Submitted:
15 May 2025
Posted:
15 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Clinical Diagnostic Approaches of C. auris
3. Screening Guidelines and Protocols of
3.1. Importance of Screening for C. auris Colonization
3.2. Identifying Individuals for Screening
3.3. Screening Methods, Timing, and Laboratory Protocols
4. Diagnostic Methods for C. auris Detection
4.1. Culture-Based Methods
4.1.1. Differential and Selective Media
4.1.2. Chromogenic Media
4.1.3. Conclusion of Culture-Based Methods
4.2. Biochemical Assimilation Tests
4.3. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
4.3.1. The evolution of MALDI-TOF on Detecting C. auris
4.3.2. Sample Preparation and Database Considerations
4.4. Molecular Methods for C auris Detection
4.4.1. Overview of DNA-Based Detection
4.4.2. PCR-Based Detection
4.4.3. Loop-Mediated Isothermal Amplification (LAMP)
4.4.4. T2 Magnetic Resonance (T2MR) Assay
4.4.5. Summary for Molecular Testing
5. Genomic Typing and Outbreak Investigation in
6. Antifungal Susceptibility Testing (AFST) for C. auris
6.1. Multidrug Resistance in C. auris
6.2. Reference Methods and Interpretive Challenges
6.3. Performance of Commercial Testing Platforms
6.4. Genotypic Resistance Mechanisms and Molecular Testing
6.5. MALDI-TOF MS-Based AFST: Emerging Innovations
6.6. Summary for AFST
7. Conclusions
Abbreviations
| AFST | Antifungal Susceptibility Testing |
| AFLP | Amplified Fragment Length Polymorphism |
| AMB | Amphotericin B |
| API 20C AUX | Analytical Profile Index 20C Auxanographic Yeast Identification System |
| API ID 32C | Analytical Profile Index ID 32C Yeast Identification System |
| ASTRA | Antibiotic Susceptibility Test Rapid Assay |
| BDG | Beta-D-Glucan |
| BMD | Broth Microdilution |
| BCID | Blood Culture Identification |
| CA System | Chromogenic Agar System (Bruker’s MALDI-TOF MS software) |
| CDC | Centers for Disease Control and Prevention |
| CCI | Composite Correlation Index |
| CE-IVD | Conformité Européenne – In Vitro Diagnostic |
| CFU | Colony Forming Unit |
| CHROMagar™ | Chromogenic Agar (commercial medium) |
| CLSI | Clinical and Laboratory Standards Institute |
| DNA | Deoxyribonucleic Acid |
| DOAJ | Directory of Open Access Journals |
| ECV | Epidemiologic Cutoff Value |
| FDA | U.S. Food and Drug Administration |
| GPI | Glycosylphosphatidylinositol |
| HIV | Human Immunodeficiency Virus |
| ICU | Intensive Care Unit |
| ITS | Internal Transcribed Spacer |
| IVD | In Vitro Diagnostic |
| LAMP | Loop-Mediated Isothermal Amplification |
| LD | Linear Dichroism |
| MALDI-TOF MS | Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry |
| MBT ASTRA | MALDI Biotyper Antibiotic Susceptibility Test Rapid Assay |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MIC | Minimum Inhibitory Concentration |
| MLST | Multilocus Sequence Typing |
| nad5 | NADH Dehydrogenase Subunit 5 |
| NCBI | National Center for Biotechnology Information |
| PCR | Polymerase Chain Reaction |
| qPCR | Quantitative Polymerase Chain Reaction |
| RAPID Yeast Plus | Enzyme-based Yeast Identification Panel (Remel/Thermo Fisher) |
| RUO | Research Use Only |
| SCA | Specific *Candida auris* Medium |
| SDA | Sabouraud Dextrose Agar |
| Se | Sensitivity |
| SNP | Single Nucleotide Polymorphism |
| Sp | Specificity |
| STR | Short Tandem Repeat |
| SYO | Sensititre YeastOne |
| T2MR | T2 Magnetic Resonance |
| TLA | Three Letter Acronym |
| VITEK 2 | VITEK 2 Biochemical Identification System |
| vSNF | Ventilator-Capable Skilled Nursing Facility |
| WHO | World Health Organization |
| WGS | Whole Genome Sequencing |
| YST | Yeast Susceptibility Testing |
References
- Eix, E.F. and J.E. Nett, Candida auris: Epidemiology and Antifungal Strategy. Annu Rev Med, 2025. 76(1): p. 57-67.
- Nelson, R. , Emergence of resistant Candida auris. Lancet Microbe, 2023. 4(6): p. e396.
- Lyman, M. , et al., Worsening Spread of Candida auris in the United States, 2019 to 2021. Ann Intern Med, 2023. 176(4): p. 489-495.
- Kohlenberg, A., D. L. Monnet, and D. Plachouras, Increasing number of cases and outbreaks caused by Candida auris in the EU/EEA, 2020 to 2021. Euro Surveill, 2022. 27(46).
- Sticchi, C. , et al., Increasing Number of Cases Due to Candida auris in North Italy, 19-December 2022. J Clin Med, 2023. 12(5).
- Ahmad, S. and W. Alfouzan, Candida auris: Epidemiology, Diagnosis, Pathogenesis, Antifungal Susceptibility, and Infection Control Measures to Combat the Spread of Infections in Healthcare Facilities. Microorganisms, 2021. 9(4).
- Shastri, P.S. , et al., Candida auris candidaemia in an intensive care unit - Prospective observational study to evaluate epidemiology, risk factors, and outcome. J Crit Care, 2020. 57: p. 42-48.
