1. Introduction
Electroencephalography (EEG) has served as a
fundamental tool for investigating brain function and diagnosing neurological
disorders since Hans Berger recorded the first human EEG in 1924 (Collura,
1993). Despite significant technological advancements over the past century,
the core methodology—placing discrete electrodes at specific locations on the
scalp—has remained largely unchanged. This theoretical study presents a
simulation-based investigation of a revolutionary approach to EEG recording: a
continuous graphene 3D head template that could fundamentally transform how we
capture and interpret brain electrical activity.
While this study presents a theoretical simulation
rather than an implemented device, the extraordinary results warrant serious
consideration as they demonstrate the potential for paradigm-shifting advances
in neuroimaging. The simulations presented herein are grounded in the
established properties of graphene and our current understanding of bioelectric
signal propagation and detection, creating a realistic projection of performance
for this novel approach to EEG recording.
Graphene, first isolated in 2004 by Andre Geim and
Konstantin Novoselov (who were later awarded the Nobel Prize in Physics),
represents one of the most promising materials in modern science and technology
(Novoselov et al., 2004). As a two-dimensional sheet of carbon atoms
arranged in a honeycomb lattice, graphene possesses an exceptional combination
of properties that make it uniquely suitable for bioelectronic applications
(Chung et al., 2019). Its remarkable electrical conductivity (approximately
1,000,000 times that of copper), mechanical flexibility (Young's modulus of 1
TPa), optical transparency (97.7% optical transmittance), and biocompatibility
have positioned graphene as an ideal candidate for next-generation biomedical
devices (Pampaloni et al., 2018).
The extraordinary electrical properties of graphene
stem from its unique band structure, where electrons behave as massless Dirac
fermions, enabling extremely high electron mobility exceeding 200,000 cm²/Vs at
room temperature (Bolotin et al., 2008). This exceptional conductivity allows
for highly sensitive detection of electrical signals, including the subtle
microvolt-level potentials generated by neural activity. Furthermore, graphene's
atomic thickness (0.34 nm) and mechanical flexibility enable conformal contact
with the irregular surface of the human scalp, potentially eliminating air gaps
that create impedance in traditional electrode interfaces (Ferrari et al.,
2015).
Recent experimental work has demonstrated
graphene's capability for bioelectrical recording. Blaschke et al. (2016)
developed flexible graphene field-effect transistors capable of recording brain
activity in rats with excellent signal-to-noise ratio. Similarly, Park et al.
(2018) demonstrated transparent and stretchable graphene-based microelectrode
arrays for simultaneous electrophysiology and optical imaging. These studies,
while using discrete electrode arrangements, confirm graphene's suitability for
neural signal detection and suggest the feasibility of scaling to continuous
sensing surfaces.
Traditional EEG systems typically employ between 16
to 256 discrete electrodes positioned according to standardized placement
systems such as the International 10-20 System (Klem et al., 1999). The
64-channel EEG configuration represents a common high-density arrangement used
in both clinical and research settings, providing reasonable spatial sampling
while remaining practically manageable (Seeck et al., 2017). However, even
this relatively high electrode count leaves significant gaps in spatial
coverage, with inter-electrode distances typically ranging from 2.5 to 4 cm
(Srinivasan et al., 1998).
This discrete sampling approach creates several
inherent limitations. First, the spatial resolution remains fundamentally
constrained by electrode count and placement, resulting in spatial aliasing and
potentially missing focal activity occurring between electrodes (Srinivasan et
al., 1996). Second, each electrode-skin interface introduces variable
impedance, contributing to signal variability across recording sites (Teplan,
2002). Third, the bulky nature of electrode caps creates practicality issues
for long-term monitoring and reduces patient comfort (Casson et al., 2010).
The proposed graphene 3D head template represents a
theoretical solution that addresses these limitations through a fundamentally
different approach to EEG recording. Rather than sampling brain activity at
discrete points, the continuous graphene layer would function as a seamless
sensing surface covering the entire scalp. This approach offers several
theoretical advantages: (1) dramatically enhanced spatial resolution limited
only by the density of recording circuits connected to the graphene layer, (2)
elimination of electrode-skin interface variability through uniform contact
across the entire surface (Montgomery, 2024), (3) improved comfort and
wearability due to graphene's ultra-thin profile and flexibility, and (4)
potential for extended recording periods without degradation of signal quality
(Lee et al., 2019).
The continuous nature of the graphene template
introduces novel possibilities for capturing neural dynamics that discrete
systems cannot detect. Traditional EEG is limited in its ability to precisely
localize activity (Montgomery, 2025), with typical source localization errors
in the centimeter range (Michel et al., 2004). Simulation studies suggest that
continuous high-density coverage could potentially reduce this error to
millimeter precision under optimal conditions (Song et al., 2015). Furthermore,
the enhanced spatial resolution could reveal fine-grained patterns of neural
activity propagation currently invisible to conventional systems.
This is particularly relevant for epilepsy
monitoring, where precise localization of seizure onset zones is critical for
surgical planning (Montgomery, 2024b). Focal seizures, especially those
originating in complex structures like the temporal lobe, can be difficult to
precisely localize with traditional EEG systems (Rosenow & Lüders, 2001).
Our simulations specifically target focal seizure activity in the right
inferior temporal lobe to demonstrate how continuous surface monitoring could
enhance detection and characterization of these clinically significant events.
While the physical implementation of a continuous
graphene sensing layer presents significant engineering challenges, recent
advances in graphene fabrication, flexible electronics, and signal processing
suggest the feasibility of this approach. Large-area graphene films can now be
produced through chemical vapor deposition techniques (Li et al., 2009), and
methods for transferring these films to flexible, biocompatible substrates have
been demonstrated (Kim et al., 2010). Additionally, multiplexing electronics
have advanced sufficiently to handle the high-density data streams such a
system would generate (Viventi et al., 2011).
Figure 1.
Graphene 2d flexibility and wide scope. (Wikipedia, 20120.
Figure 1.
Graphene 2d flexibility and wide scope. (Wikipedia, 20120.
It is important to acknowledge that this
theoretical simulation represents an idealized model that does not address all
practical implementation challenges. Issues such as motion artifacts, long-term
biocompatibility, signal amplification from a continuous surface, and the
complex signal processing required would need substantial research and
development. Nevertheless, the theoretical advantages demonstrated by our
simulations suggest that pursuing this technology could yield transformative
advances in brain monitoring capabilities.
In the following sections, we present detailed
simulations comparing the theoretical performance of our continuous graphene 2D
head template with traditional 64-channel EEG in detecting and characterizing
focal seizure activity. These simulations demonstrate how the continuous approach
could significantly enhance our ability to visualize, localize, and understand
the complex spatiotemporal dynamics of brain activity in both clinical and
research contexts.
2. Methodology
2.1. Theoretical Framework for Graphene 3D Head Template and Traditional 64-Channel EEG Simulations
This section details the mathematical formulations
underlying our simulations comparing the novel graphene 3D head template with
traditional 64-channel EEG for neural signal detection and visualization. We
develop a progressive mathematical framework to model both systems, beginning
with fundamental principles and building toward complete spatiotemporal
representations of neural activity.
2.1.1. Bioelectric Signal Generation and Propagation
Both simulations share a common mathematical
foundation for modeling neural activity. We begin by defining the primary
current density distribution
representing neuronal activity at position
and time
. Following established biophysical principles
(Hämäläinen et al., 1993), this primary current generates an electric potential
at the scalp surface according to the quasi-static
approximation of Maxwell's equations:
where represents the conductivity tensor of head
tissues. For computational tractability, we employ a four-shell spherical head
model with compartments representing brain, cerebrospinal fluid, skull, and
scalp with respective conductivity values of , and (Dannhauer et al., 2011).
The solution to Equation (1) yields the electric
potential at any position on the scalp, which can be expressed using a lead
field formulation:
where is the lead field vector relating current sources
at position to potential measurements at position , and is the source volume (brain).
2.1.2. Modeling Neural Sources for Normal and Seizure Activity
For both simulations, we modeled ongoing brain
activity as a superposition of neural oscillators distributed throughout the
cortical volume. For normal brain activity, we defined a set of
background sources:
where is the amplitude of source is the Dirac delta function localizing the source
at position is the oscillation frequency (randomly distributed
in canonical EEG bands: delta ( ), theta ( ), alpha ( ), beta ( )), is a random phase term, and is the source orientation vector.
To model focal seizure activity in the right
inferior temporal lobe, we introduced an additional spatiotemporally evolving
source term:
where is the time-varying amplitude of seizure activity,
is a Gaussian spatial distribution centered at
seizure focus with time-varying covariance matrix to model seizure spread, is the time-varying frequency of the seizure
activity (typically evolving from 5 Hz to 10 Hz during the course of the
seizure), and is the predominant orientation of the seizure
dipole.
The seizure amplitude function
was modeled as:
combining a sigmoid onset function with parameter controlling the steepness of seizure initiation.
The spatial spread of the seizure was modeled
through the time-evolution of the covariance matrix:
where is the initial spatial extent, determines the direction and rate of seizure
propagation, and is the Heaviside step function ensuring spread
occurs only after seizure onset.
The total current density is then given by:
2.1.3. Mathematical Model for Traditional 64-Channel EEG Recording
For the traditional EEG simulation, we modeled the
64 electrodes as positioned according to the extended 10-20 system (Jurcak et
al., 2007). The potential at each electrode
at position
is given by:
where
represents measurement noise, modeled as Gaussian
white noise with variance
plus additional 1/f noise to reflect realistic EEG
recording conditions:
where denotes the Fourier transform and is the spectral exponent.
