Submitted:
15 May 2025
Posted:
16 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Synthesis of Polyphosphate in LAB
3. Polyphosphate Kinases
4. Exopolyphosphatases
5. Physiological Roles of Polyphosphate in Lactic Acid Bacteria
6. Regulation of Polyphosphate Synthesis in Lactic Acid Bacteria
7. Functional Properties of Probiotic-Derived Polyphosphate: Current Evidence
7. Future Prospects
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DSS | Dextran Sodium Sulfate |
| ERK | Extracellular signal-Regulated Kinase |
| LAB | Lactic Acid Bacteria |
| MAPK | Mitogen-Activated Protein Kinase |
| PolyP | Polyphosphate |
| TNBS | 2,4,6-trinitrobenzenesulfonic acid |
References
- Dürr-Mayer, T.; Qiu, D.; Eisenbeis, V. B.; Steck, N.; Häner, M.; Hofer, A.; Mayer, A.; Siegel, J. S.; Baldridge, K. K.; Jessen, H. J. The chemistry of branched condensed phosphates. Nat. Commun. 2021, 12, 5368. [Google Scholar] [CrossRef] [PubMed]
- Racki, L. R.; Freddolino, L. Polyphosphate: The “Dark Matter” of Bacterial Chromatin Structure. Mol. Microbiol. 2025, 123, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, Y.; Watanabe, H.; Saitoh, M.; Namba, T. Volcanic production of polyphosphates and its relevance to prebiotic evolution. Nature 1991, 352, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Rao, N. N.; Gómez-García, M. R.; Kornberg, A. Inorganic Polyphosphate: Essential for Growth and Survival. Annu. Rev. Biochem. 2009, 78, 605–647. [Google Scholar] [CrossRef]
- Bowlin, M. Q.; Gray, M. J. Inorganic polyphosphate in host and microbe biology. Trends Microbiol. 2021, 29, 1013–1023. [Google Scholar] [CrossRef]
- Kulaev, I. S.; Vagabov, V. M. Polyphosphate Metabolism in Micro-Organisms. In Adv. Microb. Physiol., Rose, A. H.; Morris, J. G.; Tempest, D. W., Eds. Academic Press: 1983; Vol. 24, pp 83-171. [CrossRef]
- Beaufay, F.; Quarles, E.; Franz, A.; Katamanin, O.; Wholey, W.-Y.; Jakob, U. Polyphosphate Functions In Vivo as an Iron Chelator and Fenton Reaction Inhibitor. mBio, 2020; 11. [Google Scholar] [CrossRef]
- Kuroda, A.; Nomura, K.; Ohtomo, R.; Kato, J.; Ikeda, T.; Takiguchi, N.; Ohtake, H.; Kornberg, A. Role of Inorganic Polyphosphate in Promoting Ribosomal Protein Degradation by the Lon Protease in E. coli. Science 2001, 293, 705–708. [Google Scholar] [CrossRef]
- Gross, M. H.; Konieczny, I. Polyphosphate induces the proteolysis of ADP-bound fraction of initiator to inhibit DNA replication initiation upon stress in Escherichia coli. Nucleic Acids Res. 2020, 48, 5457–5466. [Google Scholar] [CrossRef]
- Beck, R. W. A chronology of microbiology in historical context. ASM Press: Washington, D.C., 2000.
- McGrath, J. W.; Quinn, J. P. Microbial Phosphate Removal and Polyphosphate Production from Wastewaters. In Adv. Appl. Microbiol., Academic Press: 2003; Vol. 52, pp 75-100. [CrossRef]
- Hansen–Bruhn, I.; Laura Craig, J.; Hinge, M.; Hull, T. R. Ammonium polyphosphates: Correlating structure to application. Eur. Polym. J. 2025, 223, 113644. [Google Scholar] [CrossRef]
- Müller, W. E. G.; Neufurth, M.; Wang, S.; Schröder, H. C.; Wang, X. The Physiological Inorganic Polymers Biosilica and Polyphosphate as Key Drivers for Biomedical Materials in Regenerative Nanomedicine. Int. J. Nanomedicine 2024, 19, 1303–1337. [Google Scholar] [CrossRef]
- Demling, P.; Baier, M.; Deitert, A.; Fees, J.; Blank, L. M. Biotechnological polyphosphate as an opportunity to contribute to the circularization of the phosphate economy. Curr. Opin. Biotechnol. 2024, 87, 103107. [Google Scholar] [CrossRef]
- Lemos Junior, W. J. F.; Santinello, D.; Mohammadzadeh, S.; Treu, L.; Sant’Ana, A. S.; Campanaro, S. Polyphosphate in food systems: Their roles and applications in foods and contribution to sustainable processing practices. Trends Food Sci. Technol. 2024, 152, 104696. [Google Scholar] [CrossRef]
- Segawa, S.; Fujiya, M.; Konishi, H.; Ueno, N.; Kobayashi, N.; Shigyo, T.; Kohgo, Y. Probiotic-Derived Polyphosphate Enhances the Epithelial Barrier Function and Maintains Intestinal Homeostasis through Integrin–p38 MAPK Pathway. PLoS One 2011, 6, e23278. [Google Scholar] [CrossRef] [PubMed]
- Orla-Jensen, S. The lactic acid bacteria. Fred Host and Son: Copenhagen, Denmark, 1919.
