Submitted:
09 May 2025
Posted:
12 May 2025
You are already at the latest version
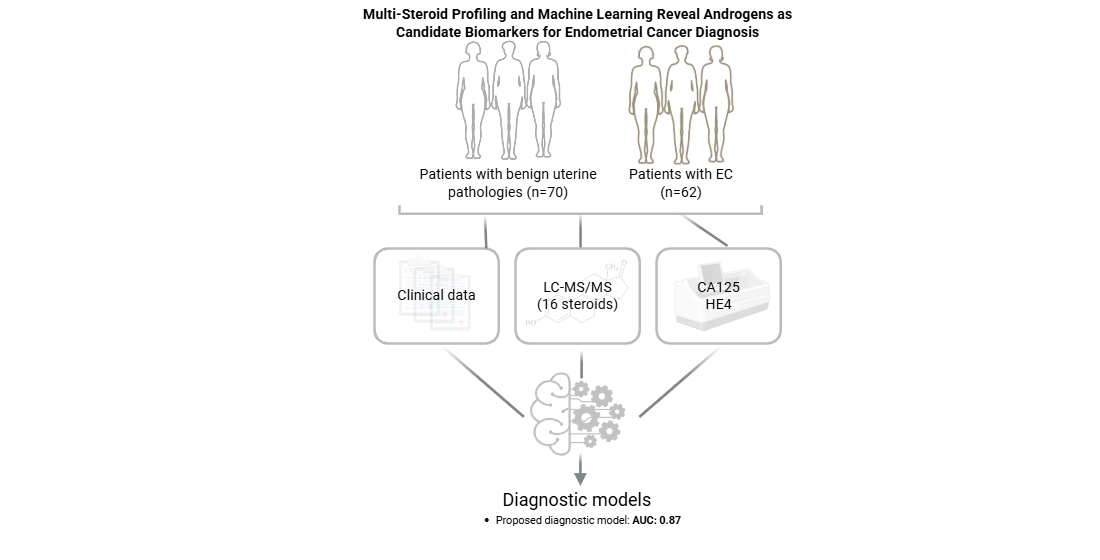
Abstract
Keywords:
1. Introduction
2. Materials and Methods
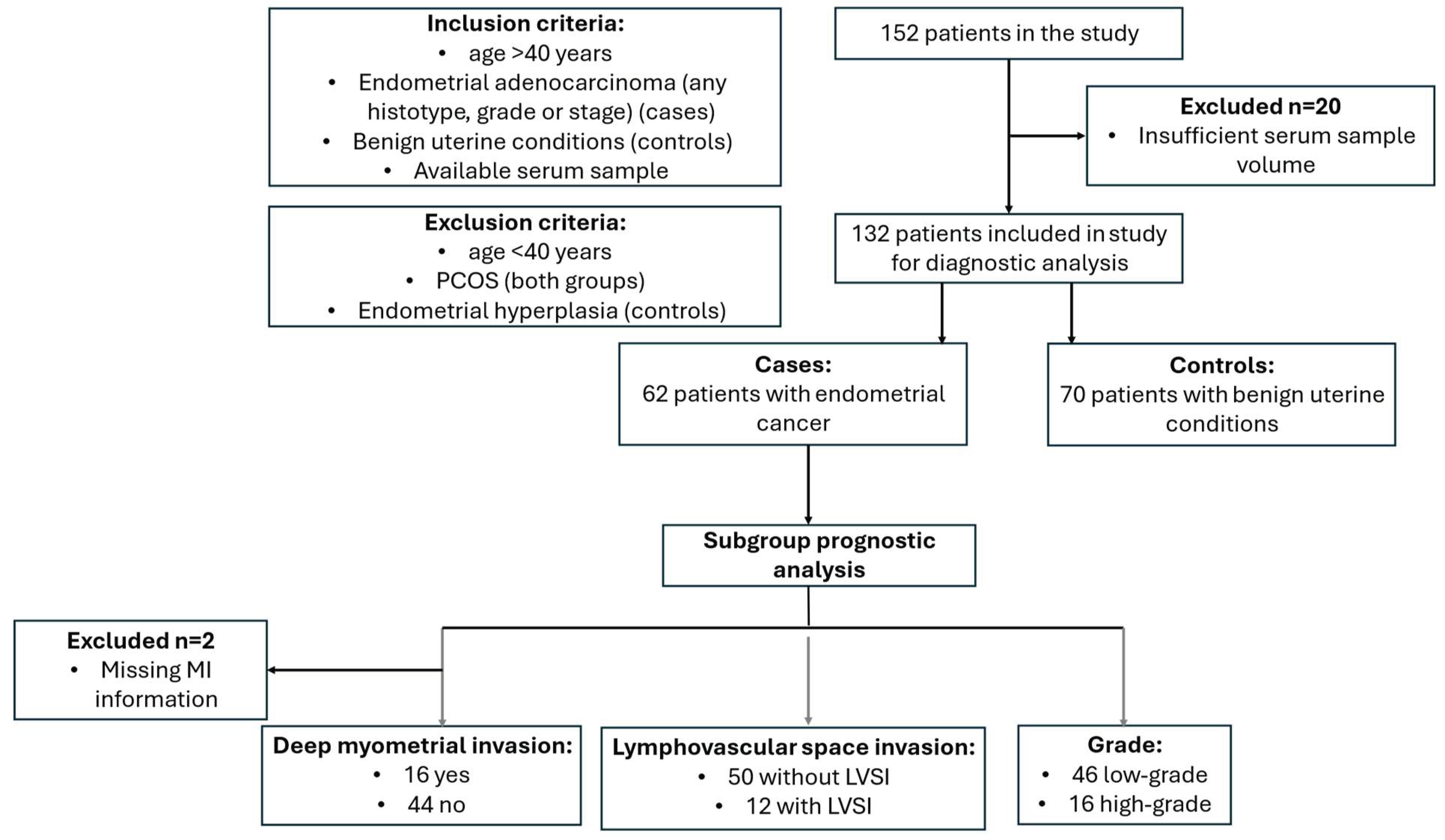
2.1. Study Population
2.2. Multi-Steroid Profiling of Serum Samples by LC-MS/MS
2.2.1. Sample Preparation
2.2.2. LC-MS/MS Analysis
2.3. Measurement of Serum CA125 and HE4 Levels
2.4. Statistical Analysis
3. Results
3.1. Description of the Cohort
3.2. Preoperative Steroid Hormone Levels Differ Between Patients with EC and Women with Benign Uterine Conditions
3.3. Preoperative 11-Oxyandrogen Levels Differ Between Tumor Grades
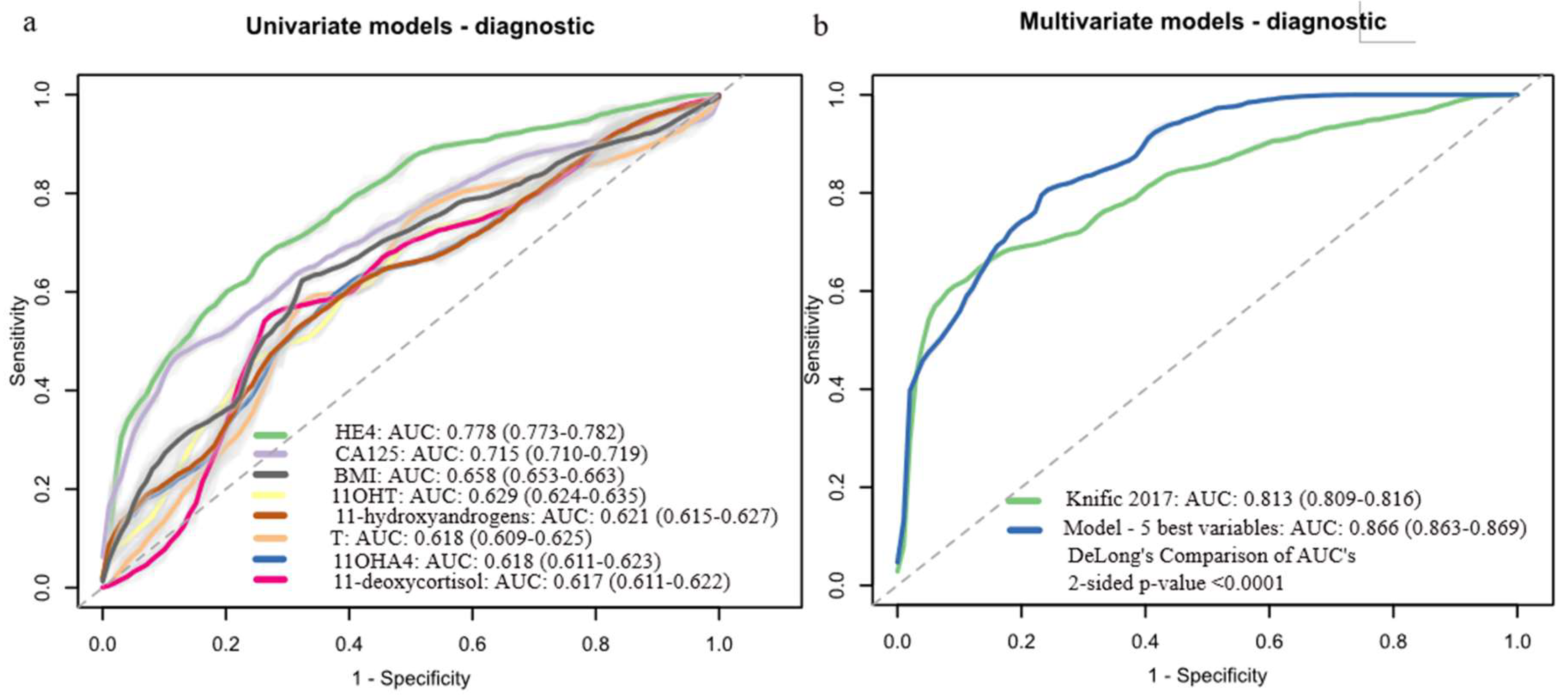
3.4. Development of Machine Learning Diagnostic Models Based on Preoperative Serum Steroid Levels
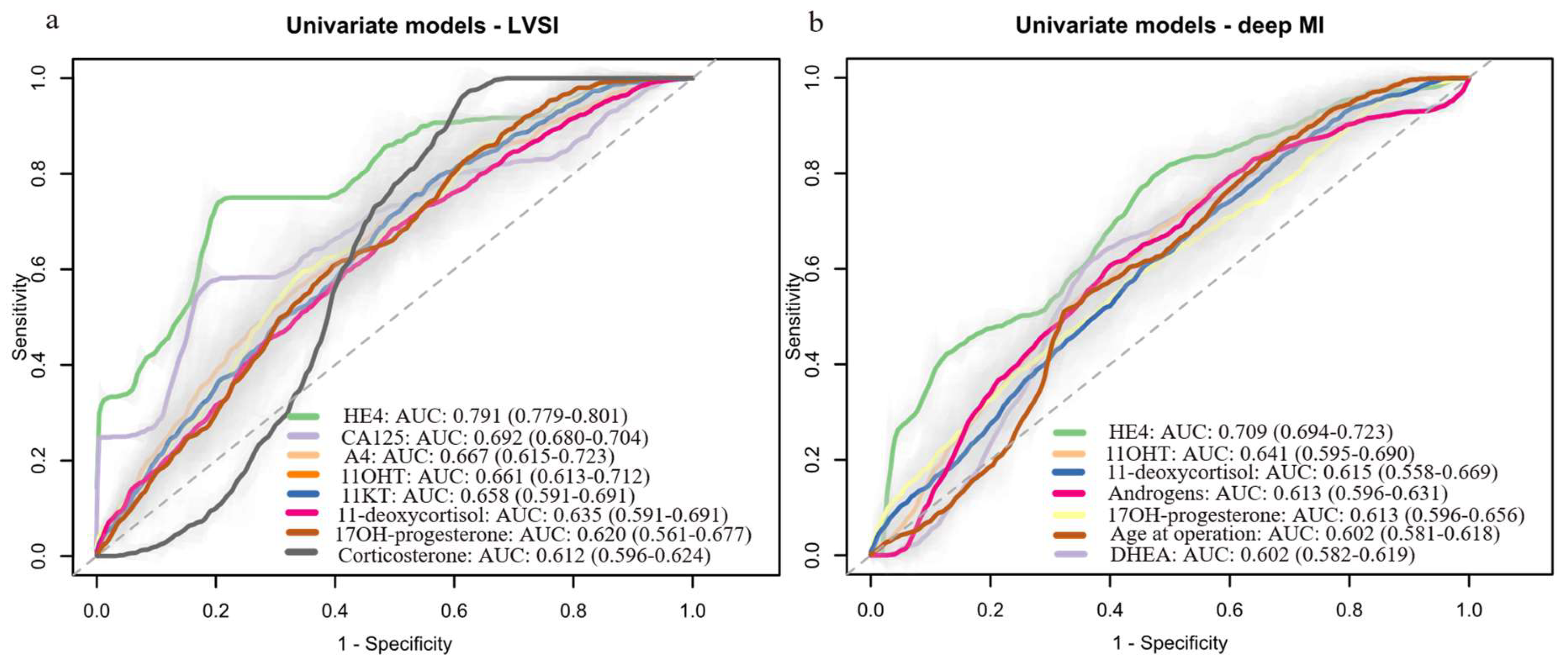
3.5. Development of Machine Learning Prognostic Models Based on Preoperative Serum Steroid Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 11KA4 | 11-keto-androstenedione |
| 11OHA4 | 11β-hydroxy-androstenedione |
| 11OHT | 11β-hydroxytestosterone |
| A4 | Androstenedione |
| AUC | Area under the receiver operating curve |
| BMI | Body mass index |
| CA125 | Cancer antigen 125 |
| DHEA | Dehydroepiandrosterone |
| DHT | 5α-dihydrotestosterone |
| dMMR | missmatch repair deficient |
| DSS | Disease-specific survival |
| EC | Endometrial cancer |
| ESI | Electrospray ionization |
| FIGO | International Federation of Gynecology and Obstetrics |
| HE4 | Human epidydymis protein 4 |
| IQR | Interquartile range |
| LC-MS/MS | Liquid chromatography tandem mass spectrometry |
| LLOQ | Lower limit of quantification |
| LVSI | Lymphovascular space invasion |
| MI | Myometrial invasion |
| MRI | Magnetic resonance imaging |
| MTBE | tert-butyl methyl ether |
| NSMP | Non-specific molecular profile |
| PCOS | Polycystic ovary syndrome |
| QC | Quality control |
| REM | Risk of Endometrial Malignancy |
| SMAC | Steorid Metabolome Analysiss Core |