- Shaukat, A. , et al., Experience of treating Candida auris cases at a general hospital in the state of Qatar. IDCases, 2021. 23: p. e01007.
- Al-Rashdi, A. , et al., Characteristics, Risk Factors, and Survival Analysis of Candida auris Cases: Results of One-Year National Surveillance Data from Oman. J Fungi (Basel), 2021. 7(1).
- Pandya, N. , et al., International Multicentre Study of Candida auris Infections. J Fungi (Basel), 2021. 7(10).
- Chen, J. , et al., Is the superbug fungus really so scary? A systematic review and meta-analysis of global epidemiology and mortality of Candida auris. BMC Infect Dis, 2020. 20(1): p. 827.
- Garcia-Bustos, V. , et al., What Do We Know about Candida auris? State of the Art, Knowledge Gaps, and Future Directions. Microorganisms, 2021. 9(10).
- Fasciana, T. , et al., Candida auris: An Overview of How to Screen, Detect, Test and Control This Emerging Pathogen. Antibiotics (Basel), 2020. 9(11).
- Welker, M. , et al., An update on the routine application of MALDI-TOF MS in clinical microbiology. Expert Rev Proteomics, 2019. 16(8): p. 695-710.
- Das, S. , et al., A Selective Medium for Isolation and Detection of Candida auris, an Emerging Pathogen. J Clin Microbiol, 2021. 59(2).
- Ibrahim, A. , et al., SCA Medium: A New Culture Medium for the Isolation of All Candida auris Clades. J Fungi (Basel), 2021. 7(6).
- Kumar, A. , et al., Simple low cost differentiation of Candida auris from Candida haemulonii complex using CHROMagar Candida medium supplemented with Pal's medium. Rev Iberoam Micol, 2017. 34(2): p. 109-111.
- Mulet Bayona, J.V. , et al., Evaluation of a novel chromogenic medium for Candida spp. identification and comparison with CHROMagar™ Candida for the detection of Candida auris in surveillance samples. Diagn Microbiol Infect Dis, 2020. 98(4): p. 115168.
- Borman, A.M., M. Fraser, and E.M. Johnson, CHROMagarTM Candida Plus: A novel chromogenic agar that permits the rapid identification of Candida auris. Med Mycol, 2021. 59(3): p. 253-258.
- de Jong, A.W. , et al., Performance of Two Novel Chromogenic Media for the Identification of Multidrug-Resistant Candida auris Compared with Other Commercially Available Formulations. J Clin Microbiol, 2021. 59(4).
- CDC. Algorithm to identify Candida auris based on phenotypic laboratory method and initial species identification. 2019 [cited April 2025. Available online: https://www.cdc.gov/candida-auris/media/pdfs/Testing-algorithm_by-Method_508.pdf.
- Kathuria, S. , et al., Multidrug-Resistant Candida auris Misidentified as Candida haemulonii: Characterization by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and DNA Sequencing and Its Antifungal Susceptibility Profile Variability by Vitek 2, CLSI Broth Microdilution, and Etest Method. J Clin Microbiol, 2015. 53(6): p. 1823-30.
- Wong, R.C. , et al., Current Updates on Molecular Diagnostic Assays Used for Detection of Candida auris: A Systematic Review. Diagnostics (Basel), 2025. 15(2).
- Sexton, D.J. , et al., Evaluation of a new T2 Magnetic Resonance assay for rapid detection of emergent fungal pathogen Candida auris on clinical skin swab samples. Mycoses, 2018. 61(10): p. 786-790.
- Welsh, R.M. , et al., Candida auris Whole-Genome Sequence Benchmark Dataset for Phylogenomic Pipelines. J Fungi (Basel), 2021. 7(3).
- de Groot, T. , et al., Development of Candida auris Short Tandem Repeat Typing and Its Application to a Global Collection of Isolates. mBio, 2020. 11(1).
- Sathi, F.A. , et al., Clonal Diversity of Candida auris, Candida blankii, and Kodamaea ohmeri Isolated from Septicemia and Otomycosis in Bangladesh as Determined by Multilocus Sequence Typing. J Fungi (Basel), 2023. 9(6).
- Vatanshenassan, M. , et al., Evaluation of Microsatellite Typing, ITS Sequencing, AFLP Fingerprinting, MALDI-TOF MS, and Fourier-Transform Infrared Spectroscopy Analysis of Candida auris. J Fungi (Basel), 2020. 6(3).
- Nguyen, M.H. , et al., Performance of Candida real-time polymerase chain reaction, β-D-glucan assay, and blood cultures in the diagnosis of invasive candidiasis. Clin Infect Dis, 2012. 54(9): p. 1240-8.
- Wang, K. , et al., Diagnostic value of Candida mannan antigen and anti-mannan IgG and IgM antibodies for Candida infection. Mycoses, 2020. 63(2): p. 181-188.
- CDC. Screening Recommendations for Healthcare Facilities. 2024 [cited 2025 April]; Available from: https://www.cdc.gov/candida-auris/hcp/screening-hcp/index.html.
- Chowdhary, A. , et al., A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother, 2018. 73(4): p. 891-899.
- Mikulska, M. , et al., The use of mannan antigen and anti-mannan antibodies in the diagnosis of invasive candidiasis: recommendations from the Third European Conference on Infections in Leukemia. Crit Care, 2010. 14(6): p. R222.
- Piedrahita, C.T. , et al., Environmental Surfaces in Healthcare Facilities are a Potential Source for Transmission of Candida auris and Other Candida Species. Infect Control Hosp Epidemiol, 2017. 38(9): p. 1107-1109.