- Makarova, K.; Slesarev, A.; Wolf, Y.; Sorokin, A.; Mirkin, B.; Koonin, E.; Pavlov, A.; Pavlova, N.; Karamychev, V.; Polouchine, N.; Shakhova, V.; Grigoriev, I.; Lou, Y.; Rohksar, D.; Lucas, S.; Huang, K.; Goodstein, D. M.; Hawkins, T.; Plengvidhya, V.; Welker, D.; Hughes, J.; Goh, Y.; Benson, A.; Baldwin, K.; Lee, J. H.; Díaz-Múñiz, I.; Dosti, B.; Smeianov, V.; Wechter, W.; Barabote, R.; Lorca, G.; Altermann, E.; Barrangou, R.; Ganesan, B.; Xie, Y.; Rawsthorne, H.; Tamir, D.; Parker, C.; Breidt, F.; Broadbent, J.; Hutkins, R.; O’Sullivan, D.; Steele, J.; Unlu, G.; Saier, M.; Klaenhammer, T.; Richardson, P.; Kozyavkin, S.; Weimer, B.; Mills, D. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 15611–6. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C. M. A. P.; Harris, H. M. B.; Mattarelli, P.; O’Toole, P. W.; Pot, B.; Vandamme, P.; Walter, J.; Watanabe, K.; Wuyts, S.; Felis, G. E.; Gänzle, M. G.; Lebeer, S. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ). Update of the list of qualified presumption of safety (QPS) recommended microbiological agents intentionally added to food or feed as notified to EFSA 21: Suitability of taxonomic units notified to EFSA until September 2024. EFSA J. 2025, 23, e9169. [CrossRef]
- Neville, N.; Roberge, N.; Jia, Z. Polyphosphate Kinase 2 (PPK2) Enzymes: Structure, Function, and Roles in Bacterial Physiology and Virulence. Int. J. Mol. Sci. 2022, 23, 670. [Google Scholar] [CrossRef]
- Bolesch, D. G.; Keasling, J. D. Polyphosphate Binding and Chain Length Recognition of Escherichia coli Exopolyphosphatase. J. Biol. Chem. 2000, 275, 33814–33819. [Google Scholar] [CrossRef]
- Hugues, D. E.; Muhammed, D. The metabolism of polyphosphate in bacteria. Colloq. Int. Centre Natl. Recherche Sci. (Paris) 1962, 106, 591–602. [Google Scholar]
- Kakefuda, T.; Holden, J. T.; Utech, N. M. Ultrastructure of the Membrane System in Lactobacillus plantarum. J. Bacteriol. 1967, 93, 472–482. [Google Scholar] [CrossRef]
- Archibald, F. S.; Fridovich, I. Investigations of the state of the manganese in Lactobacillus plantarum. Arch. Biochem. Biophys. 1982, 215, 589–596. [Google Scholar] [CrossRef]
- Aprea, G.; Mullan, W. M. A.; Mullan, A.; Murru, N.; Tozzi, M.; Cortesi, M. L. Isolation of polyphosphate-accumulating lactic acid bacteria from natural whey starters. Milchwissenschaft 2005, 60, 256–258. [Google Scholar]
- Saiki, A.; Ishida, Y.; Segawa, S.; Hirota, R.; Nakamura, T.; Kuroda, A. A Lactobacillus mutant capable of accumulating long-chain polyphosphates that enhance intestinal barrier function. Biosci. Biotechnol. Biochem. 2016, 80, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Correa Deza, M. A.; Grillo-Puertas, M.; Salva, S.; Rapisarda, V. A.; Gerez, C. L.; Font de Valdez, G. Inorganic salts and intracellular polyphosphate inclusions play a role in the thermotolerance of the immunobiotic Lactobacillus rhamnosus CRL 1505. PLoS One 2017, 12, e0179242. [Google Scholar] [CrossRef] [PubMed]