| T | Testosterone |
References
- Bray, F.; et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Siegel, R.L.; et al. Cancer statistics, 2023. CA: A Cancer Journal for Clinicians 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Bokhman, J.V. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol 1983, 15, 10–7. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, C.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [PubMed]
- Oaknin, A.; et al. Endometrial cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up<sup>☆</sup>. Annals of Oncology 2022, 33, 860–877. [Google Scholar]
- Crosbie, E.J.; et al. Endometrial cancer. Lancet 2022, 399, 1412–1428. [Google Scholar] [CrossRef]
- Concin, N.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. International Journal of Gynecologic Cancer 2021, 31, 12. [Google Scholar] [CrossRef]
- Clarke, M.A.; et al. Association of Endometrial Cancer Risk With Postmenopausal Bleeding in Women: A Systematic Review and Meta-analysis. JAMA Internal Medicine 2018, 178, 1210–1222. [Google Scholar] [CrossRef]
- Long, B.; et al. Ultrasound detection of endometrial cancer in women with postmenopausal bleeding: Systematic review and meta-analysis. Gynecologic Oncology 2020, 157, 624–633. [Google Scholar] [CrossRef]
- Plotti, F.; et al. Implementing the Risk of Endometrial Malignancy Algorithm (REM) adding obesity as a predictive factor: Results of REM-B in a single-center survey. European Journal of Obstetrics & Gynecology and Reproductive Biology 2018, 225, 51–56. [Google Scholar]
- Njoku, K.; et al. Detection of endometrial cancer in cervico-vaginal fluid and blood plasma: leveraging proteomics and machine learning for biomarker discovery. EBioMedicine 2024, 102, 105064. [Google Scholar] [CrossRef]
- Liu, W.; et al. Identification and validation of serum metabolite biomarkers for endometrial cancer diagnosis. EMBO Mol Med 2024, 16, 988–1003. [Google Scholar] [CrossRef] [PubMed]