- Welsh, R.M. , et al., Survival, Persistence, and Isolation of the Emerging Multidrug-Resistant Pathogenic Yeast Candida auris on a Plastic Health Care Surface. J Clin Microbiol, 2017. 55(10): p. 2996-3005.
- Biswal, M. , et al., Controlling a possible outbreak of Candida auris infection: lessons learnt from multiple interventions. J Hosp Infect, 2017. 97(4): p. 363-370.
- Tharp, B. , et al., Role of Microbiota in the Skin Colonization of Candida auris. mSphere, 2023. 8(1): p. e0062322.
- Kumar, J. , et al., Environmental Contamination with Candida Species in Multiple Hospitals Including a Tertiary Care Hospital with a Candida auris Outbreak. Pathog Immun, 2019. 4(2): p. 260-270.
- Magnasco, L. , et al., Frequency of Detection of Candida auris Colonization Outside a Highly Endemic Setting: What Is the Optimal Strategy for Screening of Carriage? J Fungi (Basel), 2023. 10(1).
- Chowdhary, A., C. Sharma, and J.F. Meis, Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog, 2017. 13(5): p. e1006290.
- Bergeron, G. , et al., Candida auris Colonization After Discharge to a Community Setting: New York City, 2017-2019. Open Forum Infect Dis, 2021. 8(1): p. ofaa620.
- Schwartz, I.S., S. W. Smith, and T.C. Dingle, Something wicked this way comes: What health care providers need to know about Candida auris. Can Commun Dis Rep, 2018. 44(11): p. 271-276.
- Southwick, K., E. H. Adams, and J. Greenko, 2039. New York State 2016–2018: Progression from Candida auris Colonization to Bloodstream Infection. Open Forum Infectious Diseases, 2018.
- Silva, I., I. M. Miranda, and S. Costa-de-Oliveira, Potential Environmental Reservoirs of Candida auris: A Systematic Review. J Fungi (Basel), 2024. 10(5).
- Zhang, Z. , et al., Risk of invasive candidiasis with prolonged duration of ICU stay: a systematic review and meta-analysis. BMJ Open, 2020. 10(7): p. e036452.
- Eyre, D.W. , Infection prevention and control insights from a decade of pathogen whole-genome sequencing. J Hosp Infect, 2022. 122: p. 180-186.
- Sabino, R. , et al., Candida auris, an Agent of Hospital-Associated Outbreaks: Which Challenging Issues Do We Need to Have in Mind? Microorganisms, 2020. 8(2).
- Sharp, A. , et al., Screening for Candida auris in patients admitted to eight intensive care units in England, 2017 to 2018. Euro Surveill, 2021. 26(8).
- Forsberg, K. , et al., Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med Mycol, 2019. 57(1): p. 1-12.
- Yadav, A. , et al., Colonisation and Transmission Dynamics of Candida auris among Chronic Respiratory Diseases Patients Hospitalised in a Chest Hospital, Delhi, India: A Comparative Analysis of Whole Genome Sequencing and Microsatellite Typing. J Fungi (Basel), 2021. 7(2).
- Tsay, S. , et al., Approach to the Investigation and Management of Patients With Candida auris, an Emerging Multidrug-Resistant Yeast. Clin Infect Dis, 2018. 66(2): p. 306-311.
- Ruiz-Gaitán, A. , et al., An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses, 2018. 61(7): p. 498-505.
- Caceres, D.H. , et al., Candida auris: A Review of Recommendations for Detection and Control in Healthcare Settings. J Fungi (Basel), 2019. 5(4).
- Thomas-Rüddel, D.O. , et al., Risk Factors for Invasive Candida Infection in Critically Ill Patients: A Systematic Review and Meta-analysis. Chest, 2022. 161(2): p. 345-355.
- Southwick, K. , et al., A description of the first Candida auris-colonized individuals in New York State, 2016-2017. Am J Infect Control, 2022. 50(3): p. 358-360.
- Eyre, D.W. , et al., A Candida auris Outbreak and Its Control in an Intensive Care Setting. N Engl J Med, 2018. 379(14): p. 1322-1331.
- Zhu, Y. , et al., Laboratory Analysis of an Outbreak of Candida auris in New York from 2016 to 2018: Impact and Lessons Learned. J Clin Microbiol, 2020. 58(4).
- Proctor, D.M. , et al., Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat Med, 2021. 27(8): p. 1401-1409.
- CDC, Screening: Patient Swab Collection. 2024.
- CDC. Identification of. 2024 [cited April 2025; Available from: https://www.cdc.gov/candida-auris/hcp/laboratories/identification-of-c-auris.html. /.
- Cortegiani, A. , et al., Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J Intensive Care, 2018. 6: p. 69.
- Hata, D.J., R. Humphries, and S.R. Lockhart, Candida auris: An Emerging Yeast Pathogen Posing Distinct Challenges for Laboratory Diagnostics, Treatment, and Infection Prevention. Arch Pathol Lab Med, 2020. 144(1): p. 107-114.
- Osei Sekyere, J. , Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen, 2018. 7(4): p. e00578.
- Bentz, M.L. , et al., Phenotypic switching in newly emerged multidrug-resistant pathogen Candida auris. Med Mycol, 2019. 57(5): p. 636-638.
- Du, H. , et al., Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog, 2020. 16(10): p. e1008921.
- Satoh, K. , et al., Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol, 2009. 53(1): p. 41-4.
- Mahmoudi, S. , et al., Methods for identification of Candida auris, the yeast of global public health concern: A review. J Mycol Med, 2019. 29(2): p. 174-179.