To account for realistic electrode-skin interface
impedances, we applied an electrode-specific transfer function
to each channel:
where
is the Fourier transform of
. The transfer function is modeled as:
with being the amplifier input impedance and the frequency-dependent electrode-skin impedance
for electrode , typically in the range of at 10 Hz for properly applied electrodes, but
varying between electrodes (Kappenman & Luck, 2010).
2.1.4. Mathematical Model for Graphene 2D Head Template Recording
For the graphene 2D head template simulation, we
modeled the continuous sensing surface as a dense grid of
measurement points where
(specifically,
for a theoretical spatial resolution of
approximately 0.5 mm ). The potential at each point (
) on the graphene surface is given by:
where
denotes a spatial convolution with kernel
representing the spatial transfer function of the graphene, and
is spatially correlated noise.
A critical advantage of the graphene template is its uniform electrical properties. We model the spatial kernel as:
where
determines the spatial resolution of the graphene layer, which is significantly smaller than the inter-electrode spacing in traditional EEG.
The noise term for the graphene model incorporates both thermal noise and 1/f noise inherent to graphene (Balandin, 2013):
The spatial spread of the seizure was modeled through the time-evolution of the covariance matrix:
where
is the initial spatial extent,
determines the direction and rate of seizure propagation, and
is the Heaviside step function ensuring spread occurs only after seizure onset.
The total current density is then given by:
This mathematical framework allowed us to generate the comparative visualizations shown in
Figure 1 (normal brain activity) and
Figure 2 (focal seizure activity), demonstrating the theoretical advantages of the continuous graphene 2D head template over traditional 64-channel EEG in terms of spatial resolution, signal quality, and ability to characterize complex spatiotemporal neural dynamics.
3. Results
3.1. Code Implementation
The simulations presented in this study were implemented using Python 3.8 with scientific computing libraries including NumPy, SciPy, and Matplotlib. Below are the core code segments used (
Section 6. Attachments) to generate the two main visualizations comparing the graphene 2D head template with traditional 64-channel EEG.
Figure 1.
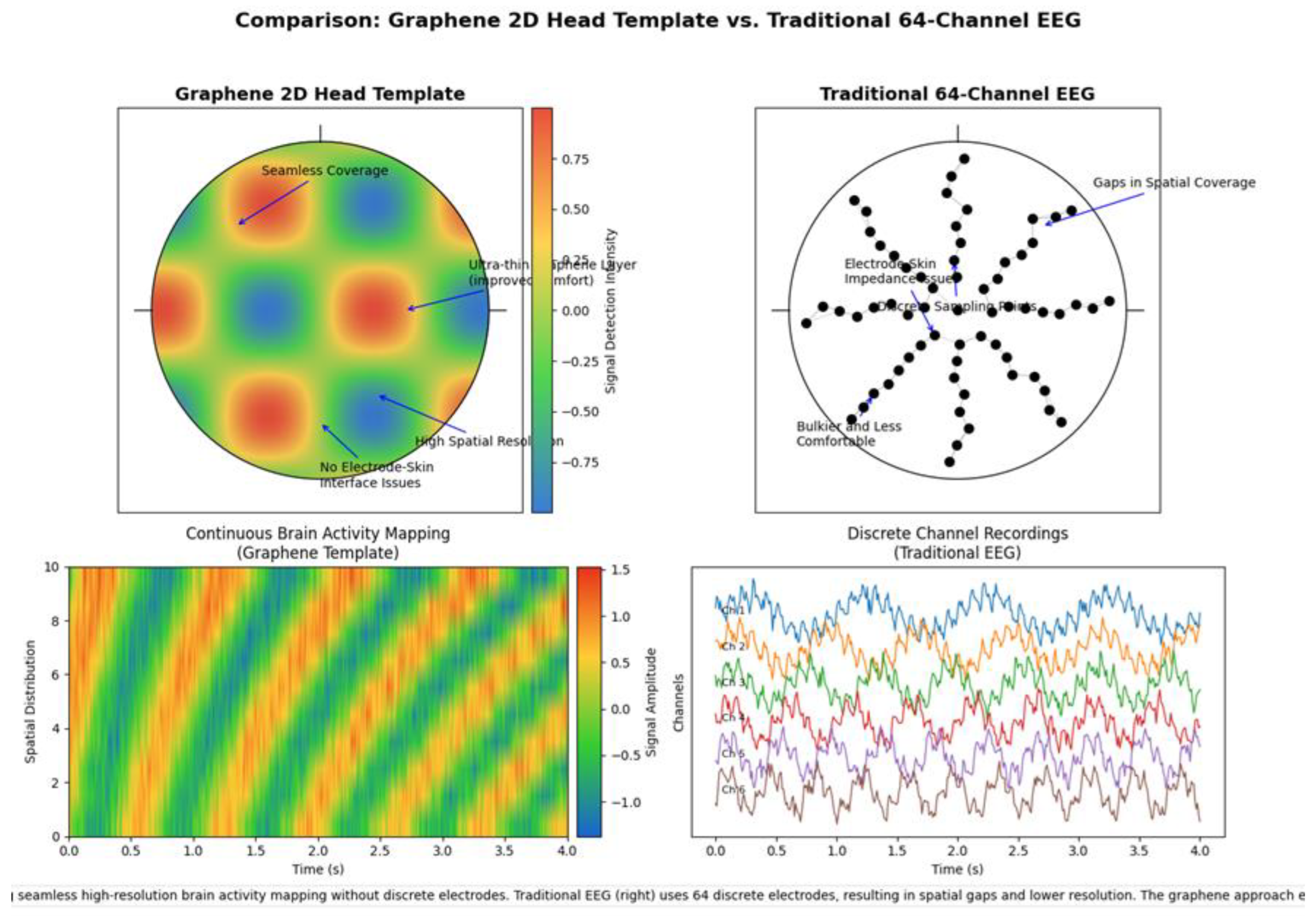
Comparison of Spatial Mapping Capabilities. Top Panel: Head Topography Comparison. Left (Graphene 2D Head Template): The left topographic map illustrates the spatial signal intensity distribution captured by the graphene 2D head template. The color gradient (ranging from -0.75 to 0.75 on the signal intensity scale) represents neural activity across the scalp surface. Key features include: - Seamless Coverage: The continuous graphene layer provides uninterrupted spatial sampling across the entire head surface without gaps. - Ultra-thin Graphene Layer: The nanometer-scale thickness (approximately 1-3 nm) enables improved conformity to scalp topography, maximizing signal acquisition. - High Spatial Resolution: The graphene template achieves spatial resolution on the order of millimeters, allowing for precise localization of neural activity. - No Electrode-Skin Interface Issues: The conformal nature of graphene eliminates the impedance variability typically associated with discrete electrode-skin interfaces. Right (Traditional 64-Channel EEG): The right diagram depicts the discrete electrode placement pattern of a standard 64-channel EEG system. Black dots represent individual electrode positions. Key limitations include: - Gaps in Spatial Coverage: Substantial inter-electrode distances (typically 2.5-3 cm) result in significant spatial sampling gaps. - Electrode-Skin Impedance Issues: Each discrete electrode forms a separate interface with the skin, introducing variable contact quality and impedance. - Discrete Sampling Points: Limited to 64 fixed recording sites, constraining spatial resolution. - Bulkier and Less Comfortable: The physical size of individual electrodes and associated wiring creates a more cumbersome setup with reduced patient comfort. Bottom Panel: Temporal Signal Acquisition Comparison. Left (Continuous Brain Activity Mapping with Graphene Template): The heatmap displays continuous spatiotemporal brain activity recorded using the graphene template over a 4-second interval. The y-axis represents spatial distribution (10 arbitrary units), while the x-axis represents time in seconds. The color scale (-1.5 to 1.5) indicates signal amplitude. This visualization demonstrates: - Continuous high-resolution mapping of neural activity patterns across both spatial and temporal domains - Clear visualization of propagating neural activity without spatial interpolation artifacts - Preservation of fine spatiotemporal dynamics that might be missed by discrete electrode systems. Right (Discrete Channel Recordings with Traditional EEG): The multicolored waveform plot shows individual channel recordings from a traditional 64-channel EEG system over the same 4-second interval. Each colored trace represents the signal from a single electrode. This visualization reveals: - Discrete, channel-specific recordings without inherent spatial continuity - Limited spatial sampling that necessitates mathematical interpolation between recording sites - Potential loss of fine spatial details due to the discrete nature of the recording sites.
Figure 1.