- Troisi, J.; et al. Development and Validation of a Serum Metabolomic Signature for Endometrial Cancer Screening in Postmenopausal Women. JAMA Netw Open 2020, 3, e2018327. [Google Scholar] [CrossRef] [PubMed]
- Knific, T.; et al. Models including plasma levels of sphingomyelins and phosphatidylcholines as diagnostic and prognostic biomarkers of endometrial cancer. J Steroid Biochem Mol Biol 2018, 178, 312–321. [Google Scholar] [CrossRef]
- Lee, M.S.; et al. Preoperative risk stratification in women with endometrial cancer: A comparison of contrast-enhanced MR imaging and diffusion-weighted MR imaging. Eur J Radiol 2022, 150, 110276. [Google Scholar] [CrossRef]
- Frumovitz, M.; et al. Predictors of final histology in patients with endometrial cancer. Gynecol Oncol 2004, 95, 463–8. [Google Scholar] [CrossRef] [PubMed]
- Visser, N.C.M.; et al. Accuracy of Endometrial Sampling in Endometrial Carcinoma: A Systematic Review and Meta-analysis. Obstetrics and gynecology 2017, 130, 803–813. [Google Scholar] [CrossRef]
- Body, N.; et al. Are preoperative histology and MRI useful for classification of endometrial cancer risk? BMC Cancer 2016, 16, 498. [Google Scholar] [CrossRef]
- Cabrera, S.; et al. Molecular classification improves preoperative risk assessment of endometrial cancer. Gynecol Oncol 2024, 189, 56–63. [Google Scholar] [CrossRef]
- Reijnen, C.; et al. Improved preoperative risk stratification with CA-125 in low-grade endometrial cancer: a multicenter prospective cohort study. J Gynecol Oncol 2019, 30, e70. [Google Scholar] [CrossRef]
- Brennan, D.J.; et al. Serum HE4 as a prognostic marker in endometrial cancer--a population based study. Gynecol Oncol 2014, 132, 159–65. [Google Scholar] [CrossRef]
- Neilson, A.; et al. Serum CA125 levels in the context of ProMisE molecular classification provides pre-operative prognostic information that can direct endometrial cancer management. Gynecologic Oncology 2025, 193, 1–11. [Google Scholar] [CrossRef]