- Zerrouki, H. , et al., Emergence of Candida auris in intensive care units in Algeria. Mycoses 2022, 65, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Gaitán, A. , et al., Usefulness of Chromogenic Media with Fluconazole Supplementation for Presumptive Identification of Candida auris. Diagnostics (Basel), 2023. 13(2).
- Komorowski, A.S. , et al., Verification, Analytical Sensitivity, Cost-effectiveness, and Comparison of 4 Candida auris Screening Methods. Open Forum Infect Dis, 2024. 11(6): p. ofae017.
- Marathe, A. , et al., Utility of CHROMagar™ Candida Plus for presumptive identification of Candida auris from surveillance samples. Mycopathologia, 2022. 187(5-6): p. 527-534.
- Tamura, T., M. M. Alshahni, and K. Makimura, Evaluation of CHROMagar™ Candida Plus chromogenic agar for the presumptive identification of Candida auris. Microbiol Immunol, 2022. 66(6): p. 292-298.
- Keighley, C. , et al., Candida auris: Diagnostic Challenges and Emerging Opportunities for the Clinical Microbiology Laboratory. Curr Fungal Infect Rep, 2021. 15(3): p. 116-126.
- Snayd, M. , et al., Misidentification of Candida auris by RapID Yeast Plus, a Commercial, Biochemical Enzyme-Based Manual Rapid Identification System, in J Clin Microbiol. 2018: United States.
- Mizusawa, M. , et al., Can Multidrug-Resistant Candida auris Be Reliably Identified in Clinical Microbiology Laboratories?, in J Clin Microbiol. 2017: United States. p. 638-640.
- Lee, W.G. , et al., First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol, 2011. 49(9): p. 3139-42.
- Ruiz Gaitán, A.C. , et al., Nosocomial fungemia by Candida auris: First four reported cases in continental Europe. Rev Iberoam Micol, 2017. 34(1): p. 23-27.
- Ding, C.H. , et al., The Pitfall of Utilizing a Commercial Biochemical Yeast Identification Kit to Detect Candida auris. Ann Clin Lab Sci, 2019. 49(4): p. 546-549.
- Ambaraghassi, G. , et al., Identification of Candida auris by Use of the Updated Vitek 2 Yeast Identification System, Version 8.01: a Multilaboratory Evaluation Study. J Clin Microbiol, 2019. 57(11).
- Tan, Y.E. , et al., Candida auris in Singapore: Genomic epidemiology, antifungal drug resistance, and identification using the updated 8.01 VITEK(Ⓡ)2 system. Int J Antimicrob Agents, 2019. 54(6): p. 709-715.
- Arnold, R.J. and J.P. Reilly, Fingerprint matching of E. coli strains with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of whole cells using a modified correlation approach. Rapid Commun Mass Spectrom, 1998. 12(10): p. 630-6.
- Marklein, G. , et al., Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J Clin Microbiol, 2009. 47(9): p. 2912-7.
- Bader, O. , et al., Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect, 2011. 17(9): p. 1359-65.
- Vatanshenassan, M. , et al., Candida auris Identification and Rapid Antifungal Susceptibility Testing Against Echinocandins by MALDI-TOF MS. Front Cell Infect Microbiol, 2019. 9: p. 20.
- Xie, T.A. , et al., Accuracy of matrix-assisted LASER desorption ionization-time of flight mass spectrometry for identification of Candida. Biosci Rep, 2019. 39(10).
- Ahmad, A. , et al., A high-throughput and rapid method for accurate identification of emerging multidrug-resistant Candida auris. Mycoses, 2019. 62(6): p. 513-518.
- Sterkel, A. , et al., Viability of Candida auris and Other Candida Species after Various Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) Mass Spectrometry-Based Extraction Protocols, in J Clin Microbiol. 2018: United States.
- Normand, A.C. , et al., Optimization of MALDI-ToF mass spectrometry for yeast identification: a multicenter study. Med Mycol, 2020. 58(5): p. 639-649.
- Ghosh, A.K. , et al., Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin Microbiol Infect, 2015. 21(4): p. 372-8.
- CDC. MicrobeNet. 2024 [cited April 2025; Available from: https://www.cdc.gov/microbenet/php/about/index.html.
- Ceballos-Garzon, A. , et al., Development and Validation of an in-House Library of Colombian Candida auris Strains with MALDI-TOF MS to Improve Yeast Identification. J Fungi (Basel), 2020. 6(2).
- Leach, L., Y. Zhu, and S. Chaturvedi, Development and Validation of a Real-Time PCR Assay for Rapid Detection of Candida auris from Surveillance Samples. J Clin Microbiol, 2018. 56(2).
- Kordalewska, M. , et al., Rapid and Accurate Molecular Identification of the Emerging Multidrug-Resistant Pathogen Candida auris. J Clin Microbiol, 2017. 55(8): p. 2445-2452.
- Hernández Felices, F.J. , et al., Evaluation of Eazyplex® LAMP test for fast Candida auris direct detection of colonized patients. Mycoses, 2024. 67(1): p. e13665.
- Ruiz-Gaitán, A.C. , et al., Molecular identification of Candida auris by PCR amplification of species-specific GPI protein-encoding genes. Int J Med Microbiol, 2018. 308(7): p. 812-818.
- Ibrahim, A. , et al., Development and standardization of a specific real-time PCR assay for the rapid detection of Candida auris. Eur J Clin Microbiol Infect Dis, 2021. 40(7): p. 1547-1551.
- Alvarado, M. , et al., Identification of Candida auris and related species by multiplex PCR based on unique GPI protein-encoding genes. Mycoses, 2021. 64(2): p. 194-202.