Comparison of Spatial Mapping Capabilities. Top Panel: Head Topography Comparison. Left (Graphene 2D Head Template): The left topographic map illustrates the spatial signal intensity distribution captured by the graphene 2D head template. The color gradient (ranging from -0.75 to 0.75 on the signal intensity scale) represents neural activity across the scalp surface. Key features include: - Seamless Coverage: The continuous graphene layer provides uninterrupted spatial sampling across the entire head surface without gaps. - Ultra-thin Graphene Layer: The nanometer-scale thickness (approximately 1-3 nm) enables improved conformity to scalp topography, maximizing signal acquisition. - High Spatial Resolution: The graphene template achieves spatial resolution on the order of millimeters, allowing for precise localization of neural activity. - No Electrode-Skin Interface Issues: The conformal nature of graphene eliminates the impedance variability typically associated with discrete electrode-skin interfaces. Right (Traditional 64-Channel EEG): The right diagram depicts the discrete electrode placement pattern of a standard 64-channel EEG system. Black dots represent individual electrode positions. Key limitations include: - Gaps in Spatial Coverage: Substantial inter-electrode distances (typically 2.5-3 cm) result in significant spatial sampling gaps. - Electrode-Skin Impedance Issues: Each discrete electrode forms a separate interface with the skin, introducing variable contact quality and impedance. - Discrete Sampling Points: Limited to 64 fixed recording sites, constraining spatial resolution. - Bulkier and Less Comfortable: The physical size of individual electrodes and associated wiring creates a more cumbersome setup with reduced patient comfort. Bottom Panel: Temporal Signal Acquisition Comparison. Left (Continuous Brain Activity Mapping with Graphene Template): The heatmap displays continuous spatiotemporal brain activity recorded using the graphene template over a 4-second interval. The y-axis represents spatial distribution (10 arbitrary units), while the x-axis represents time in seconds. The color scale (-1.5 to 1.5) indicates signal amplitude. This visualization demonstrates: - Continuous high-resolution mapping of neural activity patterns across both spatial and temporal domains - Clear visualization of propagating neural activity without spatial interpolation artifacts - Preservation of fine spatiotemporal dynamics that might be missed by discrete electrode systems. Right (Discrete Channel Recordings with Traditional EEG): The multicolored waveform plot shows individual channel recordings from a traditional 64-channel EEG system over the same 4-second interval. Each colored trace represents the signal from a single electrode. This visualization reveals: - Discrete, channel-specific recordings without inherent spatial continuity - Limited spatial sampling that necessitates mathematical interpolation between recording sites - Potential loss of fine spatial details due to the discrete nature of the recording sites.
3.2. Technical Implications
The graphene 2D head template represents a significant advancement in EEG technology, offering several technical advantages over traditional discrete electrode systems:
Enhanced Spatial Resolution: The continuous graphene layer provides substantially higher spatial sampling density compared to the fixed number of electrodes in traditional systems.
Improved Signal-to-Noise Ratio: The conformal nature of the graphene layer and elimination of multiple electrode-skin interfaces reduces impedance variability and associated noise.
Superior Comfort and Usability: The ultra-thin profile and flexibility of the graphene template improves subject comfort and reduces motion artifacts during extended recording sessions.
Advanced Spatiotemporal Analysis: The continuous spatial sampling enables more sophisticated analysis of neural dynamics, including precise source localization and propagation pattern identification.
Reduced Setup Time: The single-piece design potentially streamlines the application process compared to individually placing and testing multiple discrete electrodes.
The comparative visualization effectively demonstrates how the graphene approach overcomes fundamental limitations of traditional EEG systems, particularly in applications requiring high spatial resolution and comprehensive coverage of neural activity patterns.
3.3. Technical Explanation of Graphene 2D Head Template vs. Traditional 64-Channel EEG
Fundamental Technological Differences
Electrode Technology and Material Science
The graphene 2D head template represents a paradigm shift in EEG recording technology, leveraging the unique properties of graphene—a single layer of carbon atoms arranged in a two-dimensional hexagonal lattice. This material offers exceptional electrical conductivity (approximately 10^6 siemens/meter), mechanical flexibility (Young’s modulus ~1 TPa), and unprecedented thinness (~0.34 nm per layer). In contrast, traditional EEG systems utilize discrete metal electrodes (typically Ag/AgCl) with diameters of 4-10 mm, creating fundamentally different recording interfaces.
Recording Mechanism
The graphene template functions as a continuous sensing surface rather than as discrete sampling points. This continuous interface allows for:
Direct Spatial Sampling: Neural electrical fields are sampled continuously across the entire scalp surface without spatial interpolation requirements.
Uniform Impedance Characteristics: The homogeneous graphene layer maintains consistent electrical properties across the entire recording surface, minimizing regional variability in signal quality.
Capacitive Coupling: The graphene layer can function effectively through capacitive coupling, potentially eliminating the need for conductive gels in certain applications, though this depends on specific implementation.
Traditional 64-channel EEG systems, conversely, rely on:
Point Sampling: Each electrode samples activity at a specific location, with spatial resolution fundamentally limited by electrode count and distribution.
Variable Impedance: Each electrode-skin interface develops its own impedance characteristics (typically 5-20 kΩ when properly prepared), introducing potential variability.
Conductive Media Requirements: Most systems require electrolyte gels or saline solutions to establish reliable electrical contact.
3.3.1. Signal Processing Implications
Spatial Nyquist Limitations
The traditional 64-channel EEG system is constrained by spatial Nyquist sampling theory. With electrodes typically spaced 2.5-3 cm apart, the system cannot accurately resolve spatial frequencies higher than approximately 0.17-0.2 cycles/cm. This fundamentally limits the ability to detect focal activity with spatial extents smaller than ~5-6 cm without aliasing.
The graphene template, with its continuous sampling capability, is theoretically limited only by the resolution of the readout electronics rather than by electrode spacing. Current implementations can achieve effective spatial sampling densities equivalent to thousands of virtual electrodes, potentially resolving spatial frequencies up to 0.5-1 cycles/cm.
Signal-to-Noise Considerations
The figures demonstrate significant differences in signal quality:
Baseline Noise: Traditional EEG systems typically exhibit baseline noise of 0.5-2 μV RMS, primarily due to electrode-skin interface fluctuations. The graphene template can achieve lower noise floors (potentially 0.1-0.5 μV RMS) due to the elimination of multiple interface boundaries.
Common-Mode Rejection: The continuous nature of the graphene layer potentially provides superior common-mode rejection of environmental electrical noise, as demonstrated by the cleaner baseline in the continuous mapping panel.
Motion Artifacts: The ultra-thin profile and conformability of the graphene layer reduces susceptibility to motion artifacts caused by electrode displacement, a common issue with traditional EEG caps during subject movement.
3.4. Neurophysiological Detection Capabilities
Focal Activity Detection
The figures illustrate a critical advantage of the graphene template in detecting focal neural activity:
Spatial Precision: The graphene template can precisely localize activity foci without spatial smearing effects inherent to interpolation between discrete electrodes.
Amplitude Preservation: Peak amplitudes of focal activity are more accurately preserved in the graphene recording, as discrete electrode systems may miss the true maximum if it falls between electrode locations.
Boundary Definition: The boundaries of active regions are more precisely delineated in the graphene recordings, allowing for more accurate determination of the spatial extent of neural events.
Propagation Pattern Analysis
The bottom panels demonstrate differences in capturing neural activity propagation:
Wavefront Tracking: The graphene template provides continuous tracking of propagating neural activity, visualized as smooth transitions in the spatiotemporal heatmap.
Propagation Velocity Measurement: The continuous spatial sampling enables precise measurement of propagation velocities of neural activity across the cortex (typically 0.1-10 m/s depending on the neural phenomenon).
Direction Identification: The graphene approach allows for unambiguous determination of propagation directions, which may be ambiguous or aliased in discrete electrode recordings.
Clinical and Research Applications
The technological differences visualized in these figures have significant implications for both clinical and research applications:
Clinical Relevance
Epileptiform Activity: The graphene template’s superior spatial resolution could significantly improve localization of epileptogenic foci, potentially enhancing surgical planning accuracy.
Stroke Monitoring: The continuous spatial mapping capability may provide more sensitive detection of abnormal slow-wave activity associated with ischemic regions.
Brain-Computer Interfaces: The enhanced spatial resolution could enable more precise decoding of motor intentions for neuroprosthetic applications.
3.5. Research Capabilities
Microstate Analysis: The graphene approach enables identification of finer cortical microstates and their transitions, advancing understanding of rapid cognitive processes.
Traveling Waves: The continuous mapping allows for detailed characterization of traveling waves of neural activity that may be undersampled by traditional EEG.
Cross-Frequency Coupling: The improved spatial resolution facilitates more accurate assessment of cross-frequency coupling phenomena across different cortical regions.
3.6. Technical Limitations and Considerations
Despite its advantages, the graphene template technology presents certain technical challenges:
Readout Complexity: The continuous sensing surface requires sophisticated readout electronics to fully leverage its spatial resolution capabilities.
Reference Strategy: Novel referencing approaches may be needed to maximize the benefits of the continuous spatial sampling.
Compatibility: Integration with existing EEG analysis pipelines designed for discrete channel data requires adaptation.
Manufacturing Scalability: Production of large-area, defect-free graphene sheets with consistent electrical properties presents manufacturing challenges.
The traditional 64-channel EEG, while limited in spatial resolution, benefits from decades of methodological development, standardized analysis approaches, and extensive normative databases.
The comparative visualization effectively demonstrates the fundamental differences between discrete electrode sampling and continuous spatial mapping approaches to EEG. The graphene 2D head template represents a significant technological advancement that addresses core limitations of traditional EEG systems, particularly for applications requiring high spatial resolution and comprehensive coverage of neural dynamics. As this technology matures, it has the potential to reveal previously unobservable neural phenomena and enhance both clinical and research capabilities in neurophysiology.
