1. Introduction
In cancer cells, aerobic glycolysis, the Warburg effect, is characterized by elevated glycolytic flux and increased lactate production, even in the presence of sufficient oxygen. The biological and clinical significance of aerobic glycolysis has attracted widespread attention: a Google Scholar search for “glycolysis and cancer” yields over 436,000 results. Interest in flux control of glycolysis in cancer is also substantial, with approximately 114,000 results for the terms “flux control, glycolysis, cancer.”
Cancer cells adapt glycolytic flux according to nutrient availability. Under nutrient-rich conditions, glycolysis supports rapid growth by diverting intermediates into anabolic pathways. In nutrient-limited environments, glycolysis shifts toward energy production to maintain cell survival. Thus, glycolysis is dynamically regulated to meet varying cellular demands[
1,
2,
3].
Previous studies have primarily focused on kinetic regulation of glycolysis—examining mechanisms such as allosteric modulation, transcriptional control, feedback inhibition, and feedforward activation. These investigations have emphasized how changes in enzyme activity influence glycolytic flux. In contrast, thermodynamic considerations have typically been limited to the role of Gibbs free energy in governing reaction directionality and energy transfer within the pathway.
What remains largely unexplored is how enzyme kinetics and thermodynamics interact within the glycolytic pathway. Specifically, the coordination between glycolytic flux, the rate of each step, intermediate concentrations, and the ΔG values of individual reactions has received little attention. Another critical issue is that, although glycolysis is widely recognized as a dynamic and adaptable pathway, particularly in cancer metabolism, its capacity to maintain a stable steady state under fluctuating conditions is often overlooked.
Pyruvate kinase M2 (PKM2) is the most extensively studied glycolytic enzyme in cancer biology, with over 22,000 results retrieved from a Google Scholar search for “PKM2, glycolysis, cancer.” It is believed to orchestrate metabolic programming by shifting glycolysis between energy-generating and biosynthetic modes.
Traditionally considered a rate-limiting enzyme due to the irreversibility of its reaction, PKM2 activity is tightly regulated through multiple mechanisms: including gene expression via diverse signaling pathways[
8,
9,
10,
11,
12,
13,
14,
15,
16,
17], allosteric inhibition by amino acids (e.g., alanine, phenylalanine, proline, tryptophan, valine) [
18,
19,
20], allosteric activation by fructose-1,6-bisphosphate (FBP), serine, and SAICAR[
21,
22,
23], and post-translational modifications such as phosphorylation[
17,
24,
25,
26,
27,
28,
29,
30], acetylation[
31,
32,
33], hydroxylation[
34], lactylation[
35], oxidation[
36,
37], methylation[
38], and glycosylation[
39].
According to current understanding, this dynamic regulation allows PKM2 to modulate glycolysis in response to the metabolic needs of cancer cells. When PKM2 activity is suppressed, upstream intermediates accumulate, enabling diversion into anabolic pathways such as the pentose phosphate pathway (PPP) or serine synthesis pathway (SSP), supporting biomass production and cell growth. Conversely, activation of PKM2 depletes these intermediates and enhances ATP production.
Despite extensive study, the intermediate biochemical mechanisms, how PKM2 exerts its influence over glycolysis, remain largely unexplored.
Our prior studies [
4,
5,
6,
7] established, in living cancer cells, that in responding to large perturbations of PKM2, PGK1, GAPDH, and LDH, glycolytic flux rate remains constant, which is stabilized through interaction between enzyme kinetics and the thermodynamic properties of the glycolytic pathway. These empirical findings motivated the current work, in which I first lay out the fundamental theory of the kinetic-thermodynamic coupling in a metabolic pathway, and then formalize the experimental observations by deriving every equation directly from measured shifts in flux, ΔG, and metabolite levels and then integrate them into a coherent theoretical framework for kinetic-thermodynamic coupling.
2. Results
2.1. Fundamental Theory—Kinetic-Thermodynamic Coupling in the Glycolytic Pathway
To examine the interdependence between enzyme kinetics and thermodynamics in glycolysis, it is essential to focus on the core glycolytic pathway and exclude branching reactions (e.g., pentose phosphate pathway, serine biosynthesis pathway). This simplification enables a clearer analysis of the fundamental principles governing flux and intermediate metabolite distribution.
At steady state, the flux through every enzyme-catalyzed reaction in glycolysis is equal and constant. This raises a fundamental question: How can reaction rates remain equal when the total activity of 11 glycolytic enzymes differ by up to three orders of magnitude[
4,
5,
6,
7]?
In glycolysis, the actual catalytic activity of each enzyme depends on two factors:
Thus, for every step in the pathway to operate at the same rate, the concentrations of intermediates must be precisely tuned to compensate for differences in enzyme abundance or activity. Furthermore, these intermediate concentrations must remain stable over time to maintain this kinetic balance.
This leads to a second key question: how are these intermediate concentrations finely tuned across the pathway? This question concerns the distribution of intermediate metabolite concentrations, which is governed by the thermodynamic properties of the pathway.
Consider a linear metabolic pathway like glycolysis consisting of n sequential reactions at steady state, where each reaction k is coupled to the next via shared metabolites: the product of one step becomes the substrate for the next. Let the metabolites be labeled as X0, X1, X2, ….., Xn, where:
X0 is the initial substrate,
Xn is the final product
Xk (for k =1,2,3,…,n−1) is both the product of reaction k and the substrate of reaction k+1.