- Knific, T.; et al. Novel algorithm including CA-125, HE4 and body mass index in the diagnosis of endometrial cancer. Gynecologic Oncology 2017, 147, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Rizner, T.L.; Adamski, J. Paramount importance of sample quality in pre-clinical and clinical research—Need for standard operating procedures (SOPs). The Journal of Steroid Biochemistry and Molecular Biology 2019, 186, 1–3. [Google Scholar] [CrossRef]
- Schiffer, L.; et al. Multi-steroid profiling by UHPLC-MS/MS with post-column infusion of ammonium fluoride. Journal of Chromatography B 2022, 1209, 123413. [Google Scholar] [CrossRef] [PubMed]
- Schiffer, L.; et al. Classic and 11-oxygenated androgens in serum and saliva across adulthood: a cross-sectional study analyzing the impact of age, body mass index, and diurnal and menstrual cycle variation. European Journal of Endocrinology 2023, 188. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the caret Package. Journal of Statistical Software 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Peduzzi, P.; et al. A simulation study of the number of events per variable in logistic regression analysis. J Clin Epidemiol 1996, 49, 1373–9. [Google Scholar] [CrossRef]
- Pernigoni, N.; et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science 2021, 374, 216–224. [Google Scholar] [CrossRef]
- Devendran, S.; Méndez-García, C.; Ridlon, J.M. Identification and characterization of a 20β-HSDH from the anaerobic gut bacterium Butyricicoccus desmolans ATCC 43058. J Lipid Res 2017, 58, 916–925. [Google Scholar] [CrossRef]
- Winter, J.; et al. Mode of action of steroid desmolase and reductases synthesized by Clostridium "scindens" (formerly Clostridium strain 19). J Lipid Res 1984, 25, 1124–31. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; et al. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Poore, G.D.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef]
