Submitted:
23 May 2025
Posted:
26 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Mouse Lines and Genotyping
2.2. Tissue Processing
2.3. Bright Field Imaging
2.4. RNA-seq Data Analysis
2.5. RNA Probe Synthesis and In Situ Hybridization on Sections
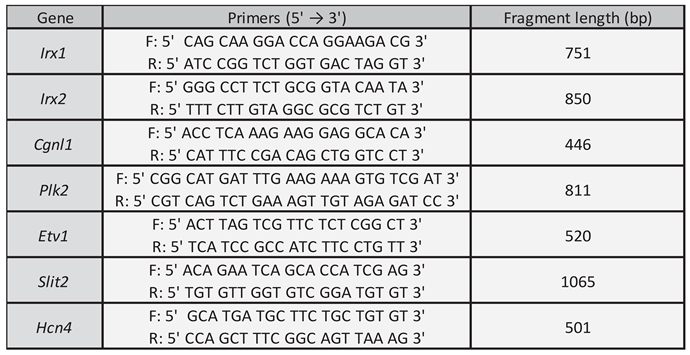
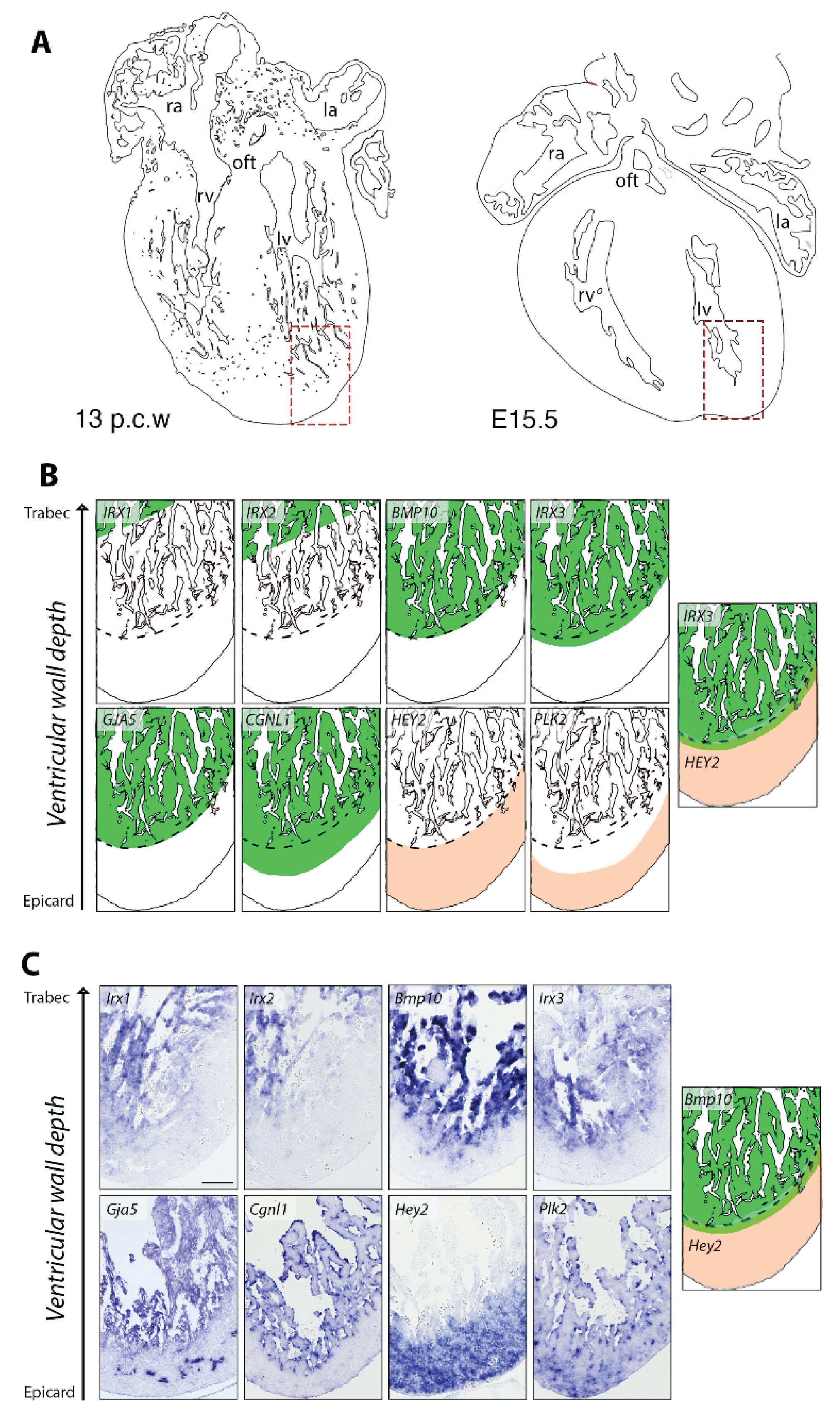
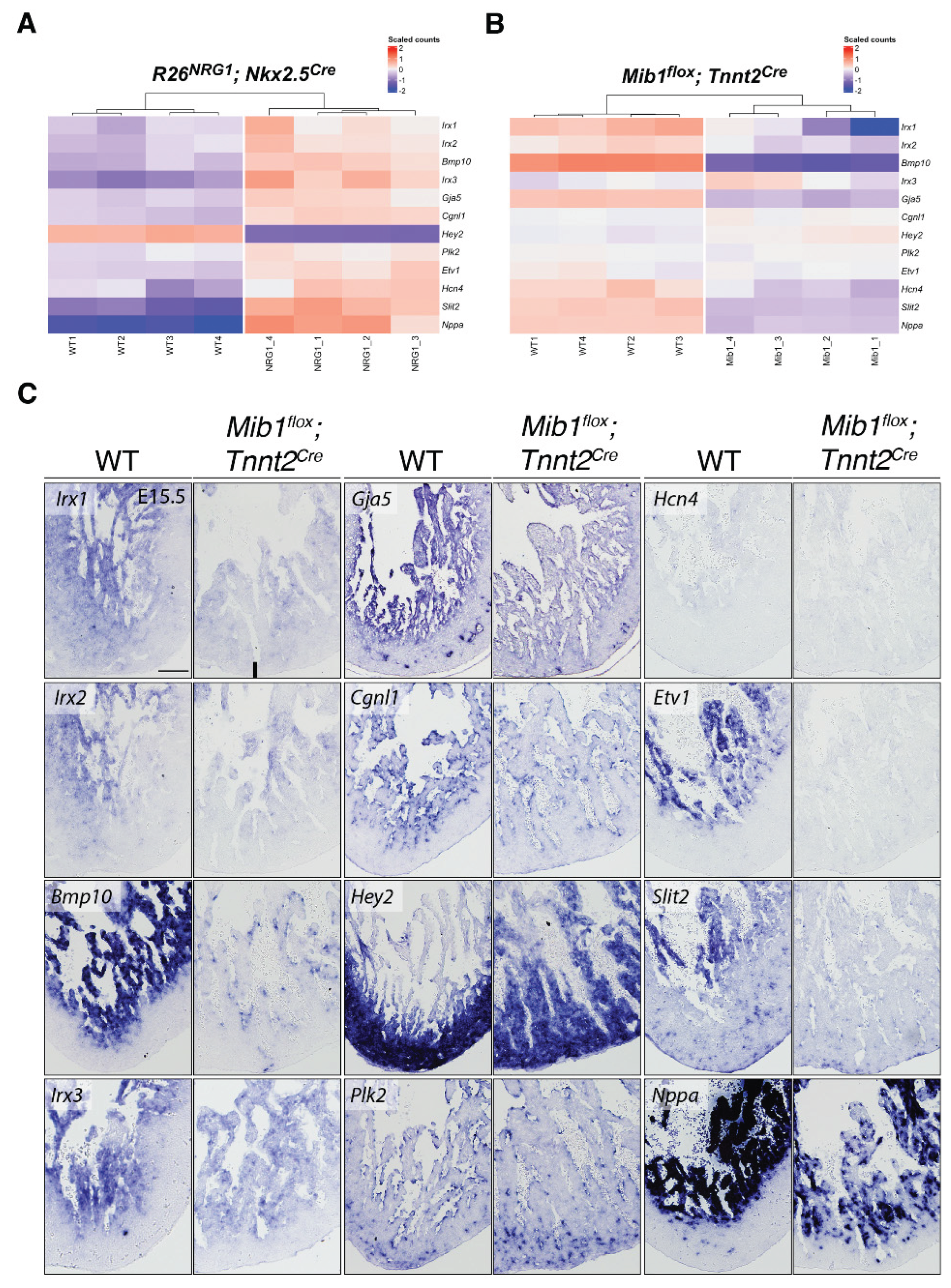
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgements
Conflicts of Interest
References
- Kelly, R.G.; Buckingham, M.E.; Moorman, A.F. Heart fields and cardiac morphogenesis. Cold Spring Harbor perspectives in medicine 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Wessels, A.; Sedmera, D. Developmental anatomy of the heart: a tale of mice and man. Physiol Genomics 2003, 15, 165–176. [Google Scholar] [CrossRef]
- Christoffels, V.M.; Habets, P.E.; Franco, D.; Campione, M.; de Jong, F.; Lamers, W.H.; Bao, Z.Z.; Palmer, S.; Biben, C.; Harvey, R.P.; et al. Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol 2000, 223, 266–278. [Google Scholar] [CrossRef]
- Moorman, A.F.M.; Christoffels, V.M. Cardiac Chamber Formation: Development, Genes, and Evolution. American Physiological Society 2003. [Google Scholar] [CrossRef] [PubMed]
- Sedmera, D.; Pexieder, T.; Vuillemin, M.; Thompson, R.P.; Anderson, R.H. Developmental patterning of the myocardium. Anat Rec 2000, 258, 319–337. [Google Scholar] [CrossRef]
- Samsa, L.A.; Yang, B.; Liu, J. Embryonic cardiac chamber maturation: Trabeculation, conduction, and cardiomyocyte proliferation. American journal of medical genetics. Part C, Seminars in medical genetics 2013, 163C, 157–168. [Google Scholar] [CrossRef]
- Miquerol, L.; Moreno-Rascon, N.; Beyer, S.; Dupays, L.; Meilhac, S.M.; Buckingham, M.E.; Franco, D.; Kelly, R.G. Biphasic development of the mammalian ventricular conduction system. Circ Res 2010, 107, 153–161. [Google Scholar] [CrossRef] [PubMed]