The Gibbs free energy (ΔG
k) for each reaction
k is given by:
Because the product of one reaction is the substrate of the next, coupling imposes the following relationships:
Reaction k produces Xk: Reaction k + 1 consumes Xk: ,
The above equations could be rearranged to
Substituting Eq. 2 into Eq. 3 yields:
Extending this sequentially across the entire pathway yields:
The above equations show that in a linear metabolic pathway at steady state, the concentrations of intermediates are interdependent and must satisfy the thermodynamic constraints imposed by each reaction's ΔG. This sequential coupling ensures that the entire pathway operates in a coordinated manner, balancing energy changes and metabolite levels.
By analogy, all the reactions and ΔG values within the glycolytic pathway are sequentially coupled to one another, and the concentrations of all the intermediates in the pathway are thermodynamically equilibrated.
From this perspective, the distribution of intermediate concentrations arises from thermodynamic equilibration. The need to maintain equal flux through each enzymatic step is satisfied by adjusting intermediate concentrations, which in turn are constrained by the overall thermodynamic landscape.
Thus, actual enzyme activity becomes a function of both:
This defines the principle of kinetic-thermodynamic coupling: the enzyme-catalyzed rates, concentrations of intermediates, and ΔG values are interdependent and co-regulated to ensure flux stability.
Taken together, steady-state glycolysis is maintained by a balance between enzyme kinetics and chemical thermodynamics. Equal flux through all steps and stable metabolite concentrations are ensured by a coordinated adjustment of enzyme activity and thermodynamic constraints. This defines the principle of kinetic-thermodynamic coupling, which explains how the system achieves stability and balance despite substantial variation in enzyme abundance.
2.2. Kinetic-Thermodynamic Coupling in the Regulation of Glycolysis by PKM2
The results below begin by summarizing the key experimental findings, followed by a series of mathematical derivations designed to explain and extend those observations.
Experimental observations
In our previous work [
4,
5,
6,
7], we observed:
Because the thermodynamic properties of glycolysis govern intermediate distributions, this profile is conserved across cell types[
4,
5,
6,
7]. Among the 11 glycolytic reactions, three catalyzed by hexokinase 2 (HK2), phosphofructokinase-1 (PFK1), and pyruvate kinase (PK) operate far from equilibrium, and thus provide the primary thermodynamic driving force for glycolysis. The remaining reactions are at near-equilibrium state. The lactate dehydrogenase (LDH) reaction is also exergonic in cancer cells, with an average ΔG of ~–7 kJ/mol. Because the ΔG profile is conserved across diverse cell types, the thermodynamic regulation of metabolite concentrations is systemically stable.
When PKM2 is knockdown by 80%, it does not change the above thermodynamic features of glycolytic pathway [
6].
However, PKM2 knockdown does induce a redistribution of Gibbs free energy in the glycolytic pathway: the value of the Gibbs free energy of PKM2-catalyzed reaction (ΔGPKM2) becomes more negative (from ~ -25 kJ/mol to ~ -28 kJ/mol); while the value of the Gibbs free energy of PFK1-catalyzed reaction (ΔGPFK1) becomes less negative (from ~ -15 kJ/mol to ~-12 kJ/mol); ΔG values for other steps remain unchanged; this redistribution of Gibbs free energy in the pathway leads to proportional increase of the intermediates (PEP, 2-PG, 3-PG, GA3P, DHAP, and FBP) in the segment between PFK1 and PKM2 increases proportionally, while does not change the concentration of F6P, G6P, and pyruvate.
Obviously, the changes of values of ΔGPKM2 and ΔGPFK1 do not change their thermodynamic nature, i.e., the reactions are still far from equilibrium.
Despite that PKM2 knockdown markedly reduces the PKM2t, it did not change PKM2a.
Glucose consumption and lactate production, i.e., the glycolytic flux, remain constant despite the 80% change of PKM2t.
These published observations [
4,
5,
6,
7] form the empirical foundation for the mathematical framework developed below. Having laid out the key experimental observations, I next derive the mathematical relationships that explain them, using PKM2 as a concrete example to illustrate how this framework fits both steady-state stability and transient responses.
Theoretical framework
To maintain clarity: the total enzyme activity of PKM2 is denoted as PKM2t, the actual catalytic activity within the pathway is denoted PKM2a, and metabolite concentrations are written in brackets (e.g., [PEP] for phosphoenolpyruvate).
Let us consider two steady states of glycolysis:
Steady state a, where PKM2t is high, and
Steady state b, where PKM2t is low.
When the system transitions from a to b, PKM2t is reduced. However, the upstream flux, such as that catalyzed by hexokinase 2 (HK2), remains constant. As a result, the substrate of PKM2, [PEP], begins to accumulate.
This increase in [PEP] drives the values of the Gibbs free energy of the PKM2 reaction (ΔGPKM2) to become more negative, i.e., the reaction becomes more exergonic. Due to thermodynamic coupling, this perturbation propagates upstream, leading to the accumulation of intermediates including 2-phosphoglycerate (2-PG), 3-phosphoglycerate (3-PG), glyceraldehyde-3-phosphate (GA3P), dihydroxyacetone phosphate (DHAP), and fructose-1,6-bisphosphate (FBP).
However, the propagation does not extend beyond the PFK1-catalyzed step. This is because the PFK1 reaction maintains a highly negative ΔG (≈ –13 kJ/mol) [
4,
5,
6,
7], creating a thermodynamic ‘barrier’ that blocks upstream diffusion of the perturbation. Even though reduced PKM2
t elevates [FBP], the resulting change in ΔG
PFK1 is modest and insufficient to bring the reaction near equilibrium. As a result, the concentrations of fructose-6-phosphate (F6P) and glucose-6-phosphate (G6P) remain essentially unchanged.