- CDC. Real-Time PCR Based Identification. 2024 [cited April 2025; Available from: https://www.cdc.gov/candida-auris/hcp/laboratories/real-time-pcr-identification.html.
- Berlau, A. , et al., Evaluation of the Eazyplex(®)Candida ID LAMP Assay for the Rapid Diagnosis of Positive Blood Cultures. Diagnostics (Basel), 2024. 14(19).
- Lin, Z.Z. , et al., A Pooled Analysis of the PCR for the Detection of Candida Auris. Clin Lab, 2022. 68(7).
- Caméléna, F. , et al., Multicenter Evaluation of the FilmArray Blood Culture Identification 2 Panel for Pathogen Detection in Bloodstream Infections. Microbiol Spectr, 2023. 11(1): p. e0254722.
- Zhang, S.X. , et al., Multicenter Evaluation of a PCR-Based Digital Microfluidics and Electrochemical Detection System for the Rapid Identification of 15 Fungal Pathogens Directly from Positive Blood Cultures. J Clin Microbiol, 2020. 58(5).
- Franco, L.C. , et al., Validation of a qualitative real-time PCR assay for the detection of Candida auris in hospital inpatient screening. J Clin Microbiol, 2024. 62(6): p. e0015824.
- Ramírez, J.D. , et al., Molecular Detection of Candida auris Using DiaSorin Molecular Simplexa(®) Detection Kit: A Diagnostic Performance Evaluation. J Fungi (Basel), 2023. 9(8).
- Rosa, R. , et al., Impact of In-house Candida auris Polymerase Chain Reaction Screening on Admission on the Incidence Rates of Surveillance and Blood Cultures With and Associated Cost Savings. Open Forum Infect Dis, 2023. 10(11): p. ofad567.
- Sattler, J. , et al., Comparison of Two Commercially Available qPCR Kits for the Detection of Candida auris. J Fungi (Basel), 2021. 7(2).
- Mulet Bayona, J.V. , et al., Validation and implementation of a commercial real-time PCR assay for direct detection of Candida auris from surveillance samples. Mycoses, 2021. 64(6): p. 612-615.
- Yamamoto, M. , et al., Rapid Detection of Candida auris Based on Loop-Mediated Isothermal Amplification (LAMP), in J Clin Microbiol. 2018: United States.
- Lim, D.H. , et al., Development of a Simple DNA Extraction Method and Candida Pan Loop-Mediated Isothermal Amplification Assay for Diagnosis of Candidemia. Pathogens, 2022. 11(2).
- Narayanan, A. , et al., ClaID: a Rapid Method of Clade-Level Identification of the Multidrug Resistant Human Fungal Pathogen Candida auris. Microbiol Spectr, 2022. 10(2): p. e0063422.
- Lockhart, S.R. , et al., Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin Infect Dis, 2017. 64(2): p. 134-140.
- CDC. GITHUB. 2021 [cited April 2025; Available from: https://github.com/CDCgov/mycosnp.
- de Groot, T. , et al., Optimization and Validation of Candida auris Short Tandem Repeat Analysis. Microbiol Spectr, 2022. 10(5): p. e0264522.
- Magobo, R. , et al., Multilocus sequence typing of azole-resistant Candida auris strains, South Africa. S Afr J Infect Dis, 2020. 35(1): p. 116.
- Biswas, C. , et al., Genetic Heterogeneity of Australian Candida auris Isolates: Insights From a Nonoutbreak Setting Using Whole-Genome Sequencing. Open Forum Infect Dis, 2020. 7(5): p. ofaa158.
- CDC. Antifungal Susceptibility Testing for. 2024 [cited April 2025; Available from: https://www.cdc.gov/candida-auris/hcp/laboratories/antifungal-susceptibility-testing.html.
- Jones, C.R. , et al., The laboratory investigation, management, and infection prevention and control of Candida auris: a narrative review to inform the 2024 national guidance update in England. J Med Microbiol, 2024. 73(5).
- Arendrup, M.C. , et al., Comparison of EUCAST and CLSI Reference Microdilution MICs of Eight Antifungal Compounds for Candida auris and Associated Tentative Epidemiological Cutoff Values. Antimicrob Agents Chemother, 2017. 61(6).
- Szekely, A., A. M. Borman, and E.M. Johnson, Candida auris Isolates of the Southern Asian and South African Lineages Exhibit Different Phenotypic and Antifungal Susceptibility Profiles In Vitro. J Clin Microbiol, 2019. 57(5).
- Kordalewska, M. , et al., Understanding Echinocandin Resistance in the Emerging Pathogen Candida auris. Antimicrob Agents Chemother, 2018. 62(6).
- Ceballos-Garzon, A. , et al., Head-to-head comparison of CLSI, EUCAST, Etest and VITEK®2 results for Candida auris susceptibility testing. Int J Antimicrob Agents, 2022. 59(4): p. 106558.
- Siopi, M. , et al., Evaluation of the Vitek 2 system for antifungal susceptibility testing of Candida auris using a representative international panel of clinical isolates: overestimation of amphotericin B resistance and underestimation of fluconazole resistance. J Clin Microbiol, 2024. 62(4): p. e0152823.
- Patwardhan, S.A. , et al., Candida auris - Comparison of sensititre YeastOne and Vitek 2 AST systems for antifungal susceptibility testing - A single centre experience. Indian J Med Microbiol, 2024. 50: p. 100618.
- Asadzadeh, M. , et al., Evaluation of Etest and MICRONAUT-AM Assay for Antifungal Susceptibility Testing of Candida auris: Underestimation of Fluconazole Resistance by MICRONAUT-AM and Overestimation of Amphotericin B Resistance by Etest. Antibiotics (Basel), 2024. 13(9).