- D'Amato, G.; Phansalkar, R.; Naftaly, J.A.; Fan, X.; Amir, Z.A.; Rios Coronado, P.E.; Cowley, D.O.; Quinn, K.E.; Sharma, B.; Caron, K.M.; et al. Endocardium-to-coronary artery differentiation during heart development and regeneration involves sequential roles of Bmp2 and Cxcl12/Cxcr4. Dev Cell 2022, 57, 2517–2532.e6. [Google Scholar] [CrossRef]
- Luxán, G.; Casanova, J.C.; Martínez-Poveda, B.; Prados, B.; D'Amato, G.; MacGrogan, D.; Gonzalez-Rajal, A.; Dobarro, D.; Torroja, C.; Martinez, F. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nature medicine 2013, 19, 193–201. [Google Scholar] [CrossRef]
- Tian, X.; Li, Y.; He, L.; Zhang, H.; Huang, X.; Liu, Q.; Pu, W.; Zhang, L.; Li, Y.; Zhao, H. Identification of a hybrid myocardial zone in the mammalian heart after birth. Nature communications 2017, 8, 87. [Google Scholar] [CrossRef]
- Chiang, I.K.; Humphrey, D.; Mills, R.J.; Kaltzis, P.; Pachauri, S.; Graus, M.; Saha, D.; Wu, Z.; Young, P.; Sim, C.B.; et al. Sox7-positive endothelial progenitors establish coronary arteries and govern ventricular compaction. EMBO Rep 2023, 24, e55043. [Google Scholar] [CrossRef] [PubMed]
- Asp, M.; Giacomello, S.; Larsson, L.; Wu, C.; Fürth, D.; Qian, X.; Wärdell, E.; Custodio, J.; Reimegård, J.; Salmén, F. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 2019, 179, 1647–1660.e19. [Google Scholar] [CrossRef]
- Holman, A.R.; Tran, S.; Destici, E.; Farah, E.N.; Li, T.; Nelson, A.C.; Engler, A.J.; Chi, N.C. Single-cell multi-modal integrative analyses highlight functional dynamic gene regulatory networks directing human cardiac development. Cell Genomics 2024, 4. [Google Scholar] [CrossRef] [PubMed]
- Litviňuková, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Worth, C.L.; Lindberg, E.L.; Kanda, M.; Polanski, K.; Heinig, M.; Lee, M. Cells of the adult human heart. Nature 2020, 588, 466–472. [Google Scholar] [CrossRef]