Based on the fundamental kinetic-thermodynamic coupling and experimental observation, we can then reason out the following equations:
During the transition from steady state a to b, ΔGPKM2 becomes more negative, while ΔGPFK1 becomes less negative. This shift leads to a proportional increase in the concentrations of intermediates between PFK1 and PKM2 (i.e., [FBP], [DHAP], [GA3P], [3-PG], [2-PG], [PEP]). Since the reactions connecting these metabolites are near equilibrium, their thermodynamic relationships are preserved, and concentrations increases proportionally. Conversely, when the system transitions from b back to a (i.e., increasing PKM2t), ΔGPKM2 becomes less negative, and ΔGPFK1 becomes more negative, resulting in a proportional decrease in those same intermediate concentrations. The concentrations of [F6P], [G6P], and pyruvate remain stable in both directions of the transition, because in the entire glycolytic pathway, only the values of values of ΔGPKM2 and ΔGPFK1 are changed.
The reciprocal changes of values of ΔG
PKM2 and ΔG
PFK1 can be quantitatively deduced from the following:
Combining (1) and (2) yield:
Now consider a shift in PKM2 total activity from state a to state b, [pyruvate], [P
i], and [F6P] remain constant, while [PEP] and [FBP] change proportionally, and thus:
And since [P
i], [F6P], and [pyruvate] stay constant:
Eq. 6 and Eq. 7 reveals the insight into the PKM2-induced redistribution of free energy in the entire glycolytic pathway, which have following implications
PKM2t changes leads to a transfer of a fraction of the free energy between PKM2 catalyzed reaction and PFK1 catalyzed reaction.
When PKM2t decreases, a fraction of free energy is transferred from to ; conversely, when PKM2t increases, a fraction of free energy is transferred from to .
The increase in the value of is equal to the decrease in the value of , and vice versa.
The changes of the concentrations of intermediates are strictly controlled by the amount of free energy transferred between PKM2-catalyzed and PFK1-catalyzed reactions.
The large negative values of ∆GPFK1 lays the theoretical basis for the concentration of [G6P] and [F6P] not to be disturbed by the downstream perturbation. The stability of [G6P] is especially important because G6P is a strong allosteric inhibitor of HK2. Preventing changes in [G6P] ensures that HK2 activity remains stable, and thus the glycolytic input is preserved. In this way, the large negative values of ΔGPFK1 functions as a thermodynamic insulator, maintaining input stability despite downstream disturbances.
As PKM2t changes, opposing changes in [PEP] and [FBP] stabilize PKM2a. Thus, despite large changes in PKM2t, PKM2a remains stable, due to the compensatory shifts in substrate concentration.
As PKM2a remains constant, glycolytic rate at steady state a and steady state b remains constant.
2.3. Quantitative Coupling of PKM2 Kinetics with Thermodynamics in the Glycolytic Pathway
Let us focus on the core glycolytic pathway and exclude branching reactions (e.g., pentose phosphate pathway, serine biosynthesis pathway). This simplification enables a clearer analysis of the intricate coordination between PKM2 activity, intermediate concentrations, Gibbs free energy values, and glycolytic rate.
Because the system is linear and lacks branching, the rate through each enzyme in the pathway is equal:
Where J
lactate is the rate of lactate generation,
Jglycolysis is the rate of glycolysis,
Jᵢ refers to the flux through any enzyme in the pathway:
JHK2, JPGI, JPFK1, Jaldolase, JTPI, JGAPDH, JPGK1, JPGAM, Jenolase, JPK, and JLDH.
Hence:
where
Jlactate could be experimentally determined and
(
) could be expressed by Michaelis-Menton kinetics [
4,
5,
6,
7]
However, Since PKM2
a in the glycolytic pathway is regulated by the FBP, and since FBP reduces the
Km without affecting
Vmax,
Km is substituted by
K0.5,
Eq. 11 is valid only when [FBP] saturates PKM2. The saturation of PKM2 by FBP is calculated based on the equation of the fractional occupancy of enzyme (θ)
Where KFBP is the dissociation constant for FBP binding to PKM2
Given:
KFBP ≈ 25.5 ± 148.1 nM (40)
[FBP] ranges from ~35 to 61 μM in cell-free glycolysis, and from ~210 to 1510 μM in cells [
4,
5,
6,
7]
Substituting into the equation of the fractional occupancy of enzyme reveals that PKM2 is nearly 100% saturated with FBP under both in vitro and in vivo conditions.
Therefore, Eq. 11 is valid under physiological and experimental conditions.
Rearranging Eq. 11 gives:
[PEP] is not an isolated variable but is constrained by the thermodynamic landscape of the glycolytic pathway. As Eq. 6 and Eq. 7 shown, changes in PKM2t activity leads to reciprocal changes in ∆GPKM2 and ∆GPFK1: when PKM2t increases, ∆GPKM2 becomes less negative while ∆GPFK1 becomes more negative; conversely, when PKM2t decreases, ∆GPKM2 becomes more negative while ∆GPFK1 becomes less negative; in either case, Gibbs free energy values of other reactions in the glycolytic pathway remains constant. Thus, [PEP] can also be expressed in terms of the actual changes of ∆GPKM2 and ∆GPFK1:
First, since:
which can be rearranged to:
for PFK1-catalyzed reaction:
which can be rearranged to:
Substituting (2) into (1) yields
where
= C
Eq. 6.
