Submitted:
13 May 2025
Posted:
15 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
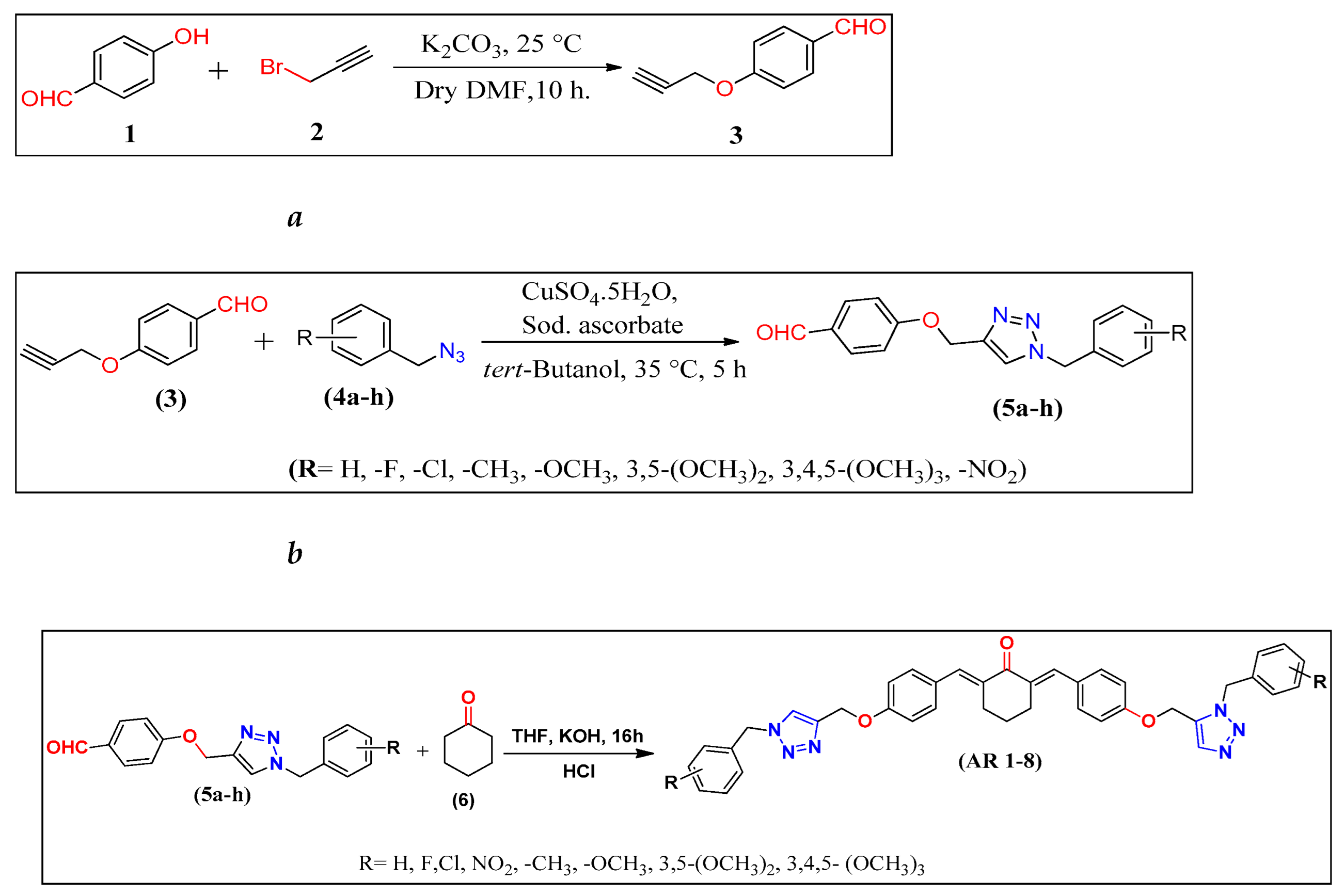