- Alcántara, C.; Blasco, A.; Zúñiga, M.; Monedero, V. Accumulation of polyphosphate in Lactobacillus spp. and its involvement in stress resistance. Appl. Environ. Microbiol. 2014, 80, 1650–1659. [Google Scholar] [CrossRef]
- Corrales, D.; Alcántara, C.; Zúñiga, M.; Monedero, V. Ppx1 putative exopolyphosphatase is essential for polyphosphate accumulation in Lacticaseibacillus paracasei. Appl. Environ. Microbiol. 2024, 90, e02290–23. [Google Scholar] [CrossRef] [PubMed]
- Ahn, K.; Kornberg, A. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 1990, 265, 11734–11739. [Google Scholar] [CrossRef]
- Akiyama, M.; Crooke, E.; Kornberg, A. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J. Biol. Chem. 1992, 267, 22556–22561. [Google Scholar] [CrossRef]
- Kornberg, A.; Kornberg, S. R.; Simms, E. S. Metaphosphate synthesis by an enzyme from Escherichia coli. Biochimica et biophysica acta 1956, 20, 215–227. [Google Scholar] [CrossRef]
- Suzuki, H.; Kaneko, T.; Ikeda, Y. Properties of polyphosphate kinase prepared from Mycobacterium smegmatis. Biochim. Biophys. Acta 1972, 268, 381–390. [Google Scholar] [CrossRef]
- Levinson, S. L.; Jacobs, L. H.; Krulwich, T. A.; Li, H.-C. Purification and characterization of a polyphosphate kinase from Arthrobacter atrocyaneus. Microbiology 1975, 88, 65–74. [Google Scholar] [CrossRef]
- Robinson, N. A.; Clark, J. E.; Wood, H. G. Polyphosphate kinase from Propionibacterium shermanii. Demonstration that polyphosphates are primers and determination of the size of the synthesized polyphosphate. J. Biol. Chem. 1987, 262, 5216–5222. [Google Scholar] [CrossRef]
- Robinson, N. A.; Wood, H. G. Polyphosphate kinase from Propionibacterium shermanii. Demonstration that the synthesis and utilization of polyphosphate is by a processive mechanism. J. Biol. Chem. 1986, 261, 4481–4485. [Google Scholar] [CrossRef]
- Tinsley, C. R.; Manjula, B. N.; Gotschlich, E. C. Purification and characterization of polyphosphate kinase from Neisseria meningitidis. Infect. Immun. 1993, 61, 3703–3710. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Huang, W.; Lee, S. S. K.; Xu, W. Crystal structure of a polyphosphate kinase and its implications for polyphosphate synthesis. EMBO Rep. 2005, 6, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Araoz, M.; Grillo-Puertas, M.; de Moreno de LeBlanc, A.; Hebert, E. M.; Villegas, J. M.; Rapisarda, V. A. Inorganic phosphate modifies stationary phase fitness and metabolic pathways in Lactiplantibacillus paraplantarum CRL 1905. Front. Microbiol. 2024, 15, 1343541. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M. H.; Rao, N. N.; Kornberg, A. Inorganic Polyphosphate Is Required for Motility of Bacterial Pathogens. J. Bacteriol. 2000, 182, 225–227. [Google Scholar] [CrossRef]