Combining Eq. 12 and Eq. 13 yields
Equations 8 – 14 together means that
PKM2 kinetics is tightly coupled with thermodynamics of the glycolytic pathway.
When [PKM2] or PKM2t decreases, ΔGPKM2 becomes more negative, ΔGPFK1 becomes less negative, and [PEP] increases.
When [PKM2] or PKM2t increases, ΔGPKM2 becomes less negative, ΔGPFK1 becomes more negative, and [PEP] decreases.
This reciprocal changes of ΔGPKM2 and ΔGPFK1 in the glycolytic pathway are the basis for the reciprocal changes of PKM2t and [PEP], that maintains PKM2a and glycolytic rate constant despite the marked change of PKM2t.
Thus, Equations 8–14 form an interdependent system that quantitatively links PKM2
t (total activity), PKM2
a (
vPKM2), [PEP], the thermodynamic landscape of the glycolytic pathway, and
Jlactate (system output) (
Figure 1B).
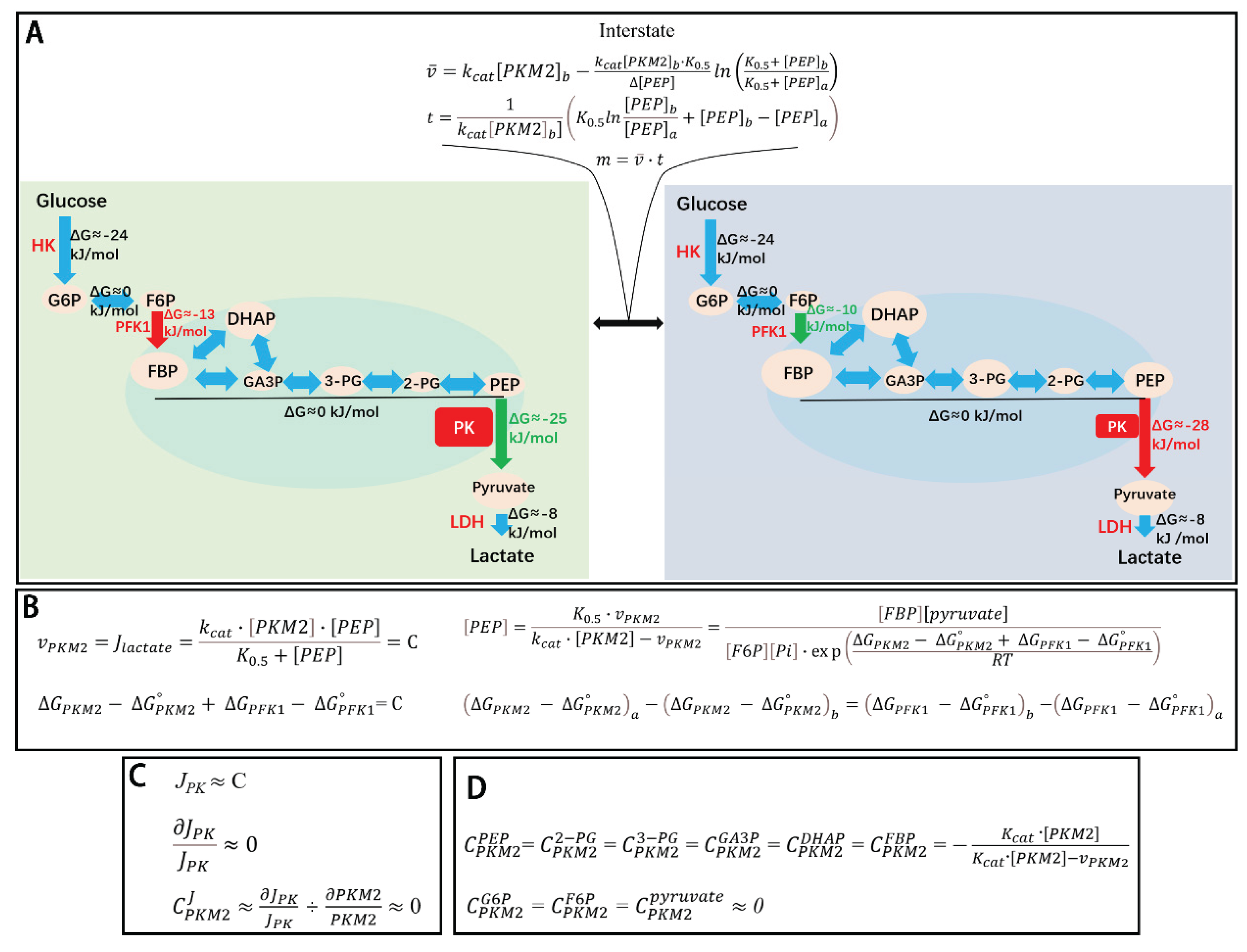
Figure 1.