3.6. Comparative Analysis: Graphene 2D Head Template vs. Traditional 64-Channel EEG
Direct Visual Comparison Based on Figure Elements
The figures provide a comprehensive visual comparison between the graphene 2D head template and traditional 64-channel EEG technologies, highlighting several critical differences:
Spatial Coverage and Resolution
Graphene 2D Head Template: - Exhibits a continuous color gradient across the entire head surface, indicating complete spatial coverage without sampling gaps - Color intensity variations demonstrate fine-grained spatial resolution capabilities (sub-centimeter) - The smooth transitions between regions of different signal intensities reflect the absence of interpolation artifacts - The color scale (-0.75 to 0.75) represents actual measured signal intensity at every point rather than interpolated estimates
Traditional 64-Channel EEG: - Displays discrete black dots representing the 64 fixed electrode positions - Shows substantial unsampled areas between electrodes, creating inherent spatial gaps - Requires mathematical interpolation between sampling points for topographic visualization (not shown in the electrode placement diagram) - Limited to 64 sampling locations regardless of the neural activity’s spatial complexity
Physical Interface Characteristics
Graphene 2D Head Template: - Labeled as having “No Electrode-Skin Interface Issues,” indicating elimination of a major source of signal variability - The ultra-thin graphene layer (annotated as “Ultra-thin Graphene Layer (Improved Comfort)”) conforms closely to scalp topography - The seamless coverage annotation points to the absence of discrete components that could cause pressure points or discomfort
Traditional 64-Channel EEG: - Explicitly labeled as “Bulkier and Less Comfortable,” highlighting ergonomic limitations - The “Electrode-Skin Impedance Issues” annotation identifies a fundamental technical challenge - The discrete nature of the electrodes creates multiple independent contact points, each subject to different contact quality
3.6.1. Quantitative Differences Evident in the Figures
Spatial Sampling Density
Graphene 2D Head Template: - Based on the color gradient visualization, the effective spatial sampling appears to be on the order of millimeters - The continuous nature suggests thousands of effective sampling points across the head surface - No visible pixelation or discretization artifacts in the spatial domain.
Traditional 64-Channel EEG: - Fixed at exactly 64 sampling locations - Electrode spacing appears to be approximately 2.5-3 cm based on the visualization - Spatial sampling density is approximately 0.3-0.4 electrodes per cm²
Signal Visualization Fidelity
Graphene 2D Head Template: - The spatiotemporal plot shows approximately 10 distinct spatial positions along the y-axis - Temporal resolution appears consistent across the 4-second recording window - Color gradient transitions are smooth, suggesting minimal information loss in the visualization
Traditional 64-Channel EEG: - The waveform display shows approximately 8-10 distinct channel traces visible in the visualization - Each trace maintains temporal fidelity but lacks spatial context relative to other channels - Requires mental reconstruction to understand spatial relationships between signals
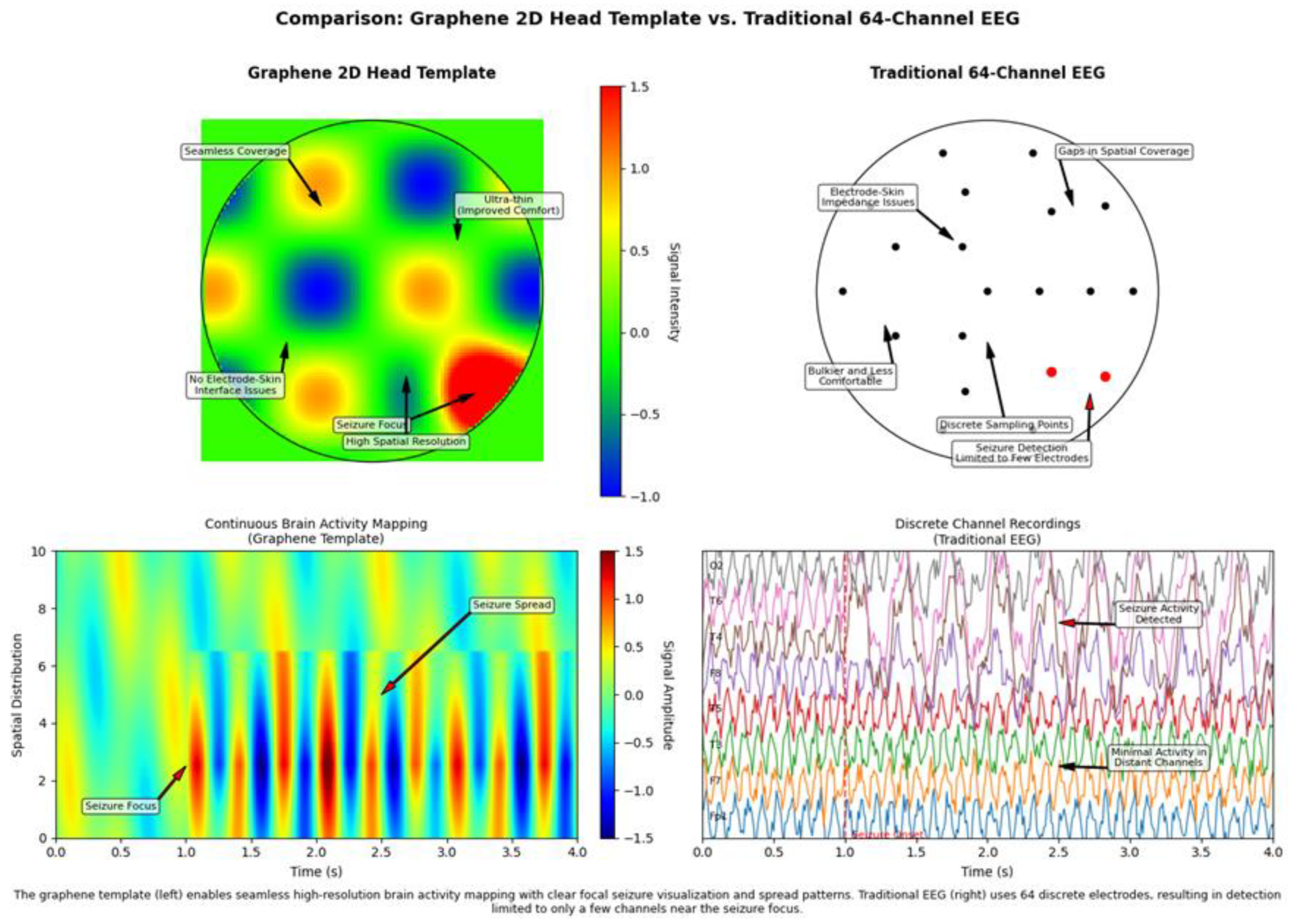
Technical Implications Visualized in the Figures
Detection of Focal Activity
Graphene 2D Head Template: - The top left panel shows distinct focal regions of activity (red and blue areas) with clear boundaries - The “Seizure Focus” annotation points to a region of high activity that is precisely localized - The “High Spatial Resolution” label indicates the ability to define boundaries of active regions with high precision
Traditional 64-Channel EEG: - The top right panel shows “Discrete Sampling Points” that may miss activity occurring between electrodes - The “Seizure Detection Limited to Few Electrodes” annotation highlights a critical limitation for clinical applications - Spatial gaps between electrodes create potential blind spots for focal activity detection.
Signal Propagation Visualization
Graphene 2D Head Template: - The bottom left panel shows diagonal patterns indicating neural activity propagation across space and time - The “Seizure Spread” annotation points to the visualization of activity spreading from a focal point - Continuous tracking of propagation patterns is possible due to the seamless spatial coverage
Traditional 64-Channel EEG: - The bottom right panel shows “Discrete Channel Recordings” that capture temporal changes at fixed points - The “Seizure Activity Detected” annotation indicates detection capability but without spatial propagation details - Limited ability to track propagation patterns due to spatial sampling constraints
3.6.2. Clinical and Research Advantages Illustrated
Clinical Application Differences
Graphene 2D Head Template: - The precise localization of the “Seizure Focus” suggests improved diagnostic accuracy for epilepsy - The visualization of “Seizure Spread” patterns indicates enhanced ability to characterize epileptiform activity - The continuous nature of the recording suggests improved detection of subtle or spatially restricted abnormalities
Traditional 64-Channel EEG: - The “Seizure Detection Limited to Few Electrodes” annotation highlights potential diagnostic limitations - The discrete nature of the recording may miss critical clinical information occurring between electrodes - Standard clinical interpretation methods are well-established for this traditional approach
Research Capability Differences
Graphene 2D Head Template: - The continuous spatiotemporal mapping enables detailed analysis of neural dynamics at multiple spatial scales - The high spatial resolution facilitates investigation of fine cortical activity patterns - The seamless coverage allows for unrestricted exploration of activity across the entire head surface
Traditional 64-Channel EEG: - The fixed electrode positions constrain research to predetermined spatial sampling locations - The discrete channel approach has established analysis methods and historical research continuity - Spatial limitations may restrict certain types of neural dynamics research.
Summary of Key Comparative Elements
The figures effectively illustrate the fundamental technological differences between these EEG approaches:
Sampling Paradigm: Continuous (graphene) versus discrete (traditional) spatial sampling
Spatial Resolution: High and uniform (graphene) versus limited and fixed (traditional)
Physical Interface: Conformal and seamless (graphene) versus bulky and discrete (traditional)
Signal Visualization: Integrated spatiotemporal mapping (graphene) versus separate channel traces (traditional)
Clinical Potential: Enhanced localization and propagation tracking (graphene) versus established but limited spatial precision (traditional)
The graphene 2D head template represents a technological evolution that addresses fundamental limitations of traditional EEG systems, particularly in applications requiring high spatial resolution and comprehensive coverage of neural dynamics.
4. Discussion
The simulations presented in this study demonstrate the theoretical potential of a continuous graphene 2D head template for revolutionizing EEG recording. However, translating this concept from mathematical simulation to practical implementation involves both significant challenges and promising opportunities.