- Farah, E.N.; Hu, R.K.; Kern, C.; Zhang, Q.; Lu, T.-Y.; Ma, Q.; Tran, S.; Zhang, B.; Carlin, D.; Monell, A. Spatially organized cellular communities form the developing human heart. Nature 2024, 627, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Choquet, C.; Boulgakoff, L.; Kelly, R.G.; Miquerol, L. New Insights into the Development and Morphogenesis of the Cardiac Purkinje Fiber Network: Linking Architecture and Function. J Cardiovasc Dev Dis 2021, 8. [Google Scholar] [CrossRef]
- D'Amato, G.; Luxán, G.; de la Pompa, J.L. Notch signalling in ventricular chamber development and cardiomyopathy. The FEBS journal 2016, 283, 4223–4237. [Google Scholar] [CrossRef]
- Grego-Bessa, J.; Gómez-Apiñaniz, P.; Prados, B.; Gómez, M.J.; MacGrogan, D.; de la Pompa, J.L. Nrg1 Regulates cardiomyocyte migration and cell cycle in ventricular development. Circulation research 2023, 133, 927–943. [Google Scholar] [CrossRef]
- Siguero-Álvarez, M.; Salguero-Jiménez, A.; Grego-Bessa, J.; de la Barrera, J.; MacGrogan, D.; Prados, B.; Sánchez-Sáez, F.; Piñeiro-Sabarís, R.; Felipe-Medina, N.; Torroja, C. A human hereditary cardiomyopathy shares a genetic substrate with bicuspid aortic valve. Circulation 2023, 147, 47–65. [Google Scholar] [CrossRef]
- Choquet, C.; Nguyen, T.H.M.; Sicard, P.; Buttigieg, E.; Tran, T.T.; Kober, F.; Varlet, I.; Sturny, R.; Costa, M.W.; Harvey, R.P.; et al. Deletion of Nkx2-5 in trabecular myocardium reveals the developmental origins of pathological heterogeneity associated with ventricular non-compaction cardiomyopathy. PLoS Genet 2018, 14, e1007502. [Google Scholar] [CrossRef]
- Jiao, K.; Kulessa, H.; Tompkins, K.; Zhou, Y.; Batts, L.; Baldwin, H.S.; Hogan, B.L.M. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes & development 2003, 17, 2362–2367. [Google Scholar]