- Ishige, K.; Zhang, H.; Kornberg, A. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 16684–16688. [Google Scholar] [CrossRef]
- Leipe, D. D.; Koonin, E. V.; Aravind, L. Evolution and Classification of P-loop Kinases and Related Proteins. J. Mol. Biol. 2003, 333, 781–815. [Google Scholar] [CrossRef]
- Motomura, K.; Hirota, R.; Okada, M.; Ikeda, T.; Ishida, T.; Kuroda, A. A New Subfamily of Polyphosphate Kinase 2 (Class III PPK2) Catalyzes both Nucleoside Monophosphate Phosphorylation and Nucleoside Diphosphate Phosphorylation. Appl. Environ. Microbiol. 2014, 80, 2602–2608. [Google Scholar] [CrossRef]
- Akiyama, M.; Crooke, E.; Kornberg, A. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 1993, 268, 633–639. [Google Scholar] [CrossRef]
- Rangarajan, E. S.; Nadeau, G.; Li, Y.; Wagner, J.; Hung, M.-N.; Schrag, J. D.; Cygler, M.; Matte, A. The Structure of the Exopolyphosphatase (PPX) from Escherichia coli O157:H7 Suggests a Binding Mode for Long Polyphosphate Chains. J. Mol. Biol. 2006, 359, 1249–1260. [Google Scholar] [CrossRef]
- Keasling, J. D.; Bertsch, L.; Kornberg, A. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc. Natl. Acad. Sci. U.S.A. 1993, 90, 7029–7033. [Google Scholar] [CrossRef] [PubMed]
- Hara, A.; Sy, J. Guanosine 5′-triphosphate, 3′-diphosphate 5′-phosphohydrolase. Purification and substrate specificity. J. Biol. Chem. 1983, 258, 1678–1683. [Google Scholar] [CrossRef]
- Mechold, U.; Potrykus, K.; Murphy, H.; Murakami, K. S.; Cashel, M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 2013, 41, 6175–6189. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Dharmasena, M. N.; Wang, C.; Shaw, G. X.; Cherry, S.; Tropea, J. E.; Jin, D. J.; Ji, X. Structure and activity of PPX/GppA homologs from Escherichia coli and Helicobacter pylori. FEBS J. 2020, 287, 1865–1885. [Google Scholar] [CrossRef]
- Kristensen, O.; Laurberg, M.; Liljas, A.; Kastrup, J. S.; Gajhede, M. Structural Characterization of the Stringent Response Related Exopolyphosphatase/Guanosine Pentaphosphate Phosphohydrolase Protein Family. Biochemistry 2004, 43, 8894–8900. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Jakob, U. Polyphosphate: a cellular Swiss army knife. Curr. Opin. Biotechnol. 2025, 93, 103303. [Google Scholar] [CrossRef]
- Schoeppe, R.; Waldmann, M.; Jessen, H. J.; Renné, T. An Update on Polyphosphate In Vivo Activities. Biomolecules 2024, 14, 937. [Google Scholar] [CrossRef]