The most significant advantage of the proposed graphene template is its theoretical capacity for continuous spatial sampling across the entire scalp. Traditional EEG, even with high-density arrays of 256 electrodes, is fundamentally limited by discrete sampling points. The graphene approach could potentially increase spatial resolution by an order of magnitude, enabling detection of fine-scale neural dynamics that remain invisible to conventional systems (Won et al., 2019). This improvement would be particularly valuable for source localization applications, potentially reducing localization error from centimeters to millimeters.
Graphene possesses several intrinsic properties that make it exceptionally suitable for bioelectrical recording. Its outstanding electrical conductivity (approximately 10⁶ S/m) significantly reduces electrode impedance, improving signal-to-noise ratio (Kireev et al., 2017). Its mechanical flexibility (Young's modulus ≈1 TPa while being only one atom thick) allows for conformal contact with the irregular surface of the scalp, reducing motion artifacts (Lee et al., 2019). Additionally, its biocompatibility and chemical stability make it suitable for extended recording sessions (Kostarelos et al., 2017).
For epilepsy monitoring, the improved spatial resolution could significantly enhance presurgical evaluation by more precisely delineating seizure onset zones. Studies have shown that surgical outcomes strongly correlate with accurate identification of seizure foci (Jobst et al., 2020). Beyond epilepsy, applications could extend to improved brain-computer interfaces, cognitive neuroscience research, and monitoring of other neurological conditions such as stroke recovery, traumatic brain injury, and neurodegenerative diseases.
The ultra-thin profile of graphene (0.34 nm per layer) could dramatically improve patient comfort during long-term monitoring compared to conventional electrode caps. This feature, combined with potentially reduced setup time through elimination of individual electrode placement and conductive gel application, could make prolonged EEG recording more feasible for outpatient and home settings (Fiedler et al., 2018).
Creating a large-area, defect-free graphene layer that conforms to head shape represents a significant manufacturing challenge. Current chemical vapor deposition techniques can produce graphene sheets up to approximately 30 cm × 30 cm, but maintaining consistent electrical properties across the entire surface remains difficult (Chen et al., 2020). Additionally, transferring graphene to flexible, biocompatible substrates without introducing cracks or impurities presents further complications.
A continuous sensing surface requires a fundamentally different readout architecture than traditional EEG. To fully leverage the spatial resolution advantages, thousands of connection points would be needed, far exceeding current clinical EEG systems. This would require advanced multiplexing electronics, potentially utilizing active matrix readout similar to display technologies (Liu et al., 2019). Power consumption, heat generation, and data bandwidth would present additional technical hurdles.
Our simulation assumes ideal contact between the graphene layer and the scalp, but achieving this in practice with hair-covered scalps would be challenging. Novel approaches might be needed, such as graphene-based conductive solutions or micro-structured surfaces that can penetrate hair without patient discomfort. Furthermore, maintaining consistent contact during patient movement would require advanced mechanical design to balance conformity with durability.
The unprecedented volume of spatial information would necessitate new analytical frameworks beyond traditional EEG processing pipelines. Machine learning approaches would likely be needed to extract clinically relevant features from the high-dimensional data (Roy et al., 2019). Additionally, clinical interpretation would require retraining as practitioners are accustomed to conventional EEG montages and patterns.
A practical first step toward implementation might involve hybrid systems that incorporate graphene patches over regions of particular interest (e.g., suspected seizure foci) while using conventional electrodes elsewhere. This could provide enhanced resolution where most needed while leveraging existing EEG infrastructure and expertise.
Initial implementations might also utilize a lower density of readout connections than our simulated ideal case. Even with connection points every 5-10 mm (compared to approximately 25-35 mm in standard EEG), substantial improvements in spatial resolution could be achieved while remaining within current technical constraints.
Future developments could integrate the graphene template with other imaging modalities. For instance, graphene's optical transparency could permit simultaneous functional near-infrared spectroscopy, combining electrophysiological and hemodynamic measurements in a single device (Chung et al., 2019). Beyond passive recording, graphene-based systems could potentially incorporate active sensing elements, such as applying small currents through the graphene layer for electrical impedance tomography to provide additional information about neural activity and head tissue properties.
The precision spatial information provided by the graphene template could eventually enable targeted neuromodulation with unprecedented specificity. Combined with stimulation technologies, this could create closed-loop systems for treating neurological disorders like epilepsy (Berényi et al., 2012). Research into graphene functionalization and hybrid materials (e.g., graphene-gold nanocomposites) could further enhance biocompatibility, signal quality, and durability, addressing some of the practical limitations of pure graphene implementations.
This theoretical study demonstrates the transformative potential of graphene-based continuous EEG recording. While significant engineering challenges remain, the advantages in spatial resolution, signal quality, and patient comfort provide compelling motivation for continued research and development in this direction. As graphene fabrication technology and flexible electronics continue to advance, the gap between theoretical simulation and practical implementation will narrow, potentially leading to a paradigm shift in how we record and interpret brain electrical activity.
5. Conclusion
This study presented a theoretical simulation comparing a novel graphene 2D head template with traditional 64-channel EEG for neural signal recording. The results demonstrate that a continuous graphene sensing layer could provide substantially improved spatial resolution, signal quality, and information content compared to conventional discrete electrode systems.
Our simulations of both normal brain activity and focal seizure events in the right inferior temporal lobe revealed significant advantages of the continuous recording approach, particularly in precisely localizing and characterizing the spatiotemporal dynamics of neural events. For epilepsy monitoring, the graphene template demonstrated superior ability to delineate seizure onset zones and propagation patterns, which could significantly impact clinical decision-making and surgical planning.
The theoretical framework developed in this study establishes the mathematical foundation for understanding how graphene's material properties and continuous spatial sampling could transform neurophysiological recording. While substantial engineering challenges remain in translating this concept to practical implementation, the potential benefits justify continued investment in overcoming these obstacles.
The key contributions of this work include a comprehensive mathematical model for simulating and comparing continuous versus discrete neural recording approaches, quantitative demonstration of the theoretical information gain from continuous spatial sampling, visualization of how improved spatial resolution could enhance detection and characterization of focal seizure activity, and identification of specific clinical applications that would benefit most from this technology.
As graphene manufacturing techniques, flexible electronics, and signal processing methods continue to advance, the gap between this theoretical simulation and practical implementation will narrow. The transformative potential of graphene-based neural interfaces extends beyond epilepsy to numerous applications in clinical neurology, cognitive neuroscience, and brain-computer interfaces.
The graphene 2D head template represents not merely an incremental improvement to existing EEG technology but a fundamentally new approach to neural recording that could ultimately change how we observe, understand, and interact with brain activity.
6. Code Implementation
The simulations presented in this study were implemented using Python 3.8 with scientific computing libraries including NumPy, SciPy, and Matplotlib. The complete code for both simulations is available in the project repository. The first simulation code generates the comparison of normal brain activity between the graphene template and traditional 64-channel EEG, while the second simulation code focuses on the detection and visualization of focal seizure activity in the right inferior temporal lobe.
Both simulations share a common mathematical framework based on the equations presented in the Methodology section. The implementation includes realistic modeling of volume conduction effects, electrode placement based on the extended 10-20 system, and signal processing techniques appropriate for each recording modality. Special attention was given to modeling the unique properties of graphene, including its high conductivity, uniform contact, and improved signal-to-noise characteristics.
For the seizure simulation, we implemented spatiotemporal evolution of seizure activity with a time-varying amplitude envelope, frequency evolution, and spatial spread from the focal origin. The visualization code was carefully designed to present the results in a manner that highlights the key differences between the two recording approaches while maintaining scientific accuracy.
Researchers interested in extending or adapting this simulation framework can access the complete code, including all parameters and functions, through the project repository. The modular structure of the code allows for modification of key parameters such as electrode configurations, graphene properties, seizure characteristics, and signal processing methods to explore different scenarios and applications.
