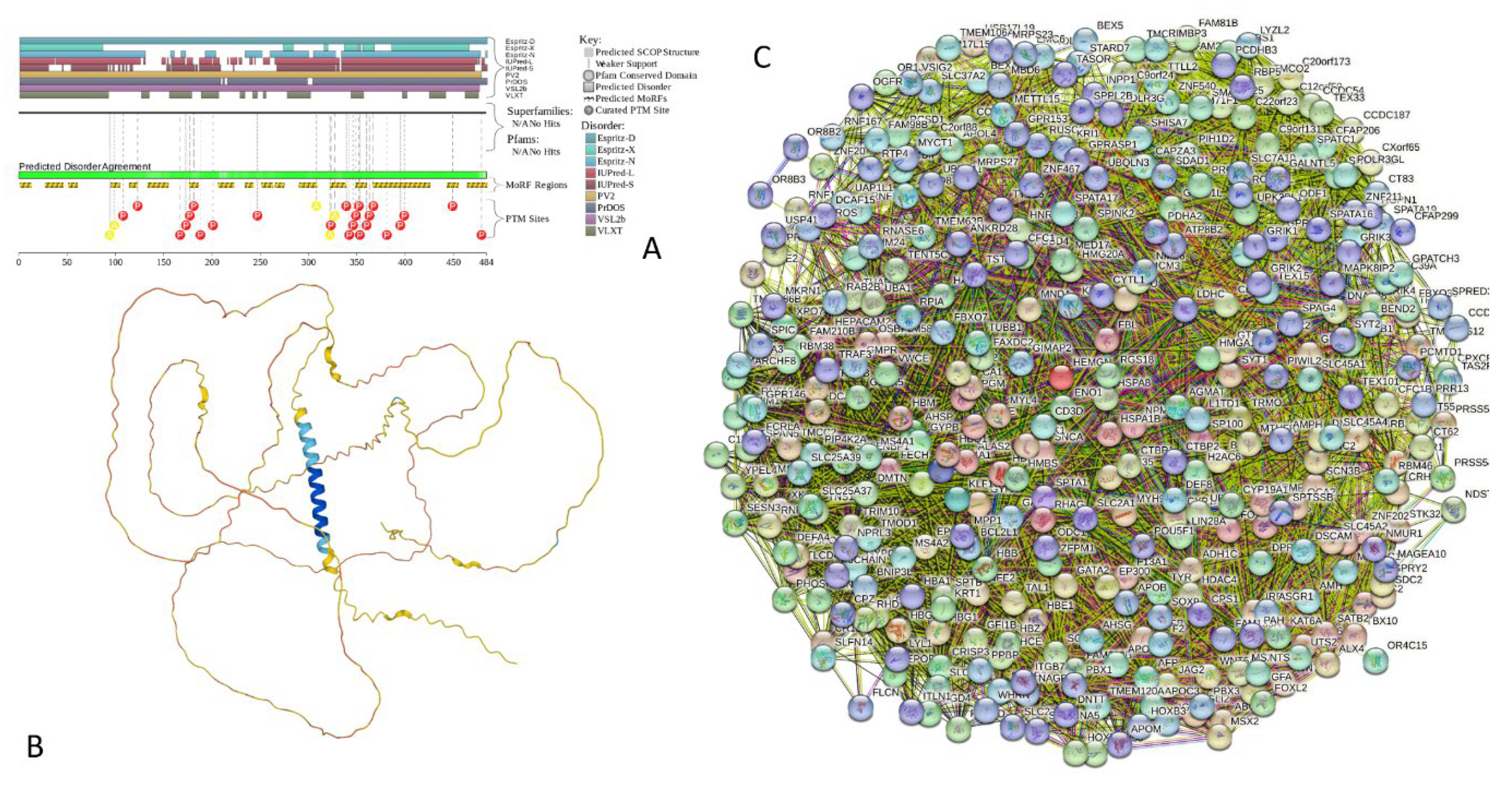
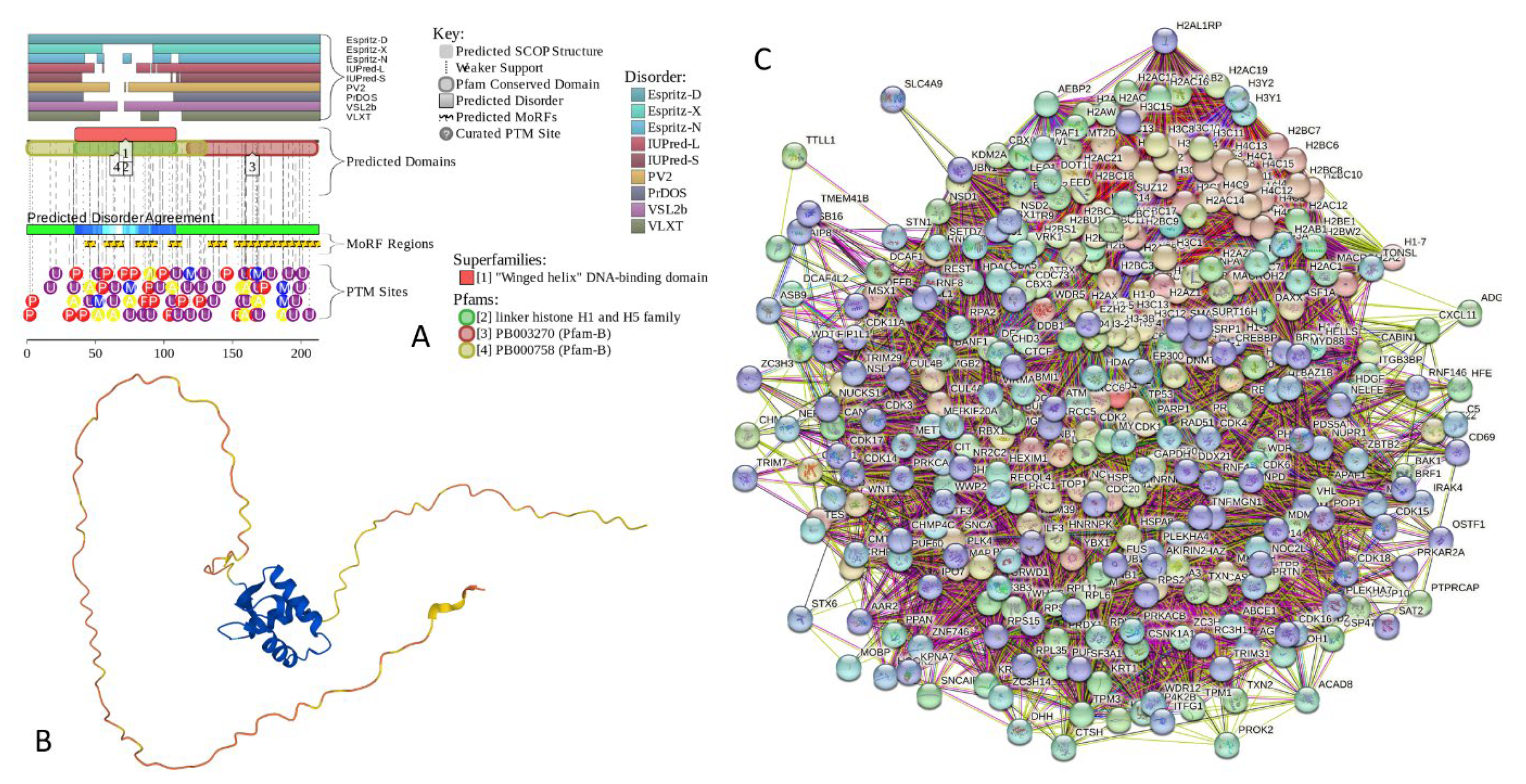
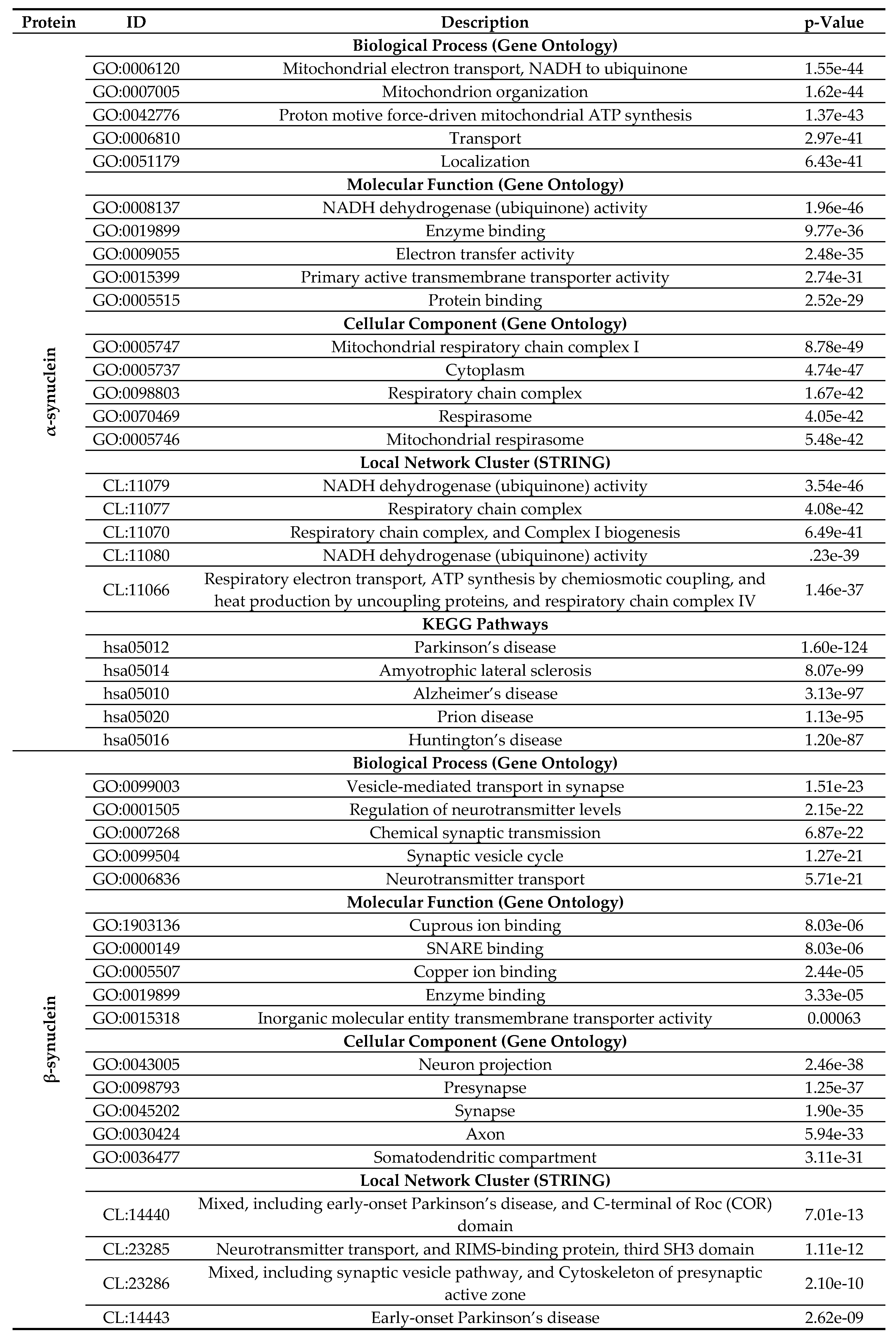
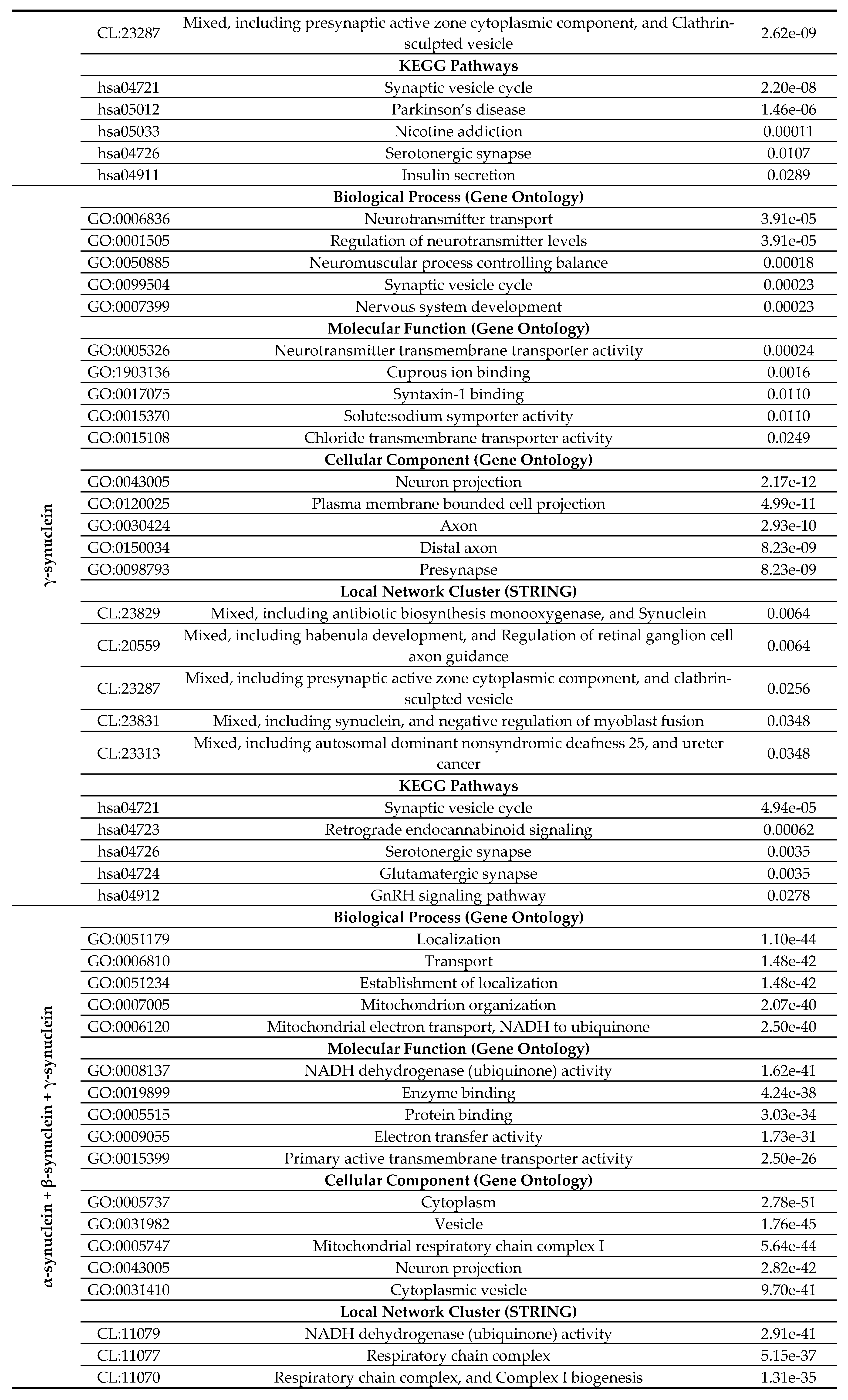
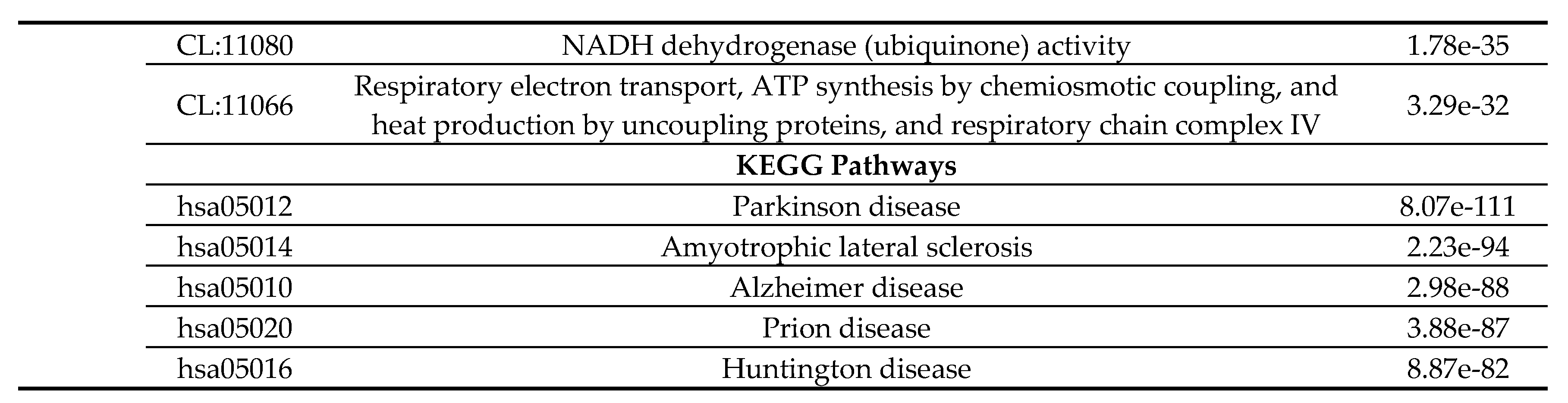
| Dataset | Protein number | PONDR® VSL2 