Coupling between enzyme kinetics and chemical thermodynamics in PKM2-mediated regulation of glycolysis. (
A) A model transition between two steady-state glycolytic conditions in Hela cells [
6]. Left panel: glycolysis at steady state with 100% PKM2 total activity (PKM2
t). Right panel: glycolysis at steady state with 20% PKM2
t. Middle panel: transient intermediate state during the transition from high to low PKM2
t. Decreasing PKM2
t from 100% to 20% redistributes the Gibbs free energy (ΔG) along the glycolytic pathway. at high PKM2
t: ΔG
PKM2 is less negative, whereas ΔG
PFK1 is more negative, at low PKM2
t: ΔG
PKM2 becomes more negative, while ΔG
PFK1 is less negative, and ΔG values of other glycolytic reactions remain largely unchanged. This redistribution of ΔG results in coordinated changes in glycolytic intermediate concentrations. At high PKM2
t: levels of FBP, GA3P, DHAP, 3-PG, 2-PG, and PEP are proportionally reduced; at low PKM2
t, levels of these intermediates proportionally increase, while G6P, F6P, and pyruvate levels are largely unaffected. The sizes of ovals represent relative concentrations of intermediates, distributed according to the thermodynamic landscape. PKM2 is saturated by its dominant allosteric activator, FBP, and is not further inhibited or activated by other allosteric effectors such as alanine, phenylalanine, proline, tryptophan, serine, or TEPP-46 [
4,
5,
6,
7]. The shift from one steady state to another requires passing through a transient, non-steady-state glycolytic state, which is mathematically tractable in terms of flux (
v̅), time (
t), and mass (
m), and contributes minimally to the overall system. (
B) Equations showing PKM2
t, PKM2
a, glycolytic flux, intermediate concentrations, and pathway thermodynamics are tightly coupled, quantitatively reflecting glycolysis steady state shift between 100% PKM2
t to 20% PKM2
t. (
C) Flux control coefficient equation indicating that PKM2 has a negligible effect on total glycolytic flux. (
D) Concentration control coefficient equation indicating that PKM2 significantly controls the levels of intermediates in the segment between PFK1 and PKM2, while negligibly the levels of G6P, F6P, and pyruvate.
Figure 1.
Coupling between enzyme kinetics and chemical thermodynamics in PKM2-mediated regulation of glycolysis. (
A) A model transition between two steady-state glycolytic conditions in Hela cells [
6]. Left panel: glycolysis at steady state with 100% PKM2 total activity (PKM2
t). Right panel: glycolysis at steady state with 20% PKM2
t. Middle panel: transient intermediate state during the transition from high to low PKM2
t. Decreasing PKM2
t from 100% to 20% redistributes the Gibbs free energy (ΔG) along the glycolytic pathway. at high PKM2
t: ΔG
PKM2 is less negative, whereas ΔG
PFK1 is more negative, at low PKM2
t: ΔG
PKM2 becomes more negative, while ΔG
PFK1 is less negative, and ΔG values of other glycolytic reactions remain largely unchanged. This redistribution of ΔG results in coordinated changes in glycolytic intermediate concentrations. At high PKM2
t: levels of FBP, GA3P, DHAP, 3-PG, 2-PG, and PEP are proportionally reduced; at low PKM2
t, levels of these intermediates proportionally increase, while G6P, F6P, and pyruvate levels are largely unaffected. The sizes of ovals represent relative concentrations of intermediates, distributed according to the thermodynamic landscape. PKM2 is saturated by its dominant allosteric activator, FBP, and is not further inhibited or activated by other allosteric effectors such as alanine, phenylalanine, proline, tryptophan, serine, or TEPP-46 [
4,
5,
6,
7]. The shift from one steady state to another requires passing through a transient, non-steady-state glycolytic state, which is mathematically tractable in terms of flux (
v̅), time (
t), and mass (
m), and contributes minimally to the overall system. (
B) Equations showing PKM2
t, PKM2
a, glycolytic flux, intermediate concentrations, and pathway thermodynamics are tightly coupled, quantitatively reflecting glycolysis steady state shift between 100% PKM2
t to 20% PKM2
t. (
C) Flux control coefficient equation indicating that PKM2 has a negligible effect on total glycolytic flux. (
D) Concentration control coefficient equation indicating that PKM2 significantly controls the levels of intermediates in the segment between PFK1 and PKM2, while negligibly the levels of G6P, F6P, and pyruvate.
2.4. Transient Interstate Between Any Two Steady States
Between any two steady states of glycolysis lies a transient intermediate state - a brief period in which both flux and metabolite concentrations are dynamically adjusting. This intermediate phase is quantifiable and reflects the coordinated shifts in enzymatic activity, metabolite pools, and ΔG distribution.
Let’s consider a situation where PKM2t decreases from an initial steady-state level (a) to a new lower level (b), resulting in a shift from steady state a to steady state b. Three key parameters define this transient transition:
m denotes the total amount of substrate processed by PKM2 during the transition
v̅ denotes the average catalytic velocity of PKM2 during the transition
t denotes the time required to complete the transition
Given the [PEP] values at the beginning and end of the transition, and FBP is saturating PKM2, the velocity of PKM2 at each steady state can be defined using a Michaelis-Menten-like expression:
At steady state
a (initial):
At steady state
b (final):
The average velocity (
) during the transition is:
Substituting the equation for
and integrating yields:
The average velocity
during the transition from
va to
vb is given by:
Substitute u =
, then du = d[PEP]
The time (t) required for the system to transition between steady states is deduced from following:
This framework can be applied to:
If PKM2 activity does not change instantaneously but transitions stepwise in n discrete steps, then the total mass (m) and total time (t) are the sums across all transient substrates:
The average velocity over the transition is then:
Notably, the cumulative effect of stepwise transitions is mathematically equivalent to that of a single instantaneous transition from [PKM2]ₐ to [PKM2]b, when integrated across all intermediate states. Therefore, these equations provide a framework for evaluating both instantaneous and gradual transitions between glycolytic steady states
As an example, consider a scenario where PKM2 activity decreases in Hela cells due to siRNA knockdown, resulting in a drop from 958301000 to 286268000 μmol/min·l cells, and during this transition, the intracellular [PEP] increases from 67 to 215 μM (
Table 1).