- Rosigkeit, H.; Kneißle, L.; Obruča, S.; Jendrossek, D. The Multiple Roles of Polyphosphate in Ralstonia eutropha and Other Bacteria. Microb. Physiol. 2021, 31, 163–177. [Google Scholar] [CrossRef]
- Reusch, R. N.; Sadoff, H. L. Putative structure and functions of a poly-beta-hydroxybutyrate/calcium polyphosphate channel in bacterial plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 1988, 85, 4176–4180. [Google Scholar] [CrossRef]
- Jendrossek, D.; Pfeiffer, D. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ. Microbiol. 2014, 16, 2357–2373. [Google Scholar] [CrossRef]
- Gray, M. J.; Jakob, U. Oxidative stress protection by polyphosphate—new roles for an old player. Curr. Opin. Microbiol. 2015, 24, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gray, M. J.; Wholey, W.-Y.; Wagner, N. O.; Cremers, C. M.; Mueller-Schickert, A.; Hock, N. T.; Krieger, A. G.; Smith, E. M.; Bender, R. A.; Bardwell, J. C. A.; Jakob, U. Polyphosphate Is a Primordial Chaperone. Mol. Cell 2014, 53, 689–699. [Google Scholar] [CrossRef]
- Kornberg, A.; Rao, N. N.; Ault-Riché, D. Inorganic Polyphosphate: A Molecule of Many Functions. Annu. Rev. Biochem. 1999, 68, 89–125. [Google Scholar] [CrossRef] [PubMed]
- Albi, T.; Serrano, A. Inorganic polyphosphate in the microbial world. Emerging roles for a multifaceted biopolymer. World J. Microbiol. Biotechnol. 2016, 32, 27. [Google Scholar] [CrossRef]
- Albi, T.; Serrano, A. Two strictly polyphosphate-dependent gluco(manno)kinases from diazotrophic Cyanobacteria with potential to phosphorylate hexoses from polyphosphates. Appl. Microbiol. Biotechnol. 2015, 99, 3887–3900. [Google Scholar] [CrossRef]
- Rojas, D.; Marcoleta, A. E.; Gálvez-Silva, M.; Varas, M. A.; Díaz, M.; Hernández, M.; Vargas, C.; Nourdin-Galindo, G.; Koch, E.; Saldivia, P.; Vielma, J.; Gan, Y.-H.; Chen, Y.; Guiliani, N.; Chávez, F. P. Inorganic Polyphosphate Affects Biofilm Assembly, Capsule Formation, and Virulence of Hypervirulent ST23 Klebsiella pneumoniae. ACS Infect. Dis. 2024, 10, 606–623. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Zhou, Y.; Liu, B.; Guan, J.; Zhang, P.; Deng, X.; Li, D.; Wang, J. Polyphosphate Kinase Is Required for the Processes of Virulence and Persistence in Acinetobacter baumannii. Microbiol. Spectr. 2022, 10, e01230–22. [Google Scholar] [CrossRef]
- Keasling, J. D. Regulation of Intracellular Toxic Metals and Other Cations by Hydrolysis of Polyphosphate. Ann. N. Y. Acad. Sci. 1997, 829, 242–249. [Google Scholar] [CrossRef]