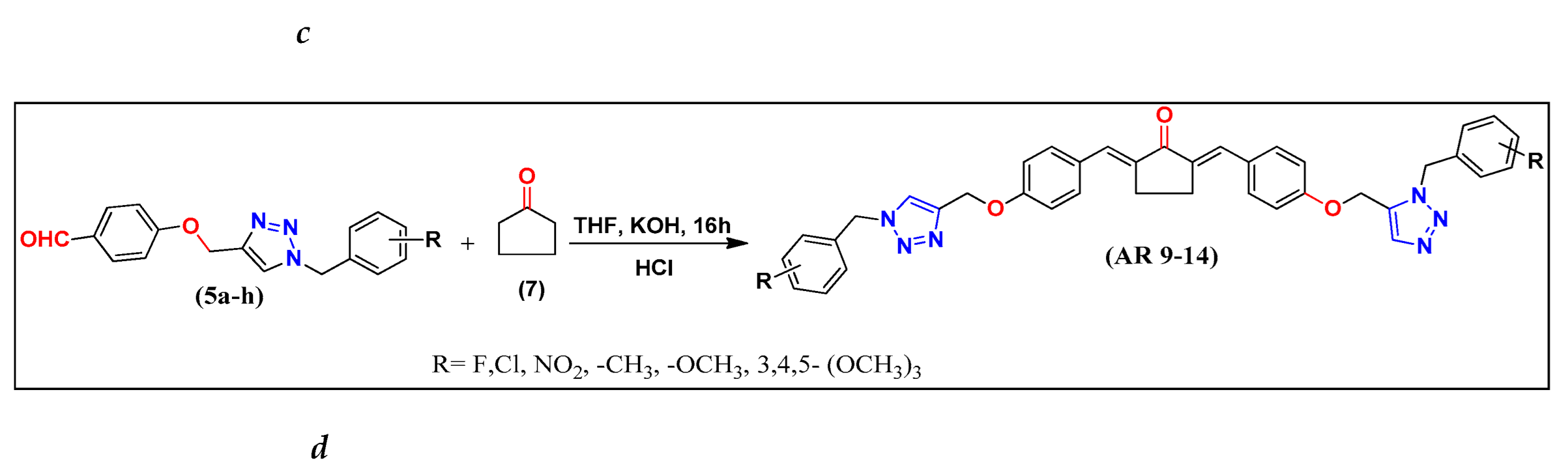
2.1. Chemistry
2.2. Biology
2.2.1. Cytotoxic Study