Table 1.
The data used to calculate v̅, t, and mass transfer (m) for the system to transition between steady states. The numbers from Hela cells [
41] are used for calculating
v̅,
t, and
m.
Table 1.
The data used to calculate v̅, t, and mass transfer (m) for the system to transition between steady states. The numbers from Hela cells [
41] are used for calculating
v̅,
t, and
m.
Kcat[PKM2]a (μmol/min∙mg protein)
(before PKM2 KD) |
3387 |
Kcat[PKM2]b (μmol/min∙mg protein)
(after PKM2 KD) |
1012 |
| mg protein/l cells |
282000 |
|
Kcat[PKM2]a (μmol/min∙l cells) |
955134000 |
|
Kcat[PKM2]b (μmol/min∙l cells) |
285666000 |
|
K0.5 (μM) |
74 |
| [PEP]a (μM) (before PKM2 KD) |
67 |
| [PEP]b (μM) (after PKM2 KD) |
215 |
| ∆[PEP] (μM) |
148 |
|
v̅ (μmol/min∙l cells) |
183159000 |
|
t (ms) |
0.049 |
|
m (μmol/l cells) |
150 |
Applying Eq. 15, 17 and 18:
= 183159000 µmol/min·l cells
= 0.049 milliseconds
= 150 μmol/l cells
This calculation demonstrates that the duration of the interstate is very short so that the total mass processed is small.
Together, transitions between steady states in glycolysis occur through a brief, quantifiable “interstate” phase (
Figure 1A, middle panel), which can be expressed by Eq. 15–18. The rapid transitions between steady states are made possible by the micromolar concentrations of glycolytic intermediates, the high catalytic efficiency of enzymes[
4,
5,
6,
7], and the interconnected thermodynamic landscape of the pathway.
2.5. PKM2’s Flux Control Coefficient and Intermediate Concentration Control Coefficient
The flux control coefficient (FCC) and concentration control coefficient (CCC) are central concepts in Metabolic Control Analysis (MCA). They describe how a particular enzyme influences the behavior of a metabolic pathway at steady state:
These are system-level properties governed by network context and do not, by themselves, reveal the biochemical mechanism underlying control. The principle of kinetic-thermodynamic coupling, however, offers mechanistic insight into why PKM2 exerts negligible control over glycolytic flux but significantly affects intermediate concentrations.
2.6. Flux Control Coefficient (FCC) of PKM2
FCC is defined as the infinitesimal fractional change in pathway flux (J) in response to an infinitesimal fractional change in enzyme activity:
Because an 80% knockdown of PKM2 does not significantly affect glycolytic flux, then:
This implies that PKM2 does not exert rate-limiting control under physiological conditions.
2.7. Concentration Control Coefficient (CCC) of PKM2
CCC is defined as the fractional change in metabolite concentration [Sⱼ] caused by an infinitesimal change in enzyme activity [Eᵢ]:
For example, the CCC of PKM2 on [PEP] is:
According to the definition of CCC, the infinitesimal fractional change of [PEP] responding to the infinitesimal fractional change of [PKM2] could be express by
Because
(3)
Combining (3) and (5), simplified as
The value of is negative, which means increasing PKM2 decreases PEP or decreasing PKM2 increases PEP.
Similarly,
could be calculated by the mathematical deduction:
As reaction quotient (Q) of enolase-catalyzed reaction in the glycolytic pathway is near equilibrium,
Inserting
into (1)
Combining (1) and (4), simplified as,
Inserting
into (5)
It is not surprising that Eq. 25 is the same as Eq 24, because it is assuming that in the reaction catalyzed by enolase, Q ≈Keq.
Likewise, the CCC of PKM2 over 3-PG, GA3P, DHAP, FBP could be mathematically deduced.
Alternatively, the CCC of PKM2 on 2-PG, 3-PG, GA3P, DHAP, and FBP can be derived from the following reasoning: because the reactions between PEP and FBP are near-equilibrium, the concentrations of all intermediates in this segment change proportionally in response to PKM2 perturbation:
therefore
In contrast, [F6P], [G6P], and [pyruvate] do not change significantly following PKM2 knockdown. This is consistent with:
Therefore:
While PKM2 exerts little control over glycolytic flux (FCC ≈ 0) (
Figure 1C), it strongly influences the concentrations of intermediates in the segment between PKM2 and PFK1 but not other intermediates (
Figure 1D). This arises from the kinetic-thermodynamic architecture of glycolysis and explains how intermediate levels are responsive to enzyme perturbations, even when the pathway output remains stable.
3. Discussion
At steady state, glycolysis maintains equal flux through all reactions and stable concentrations of intermediates by dynamically coordinating enzyme activity with thermodynamic constraints. This interdependence defines the principle of kinetic-thermodynamic coupling, wherein the kinetics of each step are governed not only by enzyme levels but also by the thermodynamically equilibrated concentrations of intermediates – the thermodynamic landscape of the pathway.
While the dynamic adaptability of glycolysis has been extensively studied, the inherent stability of its steady state has received far less attention. The principle of kinetic-thermodynamic coupling suggests that glycolysis possesses a built-in capacity to rapidly reestablish homeostasis following perturbation. This perspective reframes our understanding of glycolytic regulation: glycolysis is not merely a passive and flexible pathway that responds to external changes, but a resilient, self-stabilizing system governed by intrinsic thermodynamic and kinetic constraints.