- Alcántara, C.; Coll-Marqués, J. M.; Jadán-Piedra, C.; Vélez, D.; Devesa, V.; Zúñiga, M.; Monedero, V. Polyphosphate in Lactobacillus and its link to stress tolerance and probiotic properties. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Archibald, F. S.; Fridovich, I. Manganese, superoxide dismutase, and oxygen tolerance in some lactic acid bacteria. J. Bacteriol. 1981, 146, 928–936. [Google Scholar] [CrossRef]
- Archibald, F. Manganese: Its Acquisition by and Function in the Lactic Acid Bacteria. Crit. Rev. Microbiol. 1986, 13, 63–109. [Google Scholar] [CrossRef] [PubMed]
- Archibald, F. S.; Duong, M. N. Manganese acquisition by Lactobacillus plantarum. J. Bacteriol. 1984, 158, 1–8. [Google Scholar] [CrossRef]
- Breiland, A. A.; Flood, B. E.; Nikrad, J.; Bakarich, J.; Husman, M.; Rhee, T.; Jones, R. S.; Bailey, J. V. Polyphosphate-Accumulating Bacteria: Potential Contributors to Mineral Dissolution in the Oral Cavity. Appl. Environ. Microbiol. 2018, 84, e02440–17. [Google Scholar] [CrossRef]
- Huang, S.; Gaucher, F.; Cauty, C.; Jardin, J.; Le Loir, Y.; Jeantet, R.; Chen, X. D.; Jan, G. Growth in Hyper-Concentrated Sweet Whey Triggers Multi Stress Tolerance and Spray Drying Survival in Lactobacillus casei BL23: From the Molecular Basis to New Perspectives for Sustainable Probiotic Production. Front. Microbiol. 2018, 9, 2548. [Google Scholar] [CrossRef] [PubMed]
- Ault-Riché, D.; Fraley, C. D.; Tzeng, C.-M.; Kornberg, A. Novel Assay Reveals Multiple Pathways Regulating Stress-Induced Accumulations of Inorganic Polyphosphate in Escherichia coli. J. Bacteriol. 1998, 180, 1841–1847. [Google Scholar] [CrossRef] [PubMed]
- Shiba, T.; Tsutsumi, K.; Yano, H.; Ihara, Y.; Kameda, A.; Tanaka, K.; Takahashi, H.; Munekata, M.; Rao, N. N.; Kornberg, A. Inorganic polyphosphate and the induction of rpoS expression. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 11210–11215. [Google Scholar] [CrossRef]
- Pokhrel, A.; Lingo, J. C.; Wolschendorf, F.; Gray, M. J. Assaying for Inorganic Polyphosphate in Bacteria. J. Vis. Exp. 2019, 143, e58818. [Google Scholar] [CrossRef]
- Thakur, P. B.; Long, A. R.; Nelson, B. J.; Kumar, R.; Rosenberg, A. F.; Gray, M. J. Complex Responses to Hydrogen Peroxide and Hypochlorous Acid by the Probiotic Bacterium Lactobacillus reuteri. mSystems 2019, 4. [Google Scholar] [CrossRef]
- Gray, M. J. Inorganic Polyphosphate Accumulation in Escherichia coli Is Regulated by DksA but Not by (p)ppGpp. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef]
- Correa Deza, M. A.; Rodríguez de Olmos, A.; Suárez, N. E.; Font de Valdez, G.; Salva, S.; Gerez, C. L. Inorganic polyphosphate from the immunobiotic Lactobacillus rhamnosus CRL1505 prevents inflammatory response in the respiratory tract. Saudi J. Biol. Sci. 2021, 28, 5684–5692. [Google Scholar] [CrossRef]