- Gjorgoska, M.; Šturm, L.; Lanišnik Rižner, T. Pre-receptor regulation of 11-oxyandrogens differs between normal and cancerous endometrium and across endometrial cancer grades and molecular subtypes. Frontiers in Endocrinology 2024, 15. [Google Scholar] [CrossRef] [PubMed]
- Gjorgoska Marija, Š.L.; Lanišnik Rižner Tea. Pre-receptor regulation of 11-oxyandrogens differs between normal and cancerous endometrium and across endometrial cancer grades and molecular subtypes. Frontiers in Endocrinology 2024, 15. [Google Scholar] [CrossRef]
- Bhaskaran, K.; et al. Body-mass index and risk of 22 specific cancers: a population-based cohort study of 5·24 million UK adults. Lancet 2014, 384, 755–65. [Google Scholar] [CrossRef] [PubMed]
- Schiffer, L.; et al. Inhibition of the glucocorticoid-activating enzyme 11β-hydroxysteroid dehydrogenase type 1 drives concurrent 11-oxygenated androgen excess. Faseb j 2024, 38, e23574. [Google Scholar] [CrossRef]
- O'Reilly, M.W.; et al. AKR1C3-Mediated Adipose Androgen Generation Drives Lipotoxicity in Women With Polycystic Ovary Syndrome. J Clin Endocrinol Metab 2017, 102, 3327–3339. [Google Scholar] [CrossRef]
- Turcu, A.F.; et al. 11-Oxygenated androgens in health and disease. Nature Reviews Endocrinology 2020, 16, 284–296. [Google Scholar] [CrossRef]
- Lukanova, A.; et al. Circulating levels of sex steroid hormones and risk of endometrial cancer in postmenopausal women. International Journal of Cancer 2004, 108, 425–432. [Google Scholar] [CrossRef]
- Michels, K.A.; et al. Postmenopausal Androgen Metabolism and Endometrial Cancer Risk in the Women's Health Initiative Observational Study. JNCI Cancer Spectrum 2019, 3, pkz029. [Google Scholar] [CrossRef]
- Allen, N.E.; et al. Endogenous sex hormones and endometrial cancer risk in women in the European Prospective Investigation into Cancer and Nutrition (EPIC). Endocrine-Related Cancer 2008, 15, 485–497. [Google Scholar] [CrossRef]