- Frías-De-León, M.G. , et al., Antifungal Resistance in Candida auris: Molecular Determinants. Antibiotics (Basel), 2020. 9(9).
- Kordalewska, M. and D.S. Perlin, Molecular Diagnostics in the Times of Surveillance for Candida auris. J Fungi (Basel), 2019. 5(3).
- Marinach, C. , et al., MALDI-TOF MS-based drug susceptibility testing of pathogens: the example of Candida albicans and fluconazole. Proteomics, 2009. 9(20): p. 4627-31.
- Vella, A. , et al., Potential Use of MALDI-ToF Mass Spectrometry for Rapid Detection of Antifungal Resistance in the Human Pathogen Candida glabrata. Sci Rep, 2017. 7(1): p. 9099.
- Theparee, T., S. Das, and R.B. Thomson, Jr., Total Laboratory Automation and Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Improve Turnaround Times in the Clinical Microbiology Laboratory: a Retrospective Analysis. J Clin Microbiol, 2018. 56(1).
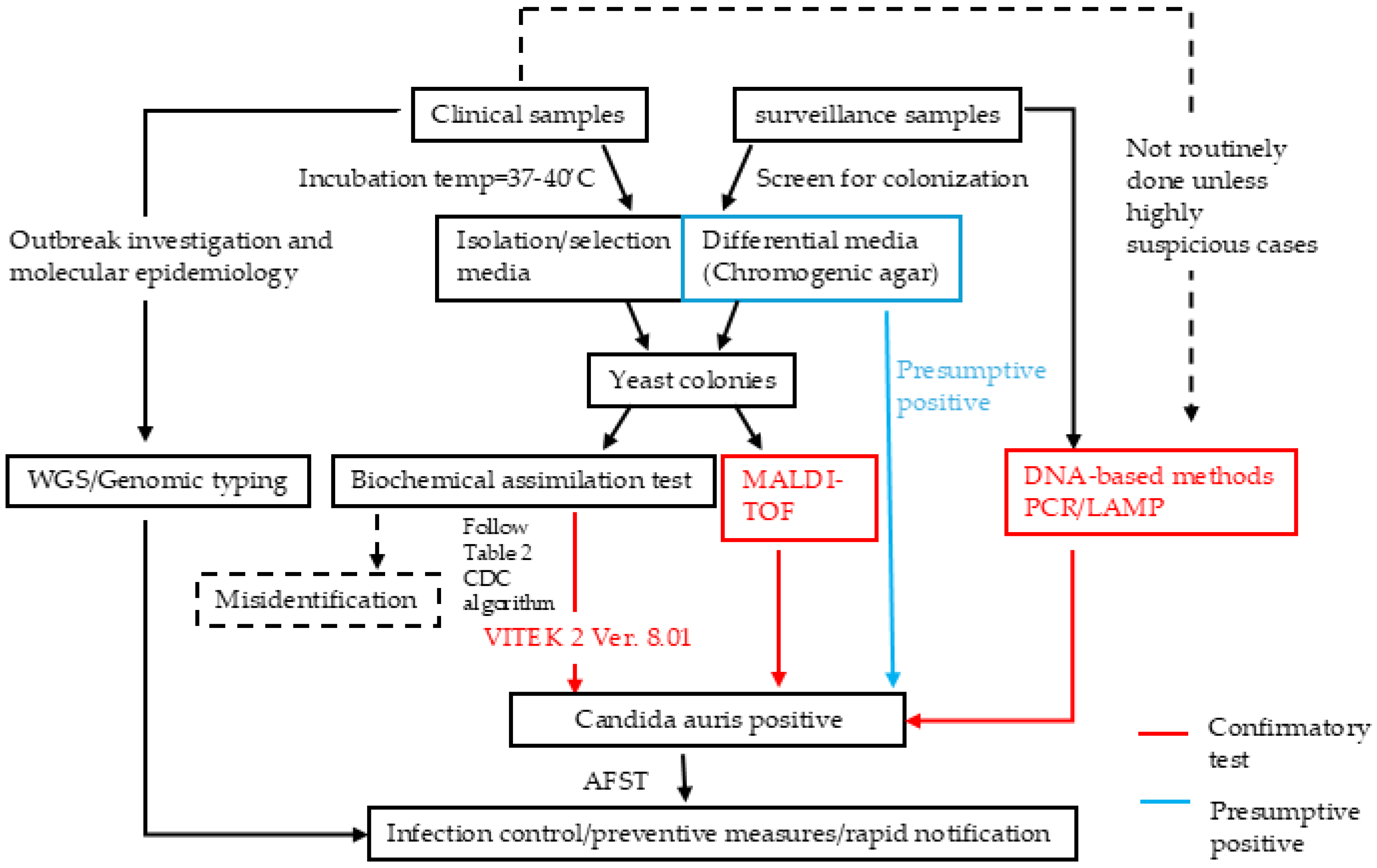
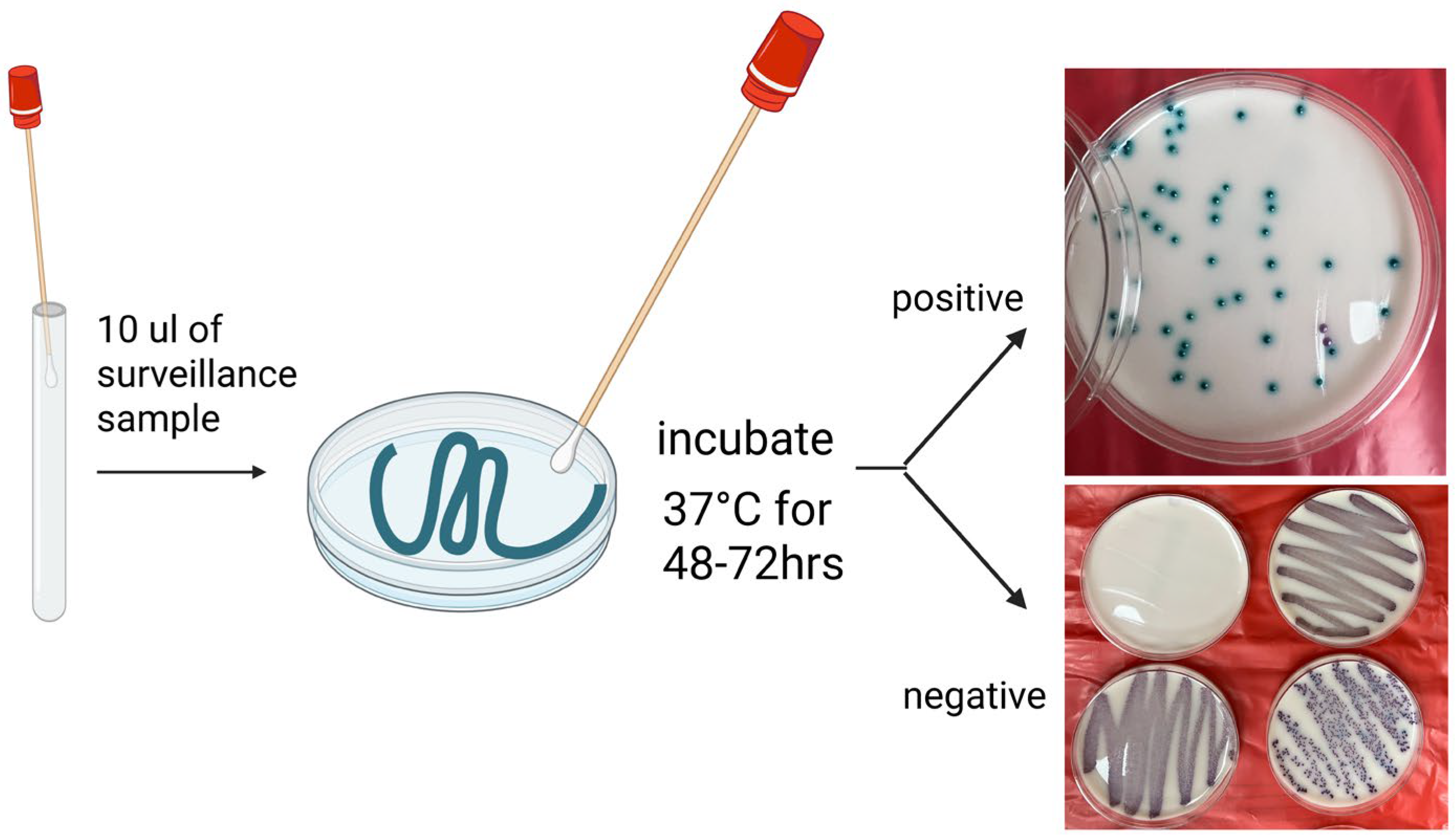
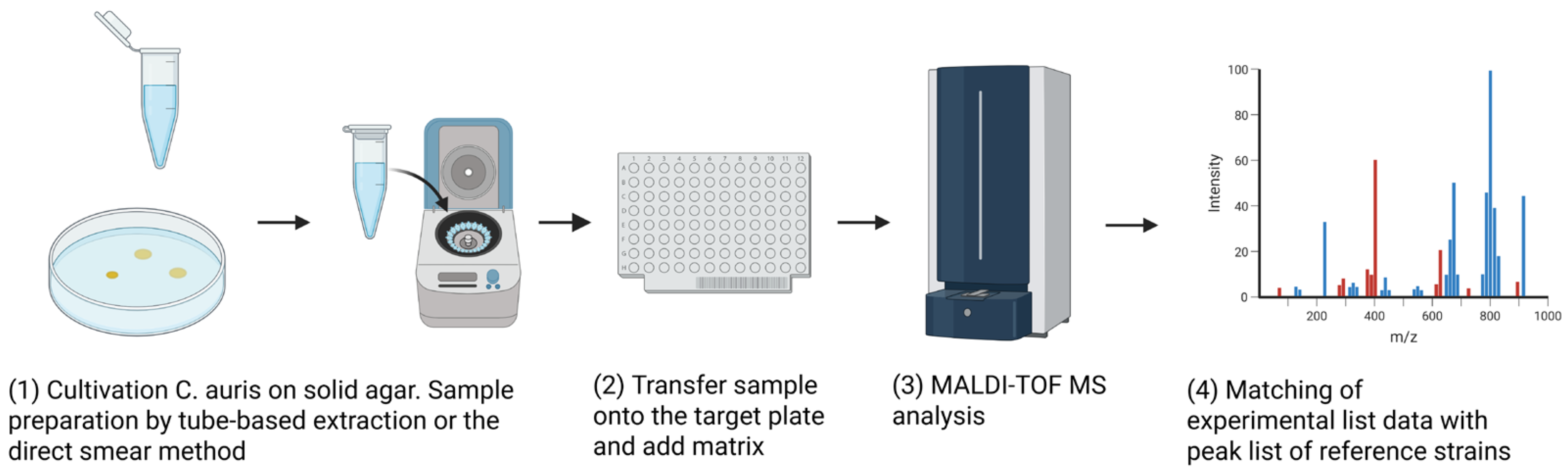
| Category | Diagnostic Method | Mechanism | Highlights | Strengths | Weaknesses/Limitations | References |
|---|---|---|---|---|---|---|
| Culture-Based | Mycological media/ Selective media/ Chromogenic media | 1. Requires higher incubation temperature(37-40'C) than routine culture. 2. Add saline, carbohydrates, or color indicators to differentiate from similar species |
1. SCA/SAM Media Se/Sp:100%/100% 2. CHROMagarTM with Pal’s medium Se/Sp: 100%/100% 3. CHROMagar™ Candida Plus Se/Sp: 90-100%/98-100% |
1. basis for culture-based methods (eg. Biochemical assimilation methods and MALDI-TOF) 2. Simple, widely available, low-cost 3. Gold-standard for diagnosis in clinical samples. Enables AFST. |
1. Slow (48-72 hrs) 2. Various accuracy among different media. Non-specific colony morphology, prone to misidentification, requires additional confirmation (MALDI-TOF MS or molecular methods) |
[15,16,17,18,19,20] |