- Christoffels, V.M.; Keijser, A.G.M.; Houweling, A.C.; Clout, D.E.W.; Moorman, A.F.M. Patterning the embryonic heart: identification of five mouse Iroquois homeobox genes in the developing heart. Developmental biology 2000, 224, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Kanzler, B.; Kuschert, S.J.; Liu, Y.H.; Mallo, M. Hoxa-2 restricts the chondrogenic domain and inhibits bone formation during development of the branchial area. Development 1998, 125, 2587–2597. [Google Scholar] [CrossRef]
- Gomez-Del Arco, P.; Isern, J.; Jimenez-Carretero, D.; Lopez-Maderuelo, D.; Pineiro-Sabaris, R.; El Abdellaoui-Soussi, F.; Torroja, C.; Vera-Pedrosa, M.L.; Grima-Terren, M.; Benguria, A.; et al. The G4 resolvase Dhx36 modulates cardiomyocyte differentiation and ventricular conduction system development. Nat Commun 2024, 15, 8602. [Google Scholar] [CrossRef]
- Cui, M.; Bezprozvannaya, S.; Hao, T.; Elnwasany, A.; Szweda, L.I.; Liu, N.; Bassel-Duby, R.; Olson, E.N. Transcription factor NFYa controls cardiomyocyte metabolism and proliferation during mouse fetal heart development. Dev Cell 2023, 58, 2867–2880.e7. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Bais, A.; He, H.; Rios, C.; Jiang, S.; Xu, J.; Chang, C.; Kostka, D.; Li, G. Single-cell transcriptomic analysis identifies murine heart molecular features at embryonic and neonatal stages. Nature Communications 2022, 13, 7960. [Google Scholar] [CrossRef]
- Boulgakoff, L.; D'Amato, G.; Miquerol, L. Molecular Regulation of Cardiac Conduction System Development. Curr Cardiol Rep 2024, 26, 943–952. [Google Scholar] [CrossRef]
- Hu, W.; Xin, Y.; Zhang, L.; Hu, J.; Sun, Y.; Zhao, Y. Iroquois Homeodomain transcription factors in ventricular conduction system and arrhythmia. International Journal of Medical Sciences 2018, 15, 808. [Google Scholar] [CrossRef]
- Kim, K.H.; Rosen, A.; Bruneau, B.G.; Hui, C.C.; Backx, P.H. Iroquois homeodomain transcription factors in heart development and function. Circ Res 2012, 110, 1513–1524. [Google Scholar] [CrossRef]