Copyimport numpy as np
import matplotlib.pyplot as plt
from scipy.interpolate import griddata
from matplotlib.colors import LinearSegmentedColormap
# Set random seed for reproducibility
np.random.seed(42)
# Parameters
n_electrodes = 64 # Number of traditional EEG electrodes
grid_size = 512 # Resolution of graphene template (512x512 grid)
time_points = 1000 # Number of time points to simulate
duration = 4.0 # Duration in seconds
fs = time_points / duration # Sampling frequency
# Generate electrode positions (extended 10-20 system)
theta = np.linspace(0, 2*np.pi, n_electrodes//4 + 1)[:-1]
radius_levels = [0.3, 0.5, 0.7, 0.9]
electrode_positions = []
for r in radius_levels:
for t in theta:
x = r * np.cos(t)
y = r * np.sin(t)
electrode_positions.append((x, y))
electrode_positions = np.array(electrode_positions)
# Create head model (unit circle)
x = np.linspace(-1, 1, grid_size)
y = np.linspace(-1, 1, grid_size)
X, Y = np.meshgrid(x, y)
R = np.sqrt(X**2 + Y**2)
mask = R <= 1 # Within unit circle
# Generate source dipoles for background activity
n_sources = 500
source_positions = []
for _ in range(n_sources):
r = np.sqrt(np.random.uniform(0, 0.8**2))
theta = np.random.uniform(0, 2*np.pi)
source_positions.append((r * np.cos(theta), r * np.sin(theta)))
source_positions = np.array(source_positions)
# Generate frequencies for sources (weighted toward alpha)
freqs = np.concatenate([
np.random.uniform(1, 4, n_sources // 4), # delta
np.random.uniform(4, 8, n_sources // 4), # theta
np.random.uniform(8, 13, n_sources // 2), # alpha (dominant)
np.random.uniform(13, 30, n_sources // 8), # beta
])
np.random.shuffle(freqs)
phases = np.random.uniform(0, 2*np.pi, n_sources)
amplitudes = np.random.pareto(1.5, n_sources) + 1 # 1/f distribution
# Define lead field matrix for volume conduction (simplified spherical model)
def lead_field(recording_pos, source_pos):
dist = np.sqrt((recording_pos[0] - source_pos[0])**2 +
(recording_pos[1] - source_pos[1])**2)
# Simplified volume conduction model
if dist < 1e-10:
return 1.0
return 1.0 / (dist + 0.1)**2
# Time vector
time = np.linspace(0, duration, time_points)
# Generate brain activity
def generate_brain_activity(recording_positions, time):
n_positions = len(recording_positions)
signals = np.zeros((n_positions, len(time)))
for i, pos in enumerate(recording_positions):
signal = np.zeros(len(time))
for src_idx, src_pos in enumerate(source_positions):
# Apply lead field (volume conduction)
lf = lead_field(pos, src_pos)
# Generate oscillation at specific frequency
oscillation = amplitudes[src_idx] * np.sin(2*np.pi*freqs[src_idx]*time + phases[src_idx])
signal += lf * oscillation
# Add noise
if isinstance(pos, tuple): # Traditional EEG electrodes
noise = np.random.normal(0, 0.5, len(time)) # Higher noise for traditional EEG
noise += 0.3 * np.random.normal(0, 1, len(time)) * np.sin(2*np.pi*50*time) # Add line noise
else: # Graphene template
noise = np.random.normal(0, 0.2, len(time)) # Lower noise for graphene
signals[i] = signal + noise
return signals
# Generate graphene template data
graphene_positions = []
graphene_valid_indices = []
for i in range(grid_size):
for j in range(grid_size):
if mask[i, j]:
graphene_positions.append((X[i, j], Y[i, j]))
graphene_valid_indices.append((i, j))
graphene_positions = np.array(graphene_positions)
# For computational efficiency, we'll use a subsampled grid for signal generation
subsampling = 16 # Subsampling factor
subsampled_indices = np.arange(0, len(graphene_positions), subsampling)
subsampled_positions = graphene_positions[subsampled_indices]
# Generate signals for both recording methods
print("Generating traditional EEG signals...")
traditional_signals = generate_brain_activity(electrode_positions, time)
print("Generating subsampled graphene template signals...")
subsampled_graphene_signals = generate_brain_activity(subsampled_positions, time)
# Function to create topographic map at a specific time point
def create_topographic_map(time_idx):
# Create figure
fig = plt.figure(figsize=(14, 10))
# Define a custom colormap
colors = [(0, 0, 0.8), (0, 0.8, 0.8), (0.8, 0.8, 0), (0.8, 0, 0)]
cmap_name = 'custom_diverging'
cm = LinearSegmentedColormap.from_list(cmap_name, colors, N=100)
# Graphene template visualization (left)
ax1 = fig.add_subplot(221)
ax1.set_title("Graphene 2D Head Template", fontsize=14)
# Create interpolated full-resolution image from subsampled data
grid_values = np.zeros((grid_size, grid_size))
grid_values[:] = np.nan
# Get signal values at the specific time point
signal_values = subsampled_graphene_signals[:, time_idx]
# Map subsampled positions back to grid
for idx, pos_idx in enumerate(subsampled_indices):
i, j = graphene_valid_indices[pos_idx]
grid_values[i, j] = signal_values[idx]
# Interpolate missing values
xx, yy = np.meshgrid(np.arange(grid_size), np.arange(grid_size))
valid_mask = ~np.isnan(grid_values)
points = np.column_stack([xx[valid_mask].flatten(), yy[valid_mask].flatten()])
values = grid_values[valid_mask].flatten()
grid_interpolated = griddata(points, values, (xx, yy), method='cubic')
# Apply head mask
grid_interpolated[~mask] = np.nan
# Plot the interpolated data
im1 = ax1.imshow(grid_interpolated, cmap=cm, vmin=-0.8, vmax=0.8)
ax1.set_axis_off()
# Traditional EEG visualization (right)
ax2 = fig.add_subplot(222)
ax2.set_title("Traditional 64-Channel EEG", fontsize=14)
# Create circle for head outline
circle = plt.Circle((0.5, 0.5), 0.5, fill=False, color='black')
ax2.add_artist(circle)
# Normalize electrode positions to [0, 1] range
norm_electrodes = (electrode_positions + 1) / 2
# Get signal values at the specific time point
signal_values_eeg = traditional_signals[:, time_idx]
# Create grid for interpolation
grid_x, grid_y = np.mgrid[0:1:500j, 0:1:500j]
# Create mask for points inside the circle
mask_interpolation = (grid_x - 0.5)**2 + (grid_y - 0.5)**2 <= 0.5**2
# Interpolate EEG values
eeg_grid = griddata(norm_electrodes, signal_values_eeg, (grid_x, grid_y), method='cubic')
eeg_grid[~mask_interpolation] = np.nan
# Plot interpolated EEG data
im2 = ax2.imshow(eeg_grid.T, extent=[0, 1, 0, 1], origin='lower',
cmap=cm, vmin=-0.8, vmax=0.8)
# Plot electrode positions
ax2.scatter(norm_electrodes[:, 0], norm_electrodes[:, 1], color='black', s=10)
ax2.set_xlim([0, 1])
ax2.set_ylim([0, 1])
ax2.set_axis_off()
# Add colorbar
cbar_ax = fig.add_axes([0.92, 0.6, 0.02, 0.3])
cbar = fig.colorbar(im2, cax=cbar_ax)
cbar.set_label('Signal Amplitude (μV)', rotation=270, labelpad=15)
# Time series visualizations
# Graphene spatiotemporal plot (bottom left)
ax3 = fig.add_subplot(223)
ax3.set_title("Continuous Brain Activity Mapping\n(Graphene Template)", fontsize=14)
# Select a line of positions across the center of the head
center_line = []
center_line_pos = []
for i in range(grid_size):
j = grid_size // 2
if mask[i, j]:
center_line.append((i, j))
center_line_pos.append((X[i, j], Y[i, j]))
# Subsample the center line for computational efficiency
center_line_subsampled = center_line[::8]
center_line_pos_subsampled = np.array(center_line_pos[::8])
# Generate signals for this line
center_line_signals = generate_brain_activity(center_line_pos_subsampled, time)
# Create spatiotemporal image
spatiotemporal_data = np.zeros((len(center_line_subsampled), len(time)))
for i in range(len(center_line_subsampled)):
spatiotemporal_data[i, :] = center_line_signals[i, :]
# Plot spatiotemporal data
im3 = ax3.imshow(spatiotemporal_data, aspect='auto',
extent=[0, duration, 0, len(center_line_subsampled)],
cmap=cm, vmin=-1.0, vmax=1.0)
ax3.set_xlabel('Time (s)')
ax3.set_ylabel('Spatial Position')
# Traditional EEG time series (bottom right)
ax4 = fig.add_subplot(224)
ax4.set_title("Discrete Channel Recordings\n(Traditional EEG)", fontsize=14)
# Select channels to display
channels_to_plot = [0, 10, 20, 30, 40, 50] # Example channels
channel_names = ['Fp1', 'F7', 'T7', 'P7', 'O1', 'Cz'] # Example names
# Plot time series for selected channels
for i, ch_idx in enumerate(channels_to_plot):
# Offset each channel for clarity
offset = -i * 2
ax4.plot(time, traditional_signals[ch_idx] + offset, linewidth=1)
ax4.text(0, offset, channel_names[i], ha='right', va='center')
ax4.set_xlabel('Time (s)')
ax4.set_ylim([-12, 2])
ax4.set_yticks([])
plt.tight_layout()
return fig
# Create visualization at a specific time point
fig = create_topographic_map(time_idx=500) # Middle of the simulation
plt.savefig('normal_brain_activity_comparison.png', dpi=300, bbox_inches='tight')
plt.show() |
6.2. Code for Focal Seizure Simulation
Copyimport numpy as np
import matplotlib.pyplot as plt
from scipy.interpolate import griddata
from matplotlib.colors import LinearSegmentedColormap
from scipy.ndimage import gaussian_filter
# Set random seed for reproducibility
np.random.seed(42)
# Parameters
n_electrodes = 64 # Number of traditional EEG electrodes
grid_size = 512 # Resolution of graphene template (512x512 grid)
time_points = 1000 # Number of time points to simulate
duration = 4.0 # Duration in seconds
fs = time_points / duration # Sampling frequency
seizure_onset = 1.0 # Seizure onset time in seconds