| Culture-Based | Biochemical Tests (VITEK, API, Microscan) | Analysis of metabolic profile through carbohydrate assimilation, nitrogen utilization, and enzymatic activity | 1. Vitek 2 Version. 8.01 is confirmatory 2. API 20C AUX or API ID 32C leads to frequent misidentification 3. MicroScan/ BD Pheonix has no C. auris database |
1. Low cost and easy to use (automated) 2. A rapid AFST tool reliable for azoles |
1. Requires database updates 2. Various misidentification rates among different commercial assays. Often requires confirmatory test following CDC algorithm (Table 2) |
[21] |
| Culture-Based | MALDI-TOF MS | Analysis of protein profiles and comparing them to reference databases. (Figure 4) | 1. Accuracy depends on databases, sample preparation, and instrument calibration 2. Limited use for AFST with no clinical breakpoint of MPCC |
1. Rapid (4-5 hrs post culture) 2. Highly specific(Sp>90%) when database is updated 3. Cost-effective |
1. Requires database updates, 2. Expensive equipment, limited access in resource-poor settings |
Kathuria [22] et al., 2015 Kwon et al., 2019 |
| Culture-Independent | DNA-Based Assays (PCR/LAMP) | DNA amplification using species-specific primers (e.g., ITS, D1/D2 regions) | 1. Both LDTs and Commercial assays demonstrate reliable accuracy(Se/ Sp>90%), LAMP has lower sensitivity 2. Current FDA-approved Commercial assays for blood culture: GenMark ePlex BCID-FP and BioFire FilmArray BCID2 |
1. Accurate and rapid, useful for colonization screening and outbreak control in healthcare settings | 1. Cannot determine antifungal susceptibility and detect DNA from both viable and non-viable C. auris cells 2. LAMP has lower sensitivity |
[23] |
| Culture-Independent | T2 MR assay | Superparamagnetic nanoparticles bind target DNA/RNA, altering T2 relaxation for MR detection | Rapid (<3 hours) and highly sensitive detection for surveillance and bloodstream infection | Research use only | [24] | |
| Culture-Independent | Whole-Genome Sequencing (WGS)/ genomic typing | High-resolution genetic analysis | Tracking outbreaks, identifying strains, and analyzing resistance, virulence, and epidemiology. Not usually used for individual diagnosis. | WGS: High resolution, gold standard for outbreaks investigation | Costly, requires specialized analysis | [25,26,27,28] |