Through the perspective of the kinetic-thermodynamic coupling in the glycolytic pathway, the following insights that have not been previously perceived can be further revealed:
Flux Stability Through Thermodynamic Buffering: When a glycolytic enzyme such as PKM2 is perturbed, the system compensates through thermodynamic adjustments by changing substrate concentrations and redistributing Gibbs free energy (ΔG) across reactions. This thermodynamic buffering maintains a constant actual enzymatic rate (v), even when total enzyme activity varies significantly. As a result, glycolytic flux remains stable across a wide range of enzymatic perturbations. This principle explains experimental observations such as the stability of glycolytic rate despite large reductions in PKM2, GAPDH, PGK1, and LDH activity, and the accompanying rise in substrate concentrations and ΔG changes [
4,
5,
6,
7].
The Interstate between 2 steady states: Transitions between steady states occur through a brief and quantifiable interstate phase. During this phase, intermediate concentrations and ΔG values adjust dynamically, but the total substrate turnover and time elapsed are minimal. The interstate is governed by the same kinetic-thermodynamic principles as the steady state, ensuring efficient and predictable transitions. For example, PKM2 perturbation induces a redistribution of [PEP] and ΔGPKM2 within fractions of a millisecond, with negligible cumulative effects on flux or pool size.
Flux and Concentration Control:While enzymes like PKM2 exert little control over overall glycolytic flux (FCC ≈ 0), they strongly influence the concentrations of intermediates between PFK1 and PKM2. This is a natural outcome of the kinetic-thermodynamic architecture: the pathway flexibly adjusts concentrations to preserve flux stability.
Pathway-Level Thermodynamic Organization:Thermodynamics in linear pathways like glycolysis does more than determine reaction direction, it shapes the entire energetic profile of the system. In contrast to isolated reactions, pathway thermodynamics governs how ΔG is distributed, how intermediate pools are stabilized, and how changes in one step influence others.
The highly exergonic ΔGPFK1 step functions as a thermodynamic barrier, preventing downstream disturbances (e.g., from PKM2, GAPDH, PGK1, LDH) from propagating backward to affect upstream metabolites like [G6P]. This preserves glycolytic input and stabilizes hexokinase (HK2) activity against feedback inhibition.
Feedforward Saturation:Fructose-1,6-bisphosphate (FBP) is a potent allosteric activator of PKM2. However, because [FBP] is present at saturating levels under both basal and perturbed conditions, its concentration fluctuations do not affect PKM2 activity. This feedforward loop is thus insulated by saturation, ensuring that changes in glycolytic rate are not attributed to FBP variation.
A Diagnostic Framework for Enzyme Regulation: Kinetic-thermodynamic coupling provides a testable signature for assessing whether an enzyme (e.g., PKM2) is functionally regulating glycolysis. If regulatory changes of PKM2 (e.g., expression, PTMs, or allosteric effectors) influence glycolysis, one should observe:
- −
Reciprocal shifts in ΔGPKM2 and ΔGPFK1
- −
Proportional changes in intermediate concentrations between PFK1 and PKM2
- −
Stability of upstream ([G6P], [F6P]) and downstream ([pyruvate]) metabolites
- −
An inverse correlation between PKM2t and [PEP] that preserves PKM2a
The flux to lactate and to side branches: Based on the proposed principle, along with experimental data, reducing PKM2
t by up to 80% does not significantly decrease PKM2
a in the pathway. Consequently, neither glucose consumption nor flux to lactate is significantly affected. Since the rate to lactate does not change significantly, implying the rate to pyruvate does not change significantly, hence the pyruvate to mitochondrial metabolism would not change significantly. Since [G6P] remains unchanged due to the restraint of ∆G
PFK1, PPP flux is not likely changing significantly. In contrast, [3PG] increases markedly, leading to approximately twofold increase in SSP flux [
6]. Nevertheless, because the flux of 3-PG to SSP is much lower than its flux to lactate, this shift has a negligible impact on overall lactate production. For example, in HeLa cells, the 3-PG flux into SSP is approximately 15 nmol/h per million cells [
41], in contrast to flux to lactate averages around 2000 nmol/h per million cells[
4,
5,
6,
7,
41], twofold increase of SSP negligibly influence the rate to lactate. The flux is schematically showing in
Figure 2.