2.2.2. In-Silico Study
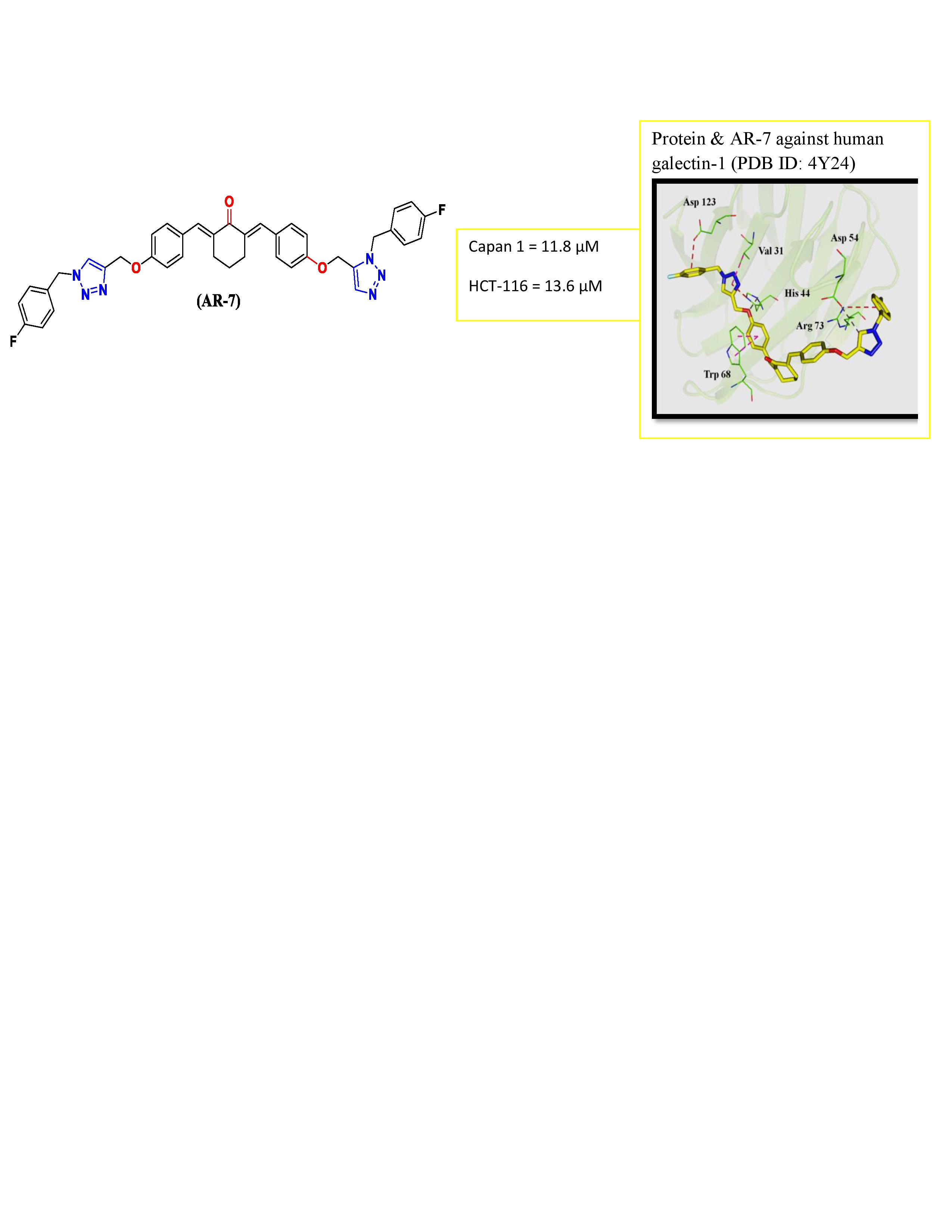
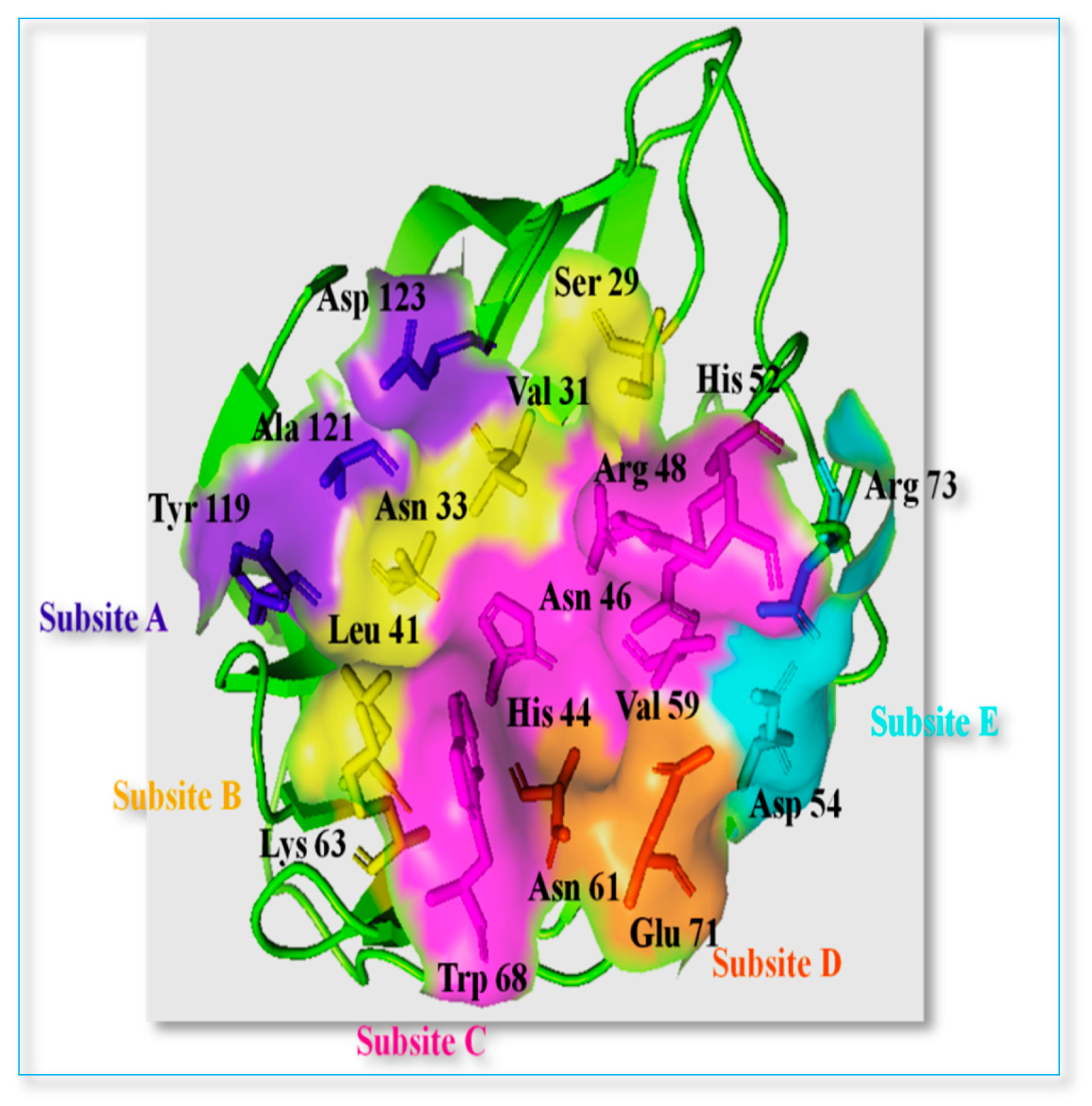
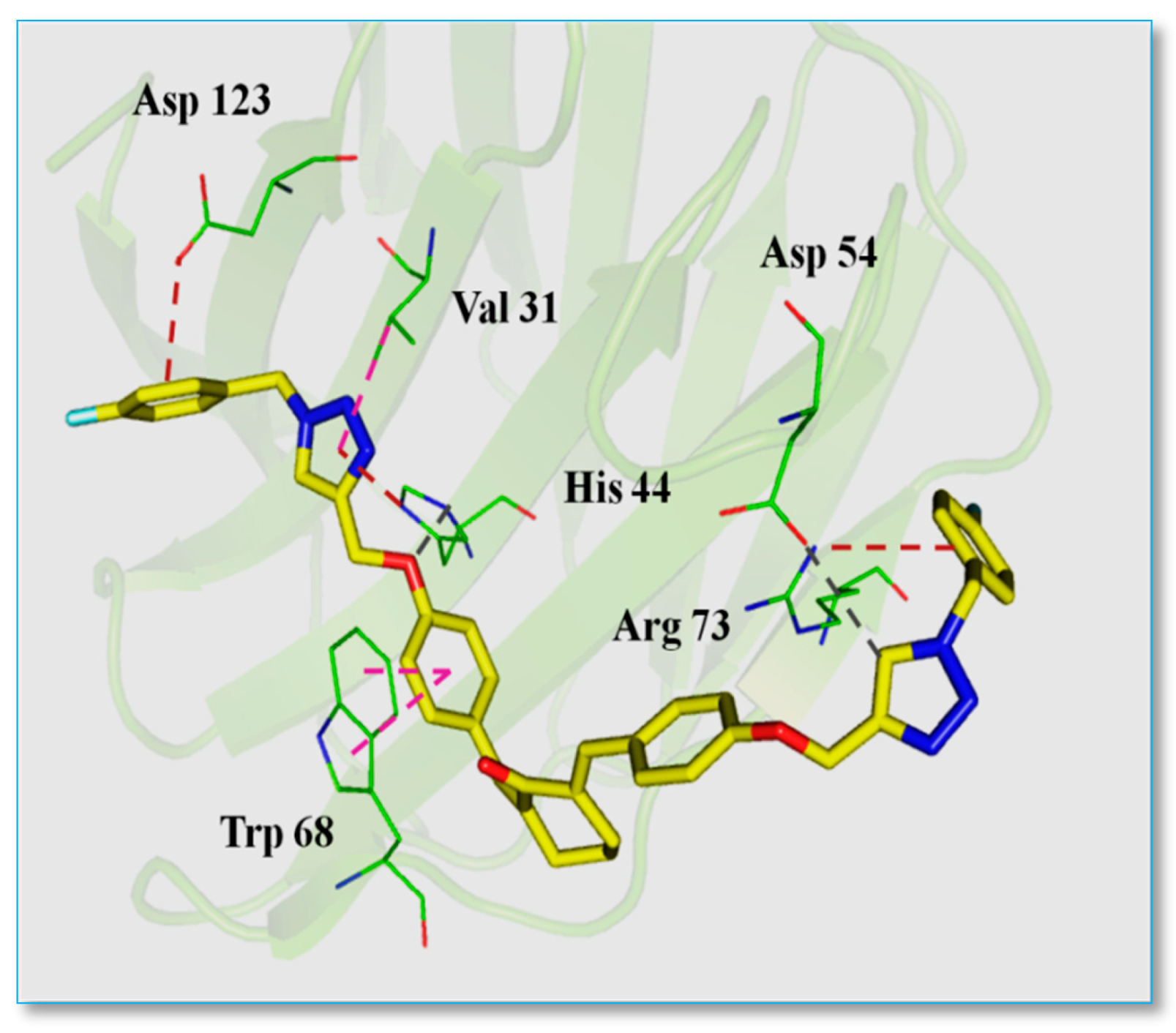
- The 4-fluorophenyl ring of AR-7 formed an electrostatic interaction with Asp123 in sub-site A.