| STR typing: High reproducibility, aligns well with WGS | Differentiate C. auris strains only if >30 SNP differences | |||||
| MLST: Differentiates C. auris, supports resistance surveillance | Low resolution within clades,limited for outbreaks | |||||
| AFLP: Rapid, cost-effectiveness | Poor reproducibility, inconsistent clustering |
|||||
| Culture-Independent | Beta-D-glucan (BDG) assays | Detection of fungal cell wall components | Non-invasive, useful for early detection for clinical samples | Limited sensitivity and low specificity for invasive candidiasis | [29,30] |
| Identification Method | Database/Software, if applicable | is confirmed if initial identification is C auris. | is possible if the following initial identifications are given. Further work-up is needed to determine if the isolate is . |
|---|---|---|---|
| Bruker Biotyper MALDI-TOF | RUO libraries (Versions 2014 [5627] and more recent) | n/a | |
| CA System library (Version Claim 4) | n/a | ||
| bioMérieux VITEK MS MALDI- TOF | RUO library (withSaccharomycetaceae update) | n/a | |
| IVD library (v3.2) | n/a | ||
| Older IVD libraries | n/a |
C. haemulonii C. lusitaniae No identification |
|
| VITEK 2 YST | Software version 8.01* |
C. haemulonii C. duobushaemulonii Candida spp. not identified |
|
| Older versions | n/a |
C. haemulonii C. duobushaemulonii Candida spp. not identified |
|
| API 20C | n/a |
Rhodotorula glutinis (without characteristic red color) C. sake Candida spp. not identified |
|
| API ID 32C | n/a |
C.intermedia C.sake Saccharomyces kluyveri |
|
| BD Phoenix | n/a |
C. catenulata C. haemulonii Candida spp. not identified |
|
| MicroScan | n/a |
C. lusitaniae** C. guilliermondii** C. parapsilosis** C. famata Candida spp. not identified |
|
| RapID Yeast Plus | n/a |
C. parapsilosis** Candida spp. not identified |
|
| GenMark ePlex BCID-FP Panel | n/a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