- Zhang, S.-S.; Kim, K.-H.; Rosen, A.; Smyth, J.W.; Sakuma, R.; Delgado-Olguín, P.; Davis, M.; Chi, N.C.; Puviindran, V.; Gaborit, N. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His–Purkinje network. Proceedings of the National Academy of Sciences 2011, 108, 13576–13581. [Google Scholar] [CrossRef]
- Shekhar, A.; Lin, X.; Liu, F.-Y.; Zhang, J.; Mo, H.; Bastarache, L.; Denny, J.C.; Cox, N.J.; Delmar, M.; Roden, D.M. Transcription factor ETV1 is essential for rapid conduction in the heart. The Journal of clinical investigation 2016, 126, 4444–4459. [Google Scholar] [CrossRef] [PubMed]
- Chrifi, I.; Hermkens, D.; Brandt, M.M.; Van Dijk, C.G.M.; Bürgisser, P.E.; Haasdijk, R.; Pei, J.; van de Kamp, E.H.M.; Zhu, C.; Blonden, L. Cgnl1, an endothelial junction complex protein, regulates GTPase mediated angiogenesis. Cardiovascular research 2017, 113, 1776–1788. [Google Scholar] [CrossRef] [PubMed]
- Lotto, J.; Cullum, R.; Drissler, S.; Arostegui, M.; Garside, V.C.; Fuglerud, B.M.; Clement-Ranney, M.; Thakur, A.; Underhill, T.M.; Hoodless, P.A. Cell diversity and plasticity during atrioventricular heart valve EMTs. Nat Commun 2023, 14, 5567. [Google Scholar] [CrossRef]
- Künzel, S.R.; Hoffmann, M.; Weber, S.; Künzel, K.; Kämmerer, S.; Günscht, M.; Klapproth, E.; Rausch, J.S.E.; Sadek, M.S.; Kolanowski, T. Diminished PLK2 induces cardiac fibrosis and promotes atrial fibrillation. Circulation research 2021, 129, 804–820. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, M.; Lorenz, V.; Ivanek, R.; Della Verde, G.; Gaudiello, E.; Marsano, A.; Pfister, O.; Kuster, G.M. Polo-Like Kinase 2 is Dynamically Regulated to Coordinate Proliferation and Early Lineage Specification Downstream of Yes-Associated Protein 1 in Cardiac Progenitor Cells. Journal of the American Heart Association 2017, 6, e005920. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