- The triazole ring exhibited a hydrophobic π-alkyl interaction with Val31 in sub-site B, Trp68 in sub-site C and an electrostatic interaction with His44 in sub-site C.
- The oxygen linker between the triazole and benzene rings participated in hydrogenbonding with His44.
- Another triazole ring formed hydrogen bonding with Asp54 in sub-site A.
- Finally, a fluorophenyl ring at the terminal end of the molecule established an electrostatic π-cation interaction with Arg73 in sub-site A (Figure 3).

3. Materials and Methods
3.1. Chemistry
3.1.1. General Information and Instrumentation
3.1.2. Synthesis of (2E,6E)-2,6-bis(4-((1-benzyl-1H-1,2,3-triazol-4-yl)methoxy)benzylidene) Cyclic Ketones (AR 1-14).
3.2. Bio-Evaluation
3.2.1. Cancer Cell Lines
3.2.2. Cytotoxicity Assays
3.2.3. In-Silico Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADT | AutoDock Tools |
| ATCC | American Type Culture Collection |
| CBS | Carbohydrate-Binding Sub-sites |
| CRD | Carbohydrate Recognition Domain |
| CuAAC | Copper(I)-catalyzed Azide-Alkyne Cycloaddition |
| DMF | Dimethyl Formamide |
| DMSO | Dimethyl Sulfoxide |
| DSMZ | Deutsche Sammlung von Mikroorganismen und Zellkulturen |
| FTIR | Fourier-Transform Infrared |
| M.p | Melting Point |
| NK | Natural Killer |
| NMR | Nuclear Magnetic Resonance |
| OD | Optical Density |
| PDB | Protein Data Bank |
| ppm | Parts Per Million |
| RMS | Root Mean Square |
| THF | Tetrahydrofuran |
| TLC | Thin-Layer Chromatography |
| TMS | Tetramethylsilane |
References
- Sathishkumar, K.; Chaturvedi, M.; Das, P.; Stephen, S.; Mathur, P. Cancer incidence estimates for 2022 & projection for 2025: Result from National Cancer Registry Programme, India. Indian J Med Res 2022, 156, 598–607. [Google Scholar] [PubMed]
- Kumar, A.; Singh, A.K.; Singh, H.; Vijayan, V.; Kumar, D.; Naik, J.; Thareja, S.; Yadav, J.P.; Pathak, P.; Grishina, M.; Verma, A.; Khalilullah, H.; Jaremko, M.; Emwas, A.H.; Kumar, P. Nitrogen containing heterocycles as anticancer Agents: a medicinal chemistry perspective. Pharmaceuticals(Basel) 2023, 14, 299. [Google Scholar] [CrossRef] [PubMed]
- Disha, P.V.; Ruturajsinh, M.V.; Hitendra, M.P. Versatile synthetic platform for 1,2,3-triazole chemistry. ACS Omega 2022, 7, 36945–36987. [Google Scholar]
- Malik, M.S.; Ahmed, S.A.; Althagafi, I.I.; Ansari, M.A.; Kamal, A. Application of triazoles as bioisosteres and linkers in the development of microtubule targeting agents. RSC Med Chem 2020, 11, 327–348. [Google Scholar] [CrossRef] [PubMed]
- Johannes, L.; Jacob, R.; Leffler, H. Galectins at a glance. J Cell Sci 2018, 131, jcs208884. [Google Scholar] [CrossRef] [PubMed]
- Camby, I.; Le Mercier, M.; Lefranc, F.; Kiss, R. Galectin-1: a small protein with major functions. Glycobiology 2006, 16, 137R–157R. [Google Scholar] [CrossRef] [PubMed]
- Cedeno-Laurent, F.; Dimitroff, C.J. Galectin-1 research in T cell immunity: past, present and future. ClinImmunol 2012, 142, 107–16. [Google Scholar] [CrossRef] [PubMed]
- Rabinovich, G. Galectin-1 as a potential cancer target. Br J Cancer 2005, 92, 1188–1192. [Google Scholar] [CrossRef] [PubMed]