score vs. PONDR® VSL2 (%) plot | CH-CDF plot | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Blue | Cyan | Dark pink | Pink | Red | Q1 | Q2 | Q3 | Q4 | ||
| Human brain proteome | 10,611 | 15 (0.15%) | 411 (3.87%) | 3,593 (33.86%) |
2,335 (22.00%) |
4,257 (40.12%) | 6,203 (58.5%) | 2,938 (27.7%) | 1,193 (11.2%) |
277 (2.6%) |
| Joint α-β-γ interactome | 467 | 0 (0.0%) |
22 (4.7%) | 172 (36.8%) |
110 (23.6%) |
163 (34.9%) |
292 (62.5%) |
105 (22.5%) |
61 (13.1%) |
9 (1.9%) |
| α-Synuclein interactome | 356 | 0 (0.0%) |
20 (5.6%) |
135 (37.9%) |
89 (25.0%) |
112 (31.5%) |
234 (65.7%) |
65 (18.3%) |
48 (13.5%) |
9 (2.5%) |
| β-Synuclein interactome | 85 | 0 (0.0%) |
0 (0.0%) |
30 (35.3%) |
19 (22.3%) |
36 (32.4%) |
48 (56.5%) |
26 (30.6%) |
11 (12.9%) |
0 (0.0%) |
| γ-Synuclein interactome | 32 | 0 (0.0%) |
0 (0.0%) |
12 (37.5%) |
4 (12.5%) |
16 (50.0%) |
14 (43.75%) |
14 (43.75%) |
4 (12.5%) |
0 (0.0%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