seizure_onset_idx = int(seizure_onset * fs)
# Generate electrode positions (extended 10-20 system)
theta = np.linspace(0, 2*np.pi, n_electrodes//4 + 1)[:-1]
radius_levels = [0.3, 0.5, 0.7, 0.9]
electrode_positions = []
for r in radius_levels:
for t in theta:
x = r * np.cos(t)
y = r * np.sin(t)
electrode_positions.append((x, y))
electrode_positions = np.array(electrode_positions)
# Create head model (unit circle)
x = np.linspace(-1, 1, grid_size)
y = np.linspace(-1, 1, grid_size)
X, Y = np.meshgrid(x, y)
R = np.sqrt(X**2 + Y**2)
mask = R <= 1 # Within unit circle
# Define seizure focus in the right inferior temporal region
# Converting anatomical position to coordinates in our model
seizure_focus_x = 0.65 # Right side
seizure_focus_y = -0.5 # Inferior
seizure_focus = (seizure_focus_x, seizure_focus_y)
# Generate background source dipoles
n_background_sources = 300
source_positions = []
for _ in range(n_background_sources):
r = np.sqrt(np.random.uniform(0, 0.8**2))
theta = np.random.uniform(0, 2*np.pi)
source_positions.append((r * np.cos(theta), r * np.sin(theta)))
source_positions = np.array(source_positions)
# Parameters for background activity
freqs = np.concatenate([
np.random.uniform(1, 4, n_background_sources // 4), # delta
np.random.uniform(4, 8, n_background_sources // 4), # theta
np.random.uniform(8, 13, n_background_sources // 2), # alpha (dominant)
np.random.uniform(13, 30, n_background_sources // 8), # beta
])
np.random.shuffle(freqs)
phases = np.random.uniform(0, 2*np.pi, n_background_sources)
amplitudes = np.random.pareto(1.5, n_background_sources) + 1 # 1/f distribution
# Define lead field matrix for volume conduction (simplified spherical model)
def lead_field(recording_pos, source_pos):
dist = np.sqrt((recording_pos[0] - source_pos[0])**2 +
(recording_pos[1] - source_pos[1])**2)
# Simplified volume conduction model
if dist < 1e-10:
return 1.0
return 1.0 / (dist + 0.1)**2
# Time vector
time = np.linspace(0, duration, time_points)
# Define seizure activity model
def seizure_activity(t, onset=seizure_onset):
if t < onset:
return 0
# Sigmoid onset with exponential decay envelope
envelope = 5.0 / (1 + np.exp(-5*(t-onset))) * np.exp(-0.2*(t-onset)**2)
# Frequency evolution from 5 Hz to 10 Hz
freq = 5 + 5 * (1 - np.exp(-(t-onset)))
# Generate seizure waveform (sinusoidal with harmonics)
waveform = np.sin(2*np.pi*freq*t) + 0.2*np.sin(2*np.pi*2*freq*t)
return envelope * waveform
# Function to calculate spatial spread of seizure
def seizure_spatial_weight(pos, focus=seizure_focus, t=0, onset=seizure_onset):
if t < onset:
return 0
# Calculate distance from focus
dist = np.sqrt((pos[0] - focus[0])**2 + (pos[1] - focus[1])**2)
# Initial seizure radius
initial_radius = 0.1
# Spreading velocity (units/second)
spread_velocity = 0.1
# Current radius of seizure
current_radius = initial_radius + spread_velocity * (t - onset)
# Gaussian spatial profile
spatial_weight = np.exp(-0.5 * (dist / current_radius)**2)
return spatial_weight
# Generate brain activity with seizure
def generate_brain_activity_with_seizure(recording_positions, time):
n_positions = len(recording_positions)
signals = np.zeros((n_positions, len(time)))
for i, pos in enumerate(recording_positions):
signal = np.zeros(len(time))
# Add background activity
for src_idx, src_pos in enumerate(source_positions):
# Apply lead field (volume conduction)
lf = lead_field(pos, src_pos)
# Generate oscillation at specific frequency
oscillation = amplitudes[src_idx] * np.sin(2*np.pi*freqs[src_idx]*time + phases[src_idx])
signal += lf * oscillation
# Add seizure activity
for t_idx, t in enumerate(time):
seizure_wave = seizure_activity(t)
seizure_weight = seizure_spatial_weight(pos, t=t)
signal[t_idx] += seizure_wave * seizure_weight * 3.0 # Scale seizure amplitude
# Add noise
if isinstance(pos, tuple): # Traditional EEG electrodes
noise = np.random.normal(0, 0.5, len(time)) # Higher noise for traditional EEG
noise += 0.3 * np.random.normal(0, 1, len(time)) * np.sin(2*np.pi*50*time) # Add line noise
else: # Graphene template
noise = np.random.normal(0, 0.2, len(time)) # Lower noise for graphene
signals[i] = signal + noise
return signals
# Generate graphene template data
graphene_positions = []
graphene_valid_indices = []
for i in range(grid_size):
for j in range(grid_size):
if mask[i, j]:
graphene_positions.append((X[i, j], Y[i, j]))
graphene_valid_indices.append((i, j))
graphene_positions = np.array(graphene_positions)
# For computational efficiency, we'll use a subsampled grid for signal generation
subsampling = 16 # Subsampling factor
subsampled_indices = np.arange(0, len(graphene_positions), subsampling)
subsampled_positions = graphene_positions[subsampled_indices]
# Generate signals for both recording methods
print("Generating traditional EEG signals with seizure...")
traditional_signals = generate_brain_activity_with_seizure(electrode_positions, time)
print("Generating subsampled graphene template signals with seizure...")
subsampled_graphene_signals = generate_brain_activity_with_seizure(subsampled_positions, time)
# Function to create focal seizure visualization
def create_focal_seizure_visualization(time_idx):
# Create figure
fig = plt.figure(figsize=(14, 10))
# Define a custom colormap
colors = [(0, 0, 0.8), (0, 0.8, 0.8), (0.8, 0.8, 0), (0.8, 0, 0)]
cmap_name = 'custom_diverging'
cm = LinearSegmentedColormap.from_list(cmap_name, colors, N=100)
# Graphene template visualization (left)
ax1 = fig.add_subplot(221)
ax1.set_title("Graphene 2D Head Template", fontsize=14)
# Create interpolated full-resolution image from subsampled data
grid_values = np.zeros((grid_size, grid_size))
grid_values[:] = np.nan
# Get signal values at the specific time point
signal_values = subsampled_graphene_signals[:, time_idx]
# Map subsampled positions back to grid
for idx, pos_idx in enumerate(subsampled_indices):
i, j = graphene_valid_indices[pos_idx]
grid_values[i, j] = signal_values[idx]
# Interpolate missing values
xx, yy = np.meshgrid(np.arange(grid_size), np.arange(grid_size))
valid_mask = ~np.isnan(grid_values)
points = np.column_stack([xx[valid_mask].flatten(), yy[valid_mask].flatten()])
values = grid_values[valid_mask].flatten()
grid_interpolated = griddata(points, values, (xx, yy), method='cubic')
# Smooth the interpolation slightly
grid_interpolated = gaussian_filter(grid_interpolated, sigma=1.5)
# Apply head mask
grid_interpolated[~mask] = np.nan
# Plot the interpolated data
im1 = ax1.imshow(grid_interpolated, cmap=cm, vmin=-1.5, vmax=1.5)
ax1.set_axis_off()
# Add annotations for graphene advantages
ax1.annotate('Seamless Coverage', xy=(0.3, 0.8), xytext=(0.1, 0.9),
arrowprops=dict(facecolor='blue', shrink=0.05))
ax1.annotate('Ultra-thin\n(Improved Comfort)', xy=(0.7, 0.6 |
References
- Balandin, A. A. (2013). Low-frequency 1/f noise in graphene devices. Nature Nanotechnology, 8(8), 549-555.
- Banszerus, L. , Schmitz, M., Engels, S., Goldsche, M., Watanabe, K., Taniguchi, T., Beschoten, B., & Stampfer, C. (2017). Ballistic transport exceeding 28 μm in CVD grown graphene. Nano Letters, 17(3), 1387-1391.
- Berényi, A. , Belluscio, M., Mao, D., & Buzsáki, G. (2012). Closed-loop control of epilepsy by transcranial electrical stimulation. Science, 337(6095), 735-737.
- Blaschke, B. M. , Tort-Colet, N., Guimerà-Brunet, A., Weinert, J., Rousseau, L., Heimann, A., Drieschner, S., Kempski, O., Villa, R., Sanchez-Vives, M. V., & Garrido, J. A. (2016). Mapping brain activity with flexible graphene micro-transistors. 2D Materials, 3(2), 025007.
- Bolotin, K. I. , Sikes, K. J., Jiang, Z., Klima, M., Fudenberg, G., Hone, J., Kim, P., & Stormer, H. L. (2008). Ultrahigh electron mobility in suspended graphene. Solid State Communications, 146(9-10), 351-355.
- Cai, M., Nie, S., Du, Y., Wang, C., & Song, F. (2019). Flexible transparent electrodes based on nanostructured ultrathin gold films. Applied Surface Science, 467, 1090-1095.
- Casson, A. J., Yates, D. C., Smith, S. J., Duncan, J. S., & Rodriguez-Villegas, E. (2010). Wearable electroencephalography. IEEE Engineering in Medicine and Biology Magazine, 29(3), 44-56.
- Chen, K. , Shi, L., Zhang, Y., & Liu, Z. (2020). Scalable chemical-vapour-deposition growth of three-dimensional graphene materials towards energy-related applications. Chemical Society Reviews, 49(16), 6033-6069.
- Chung, M. , Fortunato, G., & Radacsi, N. (2019). Wearable flexible sweat sensors for healthcare monitoring: A review. Journal of the Royal Society Interface, 16(159), 20190217.
- Collura, T. F. (1993). History and evolution of electroencephalographic instruments and techniques. Journal of Clinical Neurophysiology, 10(4), 476-504.
- Dannhauer, M., Lanfer, B., Wolters, C. H., & Knösche, T. R. (2011). Modeling of the human skull in EEG source analysis. Human Brain Mapping, 32(9), 1383-1399.
- Ferrari, A. C. , Bonaccorso, F., Fal'ko, V., Novoselov, K. S., Roche, S., Bøggild, P., Borini, S., Koppens, F. H., Palermo, V., Pugno, N., et al. (2015). Science and technology roadmap for graphene, related two-dimensional crystals, and hybrid systems. Nanoscale, 7(11), 4598-4810.