- Carlos, P.M.P; Juan, B.C.; Santiago, D.L.; Marcelo, A.M. The structural biology of Galectin-ligand recognition: current advances in modeling tools, protein engineering, and inhibitor design. Front Chem 2019, 7, 1–14. [Google Scholar]
- Sridhar, G.N. Non-carbohydrate galectin-1 inhibitors as promising anticancer agents: design strategies, structure activity relationship and mechanistic insights. Eur J MedChemRep 2024, 11, 100170. [Google Scholar]
- Cousin, J.M.; Cloninger, M.J. The role of galectin-1 in cancer progression, and synthetic multivalent systems for the study of Galectin-1. Int J MolSci 2016, 17, 1566. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, H.C.; Zhao, J.; Wu, M.H.; Shih, T.C. Immunosuppressive roles of galectin-1 in the tumor microenvironment. Biomolecules 2021, 11, 1398. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Greco, G.; Kumar, S.; Catanzaro, E.; Morigi, R.; Locatelli, A.; Schols, D.; Alici, H.; Tahtaci, H.; Ravindran, F.; Fimognari, C.; Karki, S.S. Synthesis, in-vitro cytotoxicity, molecular docking and ADME study of some indolin-2-one linked 1,2,3-triazole derivatives. ComputBiol and Chem 2022, 97, 107641. [Google Scholar] [CrossRef] [PubMed]
- Shaik, P.S.; Nayak, V.L.; Sultana, F.; SubbaRao, A.V.; Shaik, A.B.; Korrapati, S.B.; Ahmed, K. Design and synthesis of imidazo [2,1-b]thiazole linked triazole conjugates: microtubule-destabilizing agents. Eur J Med Chem 2017, 126, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Walle, T.V.; Theppawong, A.; Grootaert, C.; Jonghe, S.D.; Persoons, L.; Daelemans, D.; Hecke, K.V.; Camp, J.V.; D’hooghe, M. Synthesis and cytotoxic evaluation of monocarbonylcurcuminoids and their pyrazoline derivatives. MonatshChem 2019, 150, 2045–2051. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J ComputChem 2010, 31, 455–461. [Google Scholar]
| Compound | IC50 | ||||||||
| R | hTERT RPE-1 | Capan-1 | Hap-1 | HCT-116 | NCI-H460 | DND-41 | K-562 | Z-138 | |
| retina (non cancerous) | pancreatic adenocarcinoma | chronic myeloid leukemia | colorectal carcinoma | lung carcinoma | acute lymphoblastic leukemia | chronic myeloid leukemia | non-Hodgkin lymphoma | ||
| AR-4 | 3,4,5-tri-OCH3 | >100 | 78.2 | 89.7 | 39.9 | >100 | >100 | >100 | >100 |
| AR-5 | 4-NO2 | >100 | 58.3 | >100 | 47.3 | 24.7 | >100 | >100 | >100 |
| AR-6 | 4-CH3 | 30.9 | 31.5 | 38.2 | 27.7 | 21.2 | 24.4 | >100 | >100 |
| AR-7 | 4-F | 38.1 | 11.8 | 31.6 | 13.6 | 21.9 | >100 | >100 | >100 |
| AR-8 | 4-Cl | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| AR-9 | OCH3 | 73.8 | >100 | 84.3 | 17.4 | 76.7 | >100 | >100 | >100 |
| AR-10 | 3,4,5-tri-OCH3 | 48.5 | 31.5 | 45.6 | 80.3 | 40.5 | 68.5 | >100 | >100 |
| AR-11 | 4-CH3 | 40.3 | 25.6 | 42.5 | 29.2 | 20.8 | 37.1 | >100 | >100 |
| AR-12 | 4-NO2 | 53.8 | 20.1 | 43.1 | 20.5 | 47.4 | >100 | >100 | >100 |
| AR-13 | 4-Cl | 42.9 | 47.2 | 51.2 | 48.3 | 53.6 | >100 | >100 | >100 |
| AR-14 | 4-F | 51.8 | 50 | >100 | 67 | 58.1 | >100 | >100 | 40.8 |
| Standard | Docetaxel (nM) | 13.5 | 6.3 | 1.6 | 0.8 | 0.1 | 1.9 | 3.4 | 1.9 |
| Standard | Staurosporine(nM) | 0.4 | 4.6 | 0.3 | 0.3 | 3.2 | 6.4 | 29.8 | 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