- Corrales, D.; Alcántara, C.; Vélez, D.; Devesa, V.; Monedero, V.; Zúñiga, M. Unveiling the role of the PhoP master regulator in arsenite resistance through ackA downregulation in Lacticaseibacillus paracasei. Curr. Res. Microb. Sci. 2025, 8, 100357. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J. P.; Moon, S. J.; Lee, M.; Kwon, M. Y.; Pyo, J.-Y.; Lee, S. C.; Kim, H. J. Engineering polyphosphate-accumulating probiotics for therapeutic applications in chronic kidney disease. Food Biosci. 2025, 68, 106360. [Google Scholar] [CrossRef]
- Kus, F.; Smolenski, R. T.; Tomczyk, M. Inorganic Polyphosphate—Regulator of Cellular Metabolism in Homeostasis and Disease. Biomedicines 2022, 10, 913. [Google Scholar] [CrossRef]
- Ueno, N.; Fujiya, M.; Segawa, S.; Nata, T.; Moriichi, K.; Tanabe, H.; Mizukami, Y.; Kobayashi, N.; Ito, K.; Kohgo, Y. Heat-killed body of Lactobacillus brevis SBC8803 ameliorates intestinal injury in a murine model of colitis by enhancing the intestinal barrier function. Inflamm. Bowel Dis. 2011, 17, 2235–2250. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Fujiya, M.; Konishi, H.; Ueno, N.; Kashima, S.; Sasajima, J.; Moriichi, K.; Ikuta, K.; Tanabe, H.; Kohgo, Y. Probiotic-derived polyphosphate improves the intestinal barrier function through the caveolin-dependent endocytic pathway. Biochem. Biophys. Res. Commun. 2015, 467, 541–548. [Google Scholar] [CrossRef]
- Kashima, S.; Fujiya, M.; Konishi, H.; Ueno, N.; Inaba, Y.; Moriichi, K.; Tanabe, H.; Ikuta, K.; Ohtake, T.; Kohgo, Y. Polyphosphate, an active molecule derived from probiotic Lactobacillus brevis, improves the fibrosis in murine colitis. Transl. Res. 2015, 166, 163–175. [Google Scholar] [CrossRef]
- Li, S.; Zheng, A.; Chen, Z.; Wang, X.; Chen, J.; Zou, Z.; Liu, G. Lactobacillus plantarum-Derived Inorganic Polyphosphate Regulates Immune Function via Inhibiting M1 Polarization and Resisting Oxidative Stress in Macrophages. Antioxidants 2025, 14, 428. [Google Scholar] [CrossRef]
- Isozaki, S.; Konishi, H.; Fujiya, M.; Tanaka, H.; Murakami, Y.; Kashima, S.; Ando, K.; Ueno, N.; Moriichi, K.; Okumura, T. Probiotic-Derived Polyphosphate Accelerates Intestinal Epithelia Wound Healing through Inducing Platelet-Derived Mediators. Mediators Inflamm. 2021, 2021, 5582943. [Google Scholar] [CrossRef]
- Takauji, S.; Konishi, H.; Fujiya, M.; Ueno, N.; Tanaka, H.; Sato, H.; Isozaki, S.; Kashima, S.; Moriichi, K.; Mizukami, Y.; Okumura, T. Polyphosphate, Derived from Lactobacillus brevis, Modulates the Intestinal Microbiome and Attenuates Acute Pancreatitis. Dig. Dis. Sci. 2021, 66, 3872–3884. [Google Scholar] [CrossRef]