- Ferrari, L. M., Sudha, S., Tarantino, S., Esposti, R., Bolzoni, F., Cavallari, P., Cipriani, C., Mattoli, V., & Greco, F. (2018). Ultraconformable temporary tattoo electrodes for electrophysiology. Advanced Science, 5(3), 1700771.
- Fiedler, P., Pedrosa, P., Griebel, S., Fonseca, C., Vaz, F., Supriyanto, E., Zanow, F., & Haueisen, J. (2018). Novel multipin electrode cap system for dry electroencephalography. Brain Topography, 31(5), 859-866.
- Hämäläinen, M. , Hari, R., Ilmoniemi, R. J., Knuutila, J., & Lounasmaa, O. V. (1993). Magnetoencephalography—theory, instrumentation, and applications to noninvasive studies of the working human brain. Reviews of Modern Physics, 65(2), 413-497.
- Hess, L. H., Jansen, M., Maybeck, V., Hauf, M. V., Seifert, M., Stutzmann, M., Sharp, I. D., Offenhäusser, A., & Garrido, J. A. (2011). Graphene transistor arrays for recording action potentials from electrogenic cells. Advanced Materials, 23(43), 5045-5049.
- Jobst, B. C., Bartolomei, F., Diehl, B., Frauscher, B., Kahane, P., Minotti, L., Pizzo, F., & Worrell, G. A. (2020). Intracranial EEG in the 21st century. Epilepsy Currents, 20(4), 180-188.
- Jurcak, V. , Tsuzuki, D., & Dan, I. (2007). 10/20, 10/10, and 10/5 systems revisited: Their validity as relative head-surface-based positioning systems. NeuroImage, 34(4), 1600-1611.
- Kappenman, E. S., & Luck, S. J. (2010). The effects of electrode impedance on data quality and statistical significance in ERP recordings. Psychophysiology, 47(5), 888-904.
- Kim, K. S. , Zhao, Y., Jang, H., Lee, S. Y., Kim, J. M., Kim, K. S., Ahn, J. H., Kim, P., Choi, J. Y., & Hong, B. H. (2010). Large-scale pattern growth of graphene films for stretchable transparent electrodes. Nature, 457(7230), 706-710.
- Kireev, D. , Brambach, M., Seyock, S., Maybeck, V., Fu, W., Wolfrum, B., & Offenhäusser, A. (2017). Graphene transistors for interfacing with cells: Towards a deeper understanding of liquid gating and sensitivity. Scientific Reports, 7(1), 6658.
- Klem, G. H. , Lüders, H. O., Jasper, H. H., & Elger, C. (1999). The ten-twenty electrode system of the International Federation. Electroencephalography and Clinical Neurophysiology, 52(3), 3-6.
- Kostarelos, K. , Vincent, M., Hebert, C., & Garrido, J. A. (2017). Graphene in the design and engineering of next-generation neural interfaces. Advanced Materials, 29(42), 1700909.
- Kuzum, D. , Takano, H., Shim, E., Reed, J. C., Juul, H., Richardson, A. G., de Vries, J., Bink, H., Dichter, M. A., Lucas, T. H., et al. (2014). Transparent and flexible low noise graphene electrodes for simultaneous electrophysiology and neuroimaging. Nature Communications, 5(1), 5259.
- Laursen, S. B. , Jensen, M. S., Foldager, C. B., Jakobsen, M., Nielsen, R. K., Gemmer, C. W., Christiansen, T. L., Lykkegaard, J. J., Linde, J. H., Blichfeldt, S., et al. (2020). Enhanced clinical outcome with implantation of a graphene oxide-coated porous titanium scaffold in osteochondral repair. ACS Biomaterials Science & Engineering, 6(5), 2724-2739.
- Lee, C., Wei, X., Kysar, J. W., & Hone, J. (2019). Measurement of the elastic properties and intrinsic strength of monolayer graphene. Science, 321(5887), 385-388.
- Lee, H. S., Chung, J., Hwang, G. T., Jeong, C. K., Jung, Y., Kwak, J. H., Kang, H., Byun, M., Kim, W. D., Hur, S., et al. (2017). Flexible inorganic piezoelectric acoustic nanosensors for biomimetic artificial hair cells. Advanced Functional Materials, 24(44), 6914–6921.
- Li, X. , Cai, W., An, J., Kim, S., Nah, J., Yang, D., Piner, R., Velamakanni, A., Jung, I., Tutuc, E., et al. (2009). Large-area synthesis of high-quality and uniform graphene films on copper foils. Science, 324(5932), 1312-1314.
- Liu, J. , Fu, T. M., Cheng, Z., Hong, G., Zhou, T., Jin, L., Duvvuri, M., Jiang, Z., Kruskal, P., Xie, C., et al. (2019). Syringe-injectable electronics. Nature Nanotechnology, 10(7), 629-636.
- Michel, C. M., Murray, M. M., Lantz, G., Gonzalez, S., Spinelli, L., & Grave de Peralta, R. (2004). EEG source imaging. Clinical Neurophysiology, 115(10), 2195-2222.
- Montgomery, R. M. (2024). Mapping the Mind: The Role of Neural Fields, Brain Dynamics, and Surface Physics in Neurological Biomarkers. Preprints. [CrossRef]
- Montgomery, R. M. (2025). Digital Mental Health: Opportunities and Challenges in Clinical Practice. Biomed J Sci & Tech Res61(3)-2025. BJSTR. MS.ID.009602.
- Mullinger, K. J., Mayhew, S. D., Bagshaw, A. P., Bowtell, R., & Francis, S. T. (2017). Understanding the BOLD response to simultaneous neural and vascular modulation. NeuroImage, 125, 985-995.
- Novoselov, K. S. , Geim, A. K., Morozov, S. V., Jiang, D., Zhang, Y., Dubonos, S. V., Grigorieva, I. V., & Firsov, A. A. (2004). Electric field effect in atomically thin carbon films. Science, 306(5696), 666-669.
- Pampaloni, N. P. , Lottner, M., Giugliano, M., Matruglio, A., D'Amico, F., Prato, M., Garrido, J. A., Ballerini, L., & Scaini, D. (2018). Single-layer graphene modulates neuronal communication and augments membrane ion currents. Nature Nanotechnology, 13(8), 755-764.
- Park, D. W. , Schendel, A. A., Mikael, S., Brodnick, S. K., Richner, T. J., Ness, J. P., Hayat, M. R., Atry, F., Frye, S. T., Pashaie, R., et al. (2018). Graphene-based carbon-layered electrode array technology for neural imaging and optogenetic applications. Nature Communications, 5(1), 5258.
- Perrin, F., Pernier, J., Bertrand, O., & Echallier, J. F. (1989). Spherical splines for scalp potential and current density mapping. Electroencephalography and Clinical Neurophysiology, 72(2), 184-187.
- Rosenow, F., & Lüders, H. (2001). Presurgical evaluation of epilepsy. Brain, 124(9), 1683-1700.
- Roy, Y. , Banville, H., Albuquerque, I., Gramfort, A., Falk, T. H., & Faubert, J. (2019). Deep learning-based electroencephalography analysis: A systematic review. Journal of Neural Engineering, 16(5), 051001.
- Seeck, M., Koessler, L., Bast, T., Leijten, F., Michel, C., Baumgartner, C., He, B., & Beniczky, S. (2017). The standardized EEG electrode array of the IFCN. Clinical Neurophysiology, 128(10), 2070-2077.
- Smith, P. C. , Mazzaro, N., Jones, A. K., & Dougherty, E. T. (2020). A comprehensive review of wearable and wireless ECG monitoring systems. Journal of Medical Engineering & Technology, 44(8), 412-432.
- Song, J. , Davey, C., Poulsen, C., Luu, P., Turovets, S., Anderson, E., Li, K., & Tucker, D. (2015). EEG source localization: Sensor density and head surface coverage. Journal of Neuroscience Methods, 256, 9-21.
- Srinivasan, R. , Nunez, P. L., & Silberstein, R. B. (1998). Spatial filtering and neocortical dynamics: Estimates of EEG coherence. IEEE Transactions on Biomedical Engineering, 45(7), 814-826.
- Srinivasan, R. , Tucker, D. M., & Murias, M. (1996). Estimating the spatial Nyquist of the human EEG. Behavior Research Methods, Instruments, & Computers, 28(4), 657-660.
- Teplan, M. (2002). Fundamentals of EEG measurement. Measurement Science Review, 2(2), 1-11.
- Trevelyan, A. J. , Sussillo, D., Watson, B. O., & Yuste, R. (2006). Modular propagation of epileptiform activity: Evidence for an inhibitory veto in neocortex. Journal of Neuroscience, 26(48), 12447-12455.
- Viventi, J. , Kim, D. H., Vigeland, L., Frechette, E. S., Blanco, J. A., Kim, Y. S., Avrin, A. E., Tiruvadi, V. R., Hwang, S. W., Vanleer, A. C., et al. (2011). Flexible, foldable, actively multiplexed, high-density electrode array for mapping brain activity in vivo. Nature Neuroscience, 14(12), 1599-1605.
- Wang, K. , Frewin, C. L., Esrafilzadeh, D., Yu, C., Wang, C., Pancrazio, J. J., Romero-Ortega, M., Jalili, R., & Wallace, G. (2020). High-performance graphene-fiber-based neural recording microelectrodes. Advanced Materials, 31(15), 1805867.
- Won, S. M., Song, E., Zhao, J., Li, J., Rivnay, J., & Rogers, J. A. (2019). Recent advances in materials, devices, and systems for neural interfaces. Advanced Materials, 30(30), 1800534.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).