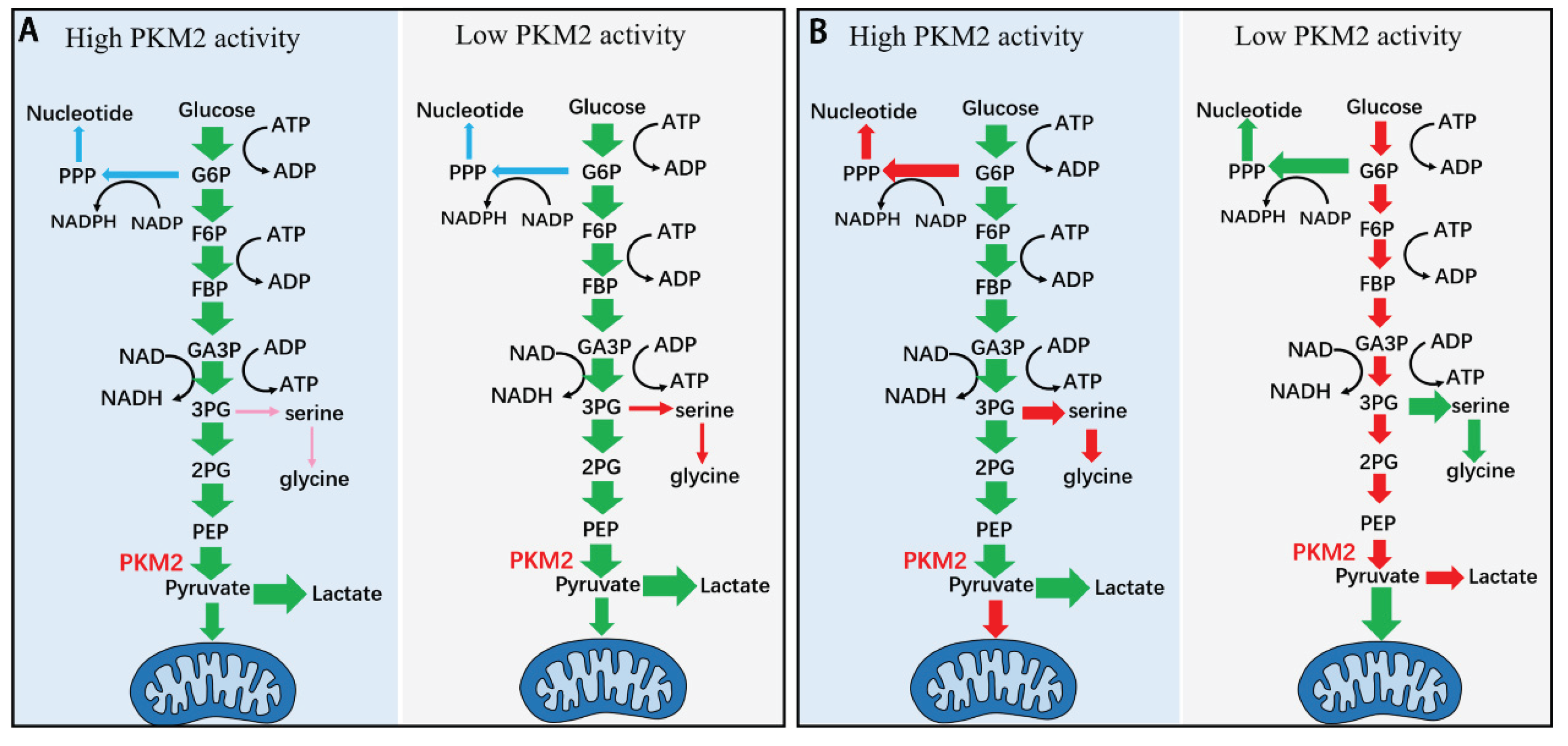
Figure 2.
Schematic diagram showing the model of kinetic-thermodynamic coupling in the regulation of glycolysis by PKM2 (A) and the model of current understanding of regulation of glycolysis by PKM2 (B). (
A) Changes in the total activity impact glycolysis through kinetic-thermodynamic coupling in the pathway (see
Figure 1 and text). Increase or decrease in the PKM2
t does not significantly change PKM2
a, does not significantly change the rate to PPP, the rate to lactate, nor the rate to mitochondria metabolism. Increase in PKM2
t decreases the rate to serine synthesis pathway, while decreases in PKM2
t increases the rate to SSP, yet the rate to SSP is negligible as compared with the rate to lactate (see also
Figure 1). (
B) Changes in the total activity of PKM2 directly exert effect on glycolysis. Increase in the total activity of PKM2 increases the glycolytic rate and depletes its upstream intermediates; the depletion of G6P and 3-PG reduces their flux to PPP and SSP, respectively; and the rate of pyruvate to mitochondria metabolism decreases while the rate of pyruvate to lactate increases. Conversely, decrease in the total activity of PKM2 reduces glycolytic rate and accumulates its upstream intermediates; the accumulation of G6P and 3-PG enhances their flux to PPP and SSP, respectively; the rate of pyruvate to mitochondria metabolism increases while the rate of pyruvate to lactate decreases. The two models are different and deserve attention.
Figure 2.
Schematic diagram showing the model of kinetic-thermodynamic coupling in the regulation of glycolysis by PKM2 (A) and the model of current understanding of regulation of glycolysis by PKM2 (B). (
A) Changes in the total activity impact glycolysis through kinetic-thermodynamic coupling in the pathway (see
Figure 1 and text). Increase or decrease in the PKM2
t does not significantly change PKM2
a, does not significantly change the rate to PPP, the rate to lactate, nor the rate to mitochondria metabolism. Increase in PKM2
t decreases the rate to serine synthesis pathway, while decreases in PKM2
t increases the rate to SSP, yet the rate to SSP is negligible as compared with the rate to lactate (see also
Figure 1). (
B) Changes in the total activity of PKM2 directly exert effect on glycolysis. Increase in the total activity of PKM2 increases the glycolytic rate and depletes its upstream intermediates; the depletion of G6P and 3-PG reduces their flux to PPP and SSP, respectively; and the rate of pyruvate to mitochondria metabolism decreases while the rate of pyruvate to lactate increases. Conversely, decrease in the total activity of PKM2 reduces glycolytic rate and accumulates its upstream intermediates; the accumulation of G6P and 3-PG enhances their flux to PPP and SSP, respectively; the rate of pyruvate to mitochondria metabolism increases while the rate of pyruvate to lactate decreases. The two models are different and deserve attention.
Beyond PKM2: The same principle applies to other glycolytic enzymes such as GAPDH and PGK1, which also demonstrate thermodynamic redistribution and flux buffering upon perturbation[
4,
5,
6,
7]. More broadly, in any linear or branched metabolic pathway, the interplay between kinetics and thermodynamics is inevitable. Thus, kinetic-thermodynamic coupling may represent a generalizable design principle underlying metabolism.