- Sakatani, A.; Fujiya, M.; Ueno, N.; Kashima, S.; Sasajima, J.; Moriichi, K.; Ikuta, K.; Tanabe, H.; Kohgo, Y. Polyphosphate Derived from Lactobacillus brevis Inhibits Colon Cancer Progression Through Induction of Cell Apoptosis. Anticancer Res. 2016, 36, 591–598. [Google Scholar]
- Moon, S. J.; Hwang, J.; Kang, W. K.; Ahn, J.-P.; Kim, H. J. Administration of the probiotic Lactiplantibacillus paraplantarum is effective in controlling hyperphosphatemia in 5/6 nephrectomy rat model. Life Sci. 2022, 306, 120856. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Viso, P.; Domene, A.; Sánchez, A.; Vélez, D.; Monedero, V.; Devesa, V.; Zúñiga, M. Challenges and strategies for preventing intestinal damage associated to mercury dietary exposure. Toxicology 2023, 494, 153580. [Google Scholar] [CrossRef] [PubMed]
- Chiocchetti, G. M.; Jadán-Piedra, C.; Monedero, V.; Zúñiga, M.; Vélez, D.; Devesa, V. Use of lactic acid bacteria and yeasts to reduce exposure to chemical food contaminants and toxicity. Crit. Rev. Food Sci. Nutr. 2018, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
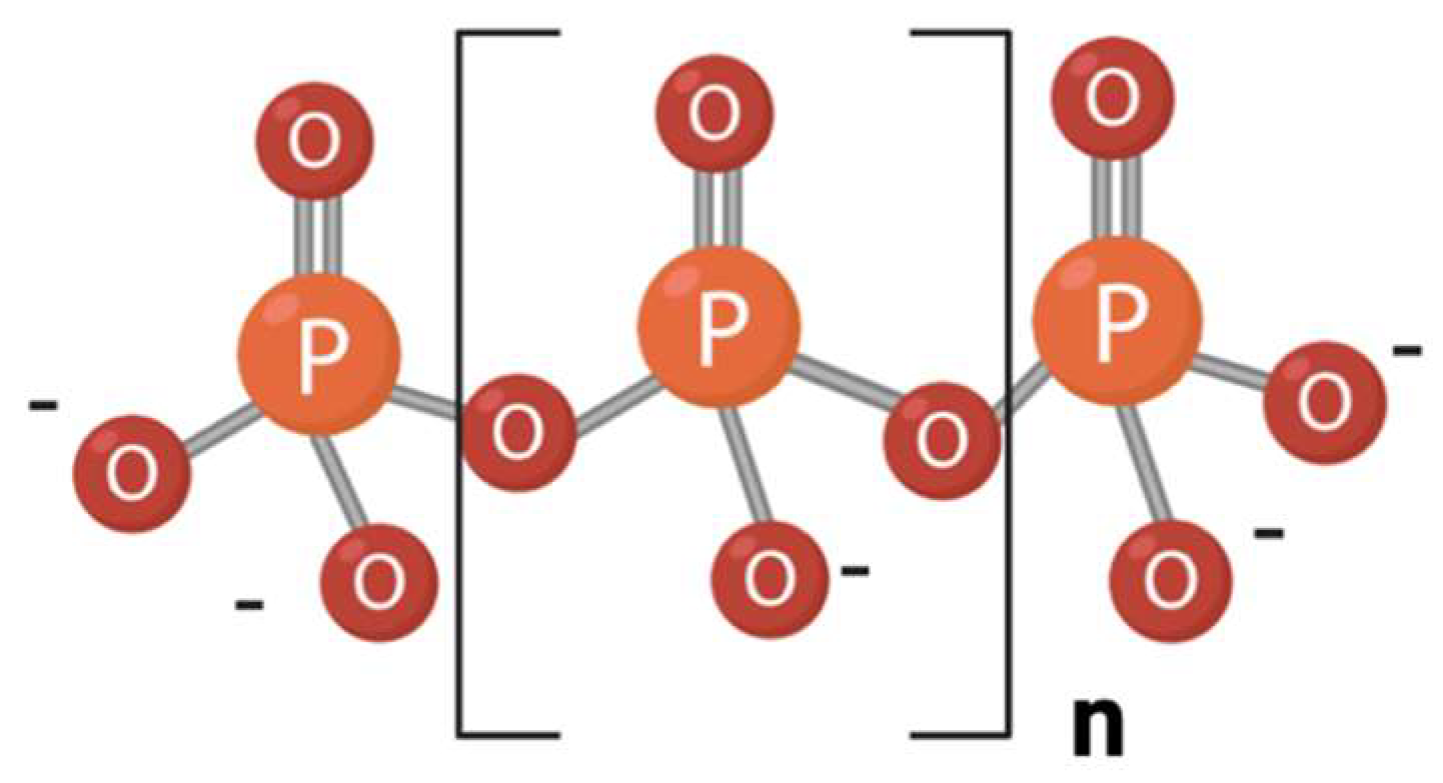
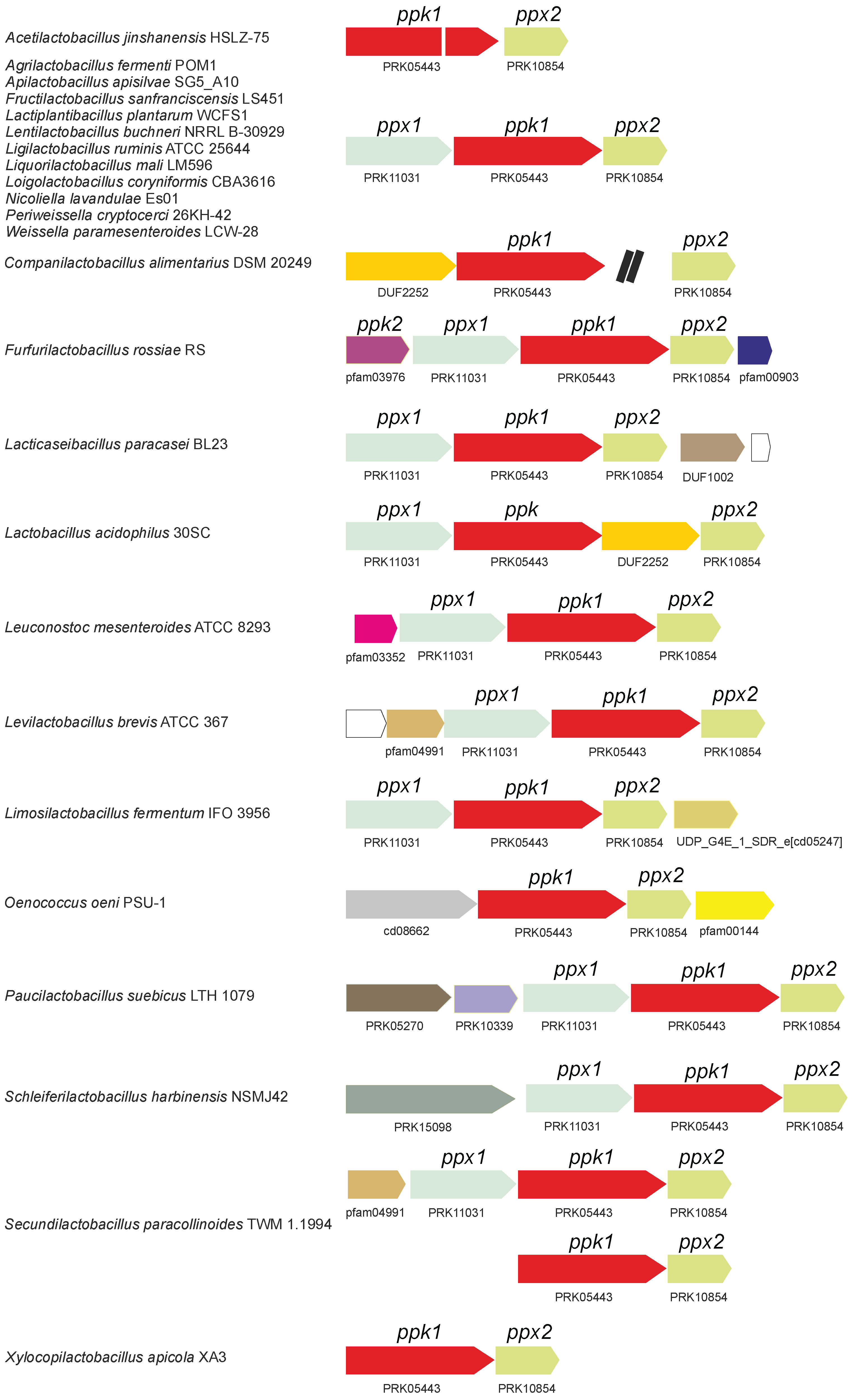
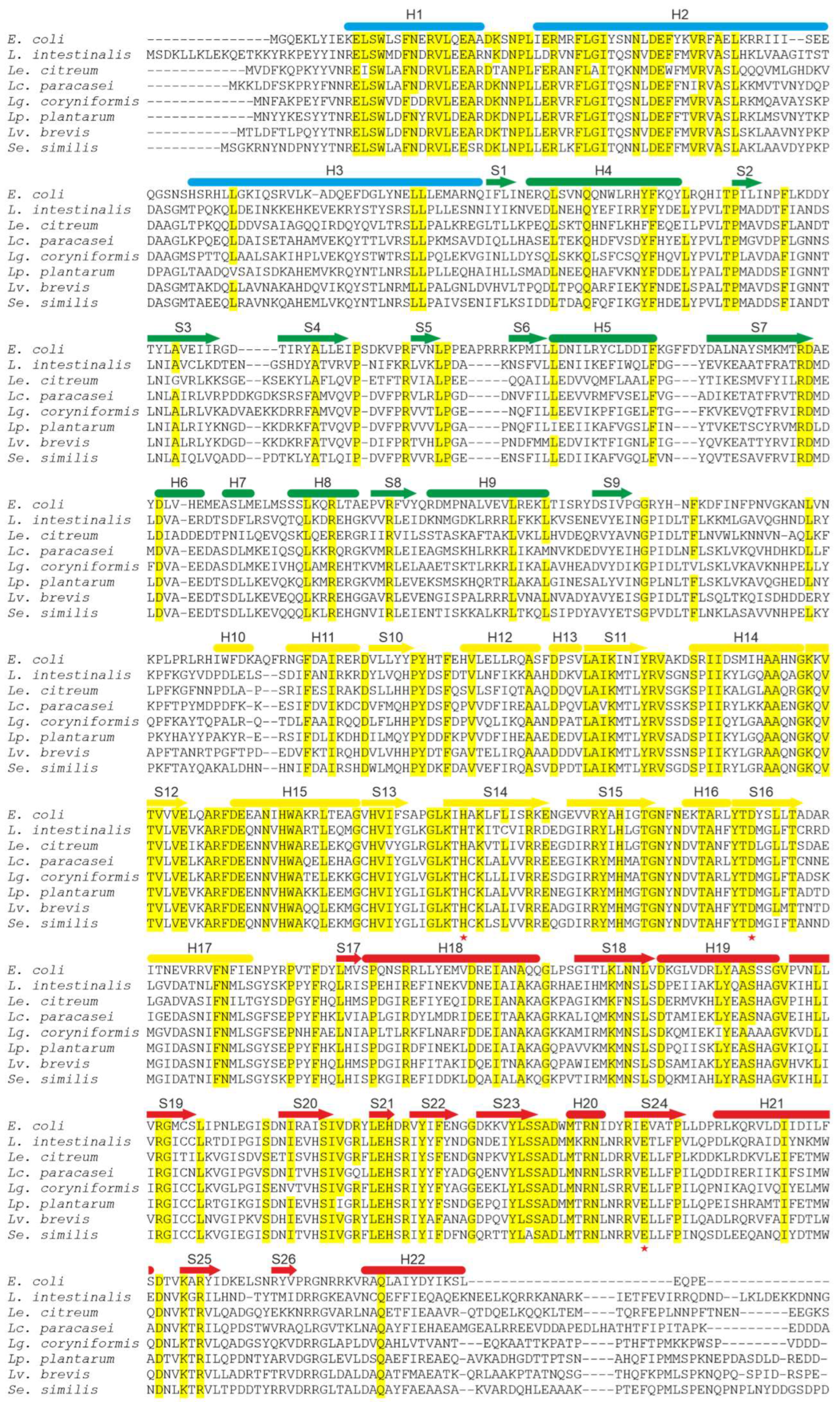
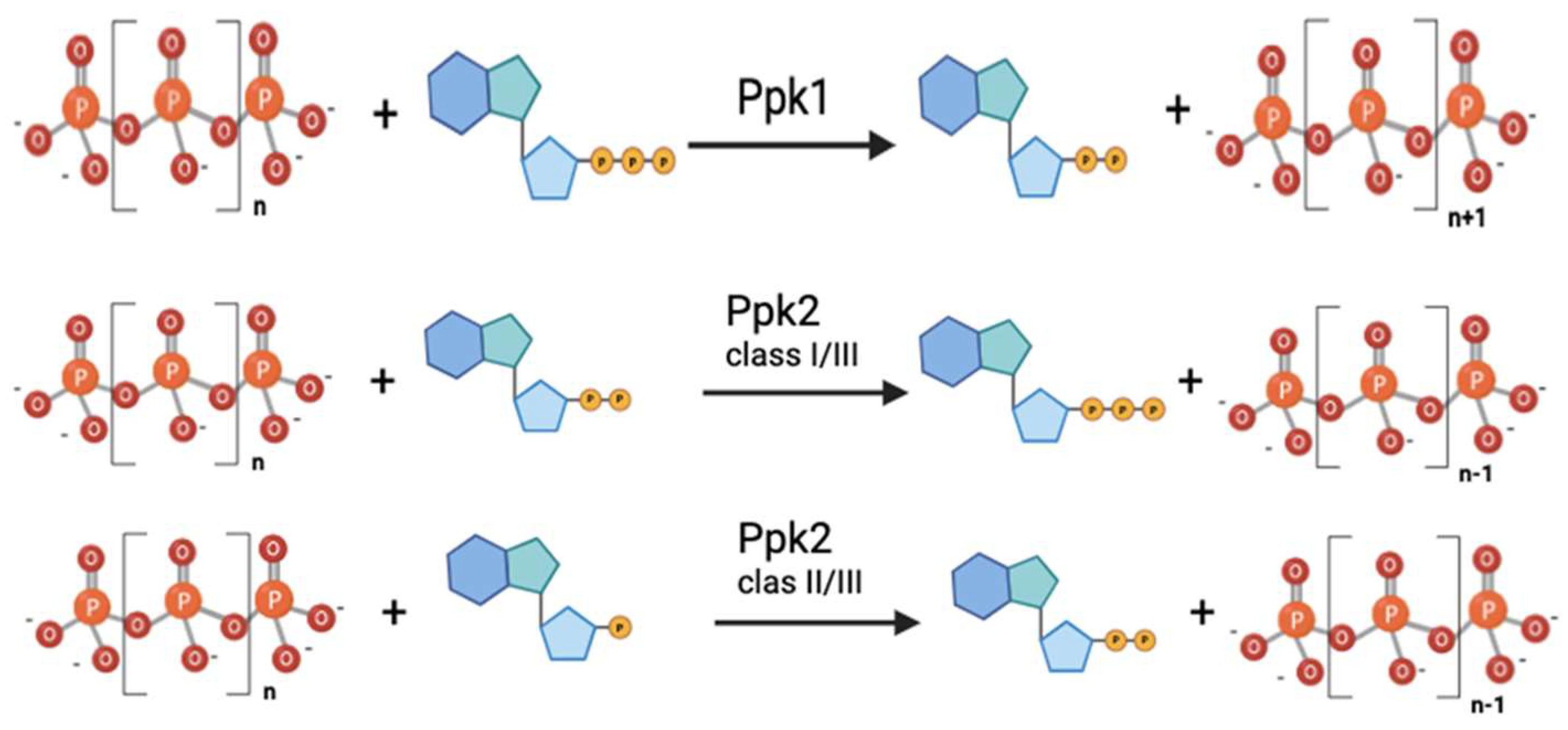
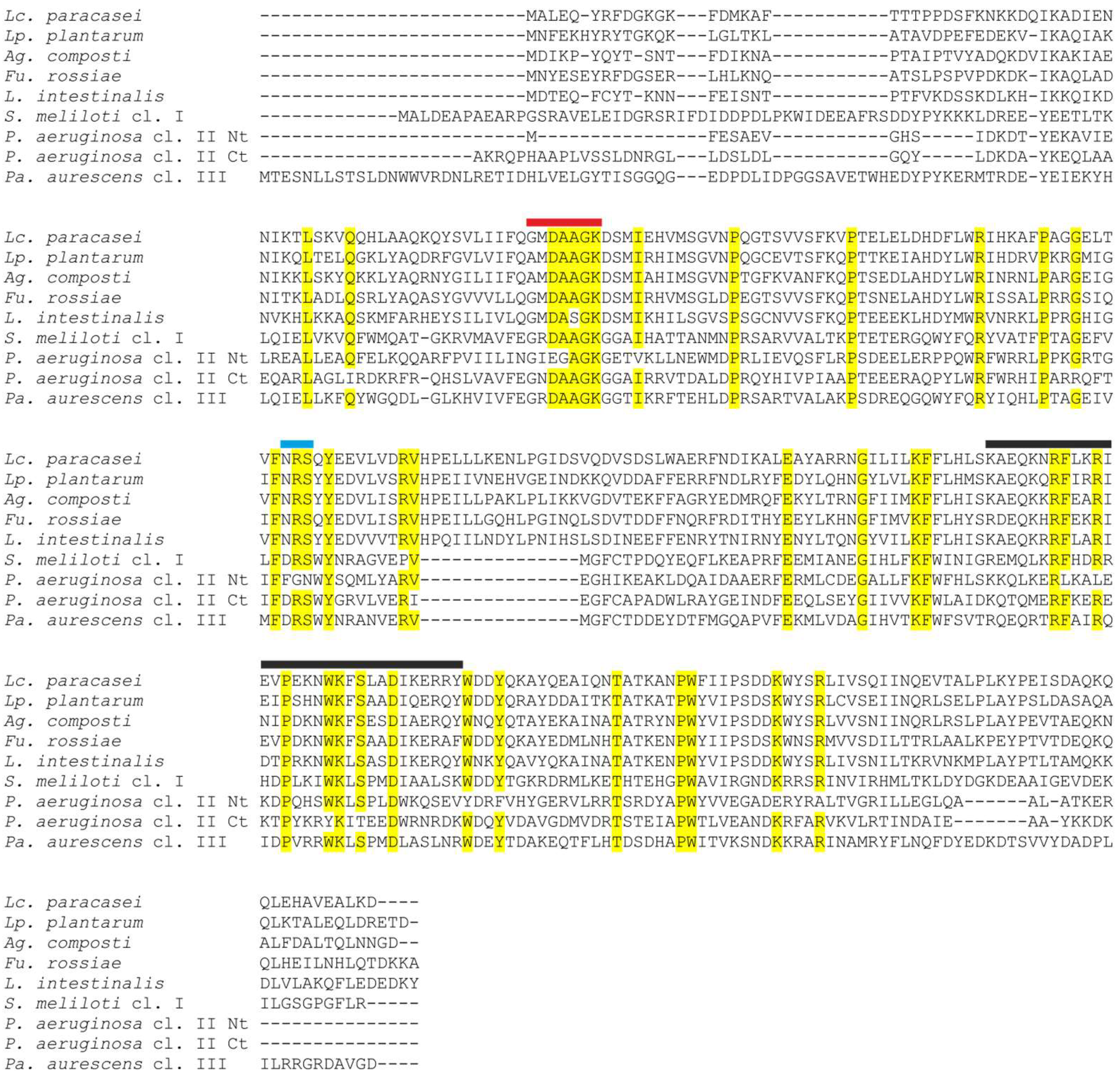
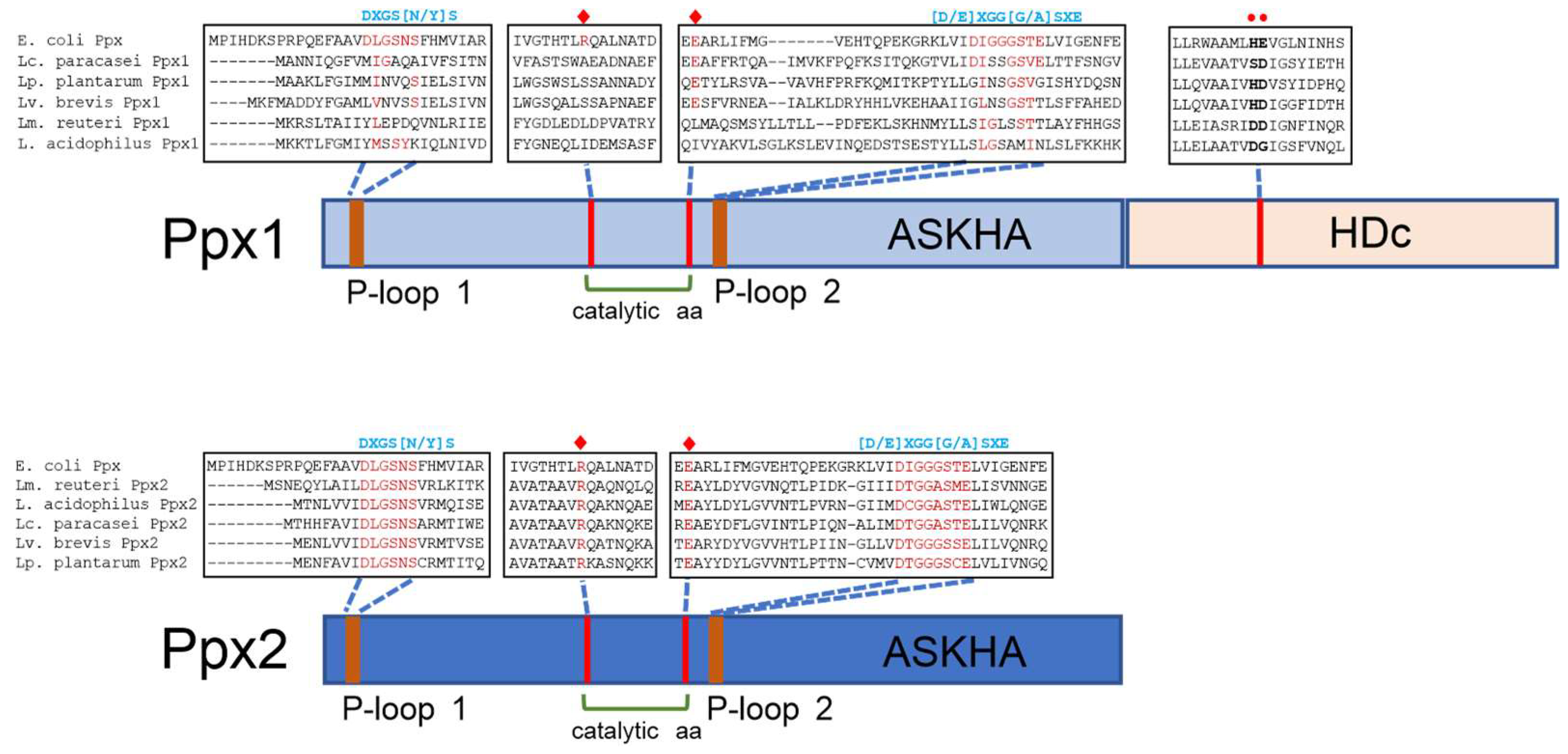
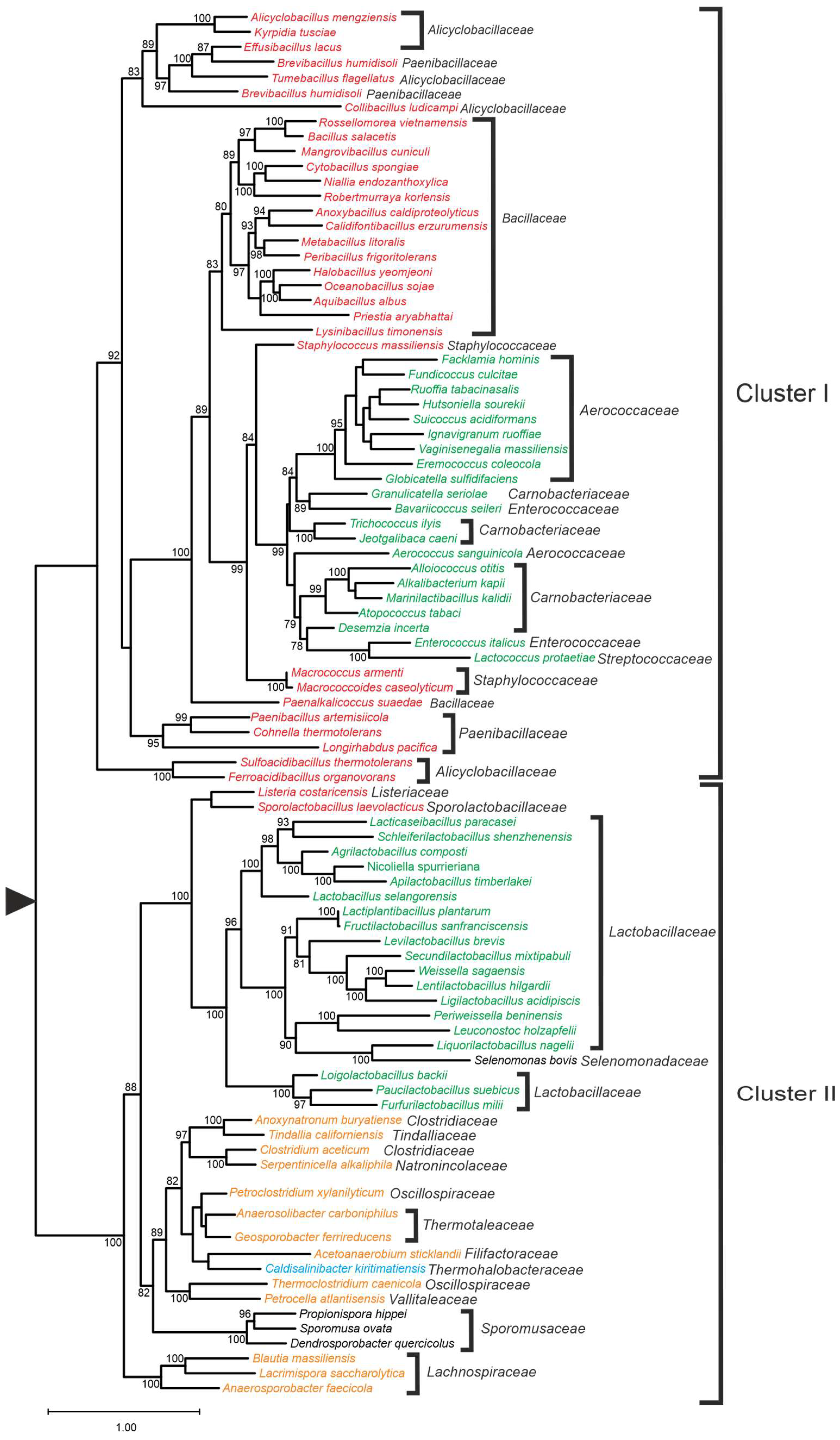
| Genus | ppk1 | ppk2 | ppx1 | ppx2 |
|---|---|---|---|---|
| Acetilactobacillus | + | - | - | + |
| Agrilactobacillus | + | ± | + | + |
| Amylolactobacillus | - | - | - | - |
| Apilactobacillus | ± | - | ± | ± |
| Bombilactobacillus | - | - | - | ± |
| Companilactobacillus | + | - | - | + |
| Convivina | - | - | - | + |
| Dellaglioa | - | - | - | + |
| Eupransor | + | - | - | + |
| Fructilactobacillus | + | - | ± | + |
| Fructobacillus | ± | - | - | + |
| Furfurilactobacillus | + | + | + | + |
| Holzapfeliella | - | - | - | - |
| Lacticaseibacillus | + | ± | + | + |
| Lactiplantibacillus | + | + | + | + |
| Lactobacillus | ± | ± | ± | ± |
| Lapidilactobacillus | - | - | - | - |
| Latilactobacillus | - | - | - | - |
| Lentilactobacillus | ± | ± | ± | + |
| Leuconostoc | + | - | + | + |
| Levilactobacillus | + | + | + | + |
| Ligilactobacillus | ± | - | ± | ± |
| Limosilactobacillus | + | + | + | + |
| Liquorilactobacillus | + | ± | + | + |
| Loigolactobacillus | ± | ± | ± | ± |
| Nicoliella | + | - | + | + |
| Oenococcus | + | - | - | + |
| Paucilactobacillus | ± | - | ± | + |
| Pediococcus | - | - | - | + |
| Periweissella | + | - | + | + |
| Philodulcilactobacillus | + | - | - | + |
| Schleiferilactobacillus | + | + | + | + |
| Secundilactobacillus | ± | - | ± | + |
| Weissella | ± | - | ± | + |
| Xylocopilactobacillus | + | - | - | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