- Clendenen, T.V.; et al. Premenopausal Circulating Androgens and Risk of Endometrial Cancer: results of a Prospective Study. Hormones and Cancer 2016, 7, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Pavlič, R.; et al. Altered Profile of E1-S Transporters in Endometrial Cancer: Lower Protein Levels of ABCG2 and OSTβ and Up-Regulation of SLCO1B3 Expression. International Journal of Molecular Sciences 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Sinreih, M.; Hevir, N.; Rižner, T.L. Altered expression of genes involved in progesterone biosynthesis, metabolism and action in endometrial cancer. Chemico-Biological Interactions 2013, 202, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Hojnik, M.; et al. AKR1C3 Is Associated with Better Survival of Patients with Endometrial Carcinomas. Journal of Clinical Medicine 2020, 9. [Google Scholar] [CrossRef]
- Ito, K.; et al. Expression of androgen receptor and 5α-reductases in the human normal endometrium and its disorders. International Journal of Cancer 2002, 99, 652–657. [Google Scholar] [CrossRef]
- Šmuc, T.; Rižner, T.L. Aberrant pre-receptor regulation of estrogen and progesterone action in endometrial cancer. Molecular and Cellular Endocrinology 2009, 301, 74–82. [Google Scholar] [CrossRef]
- Kamal, A.M.; et al. Androgen receptors are acquired by healthy postmenopausal endometrial epithelium and their subsequent loss in endometrial cancer is associated with poor survival. British Journal of Cancer 2016, 114, 688–696. [Google Scholar] [CrossRef]
- Tangen, I.L.; et al. Androgen receptor as potential therapeutic target in metastatic endometrial cancer. Oncotarget 2016, 7. [Google Scholar] [CrossRef]
- Pavlič, R.; et al. In the Model Cell Lines of Moderately and Poorly Differentiated Endometrial Carcinoma, Estrogens Can Be Formed via the Sulfatase Pathway. Frontiers in Molecular Biosciences 2021, 8. [Google Scholar] [CrossRef]
- Barr, C.E.; et al. Serum CA125 and HE4 as Biomarkers for the Detection of Endometrial Cancer and Associated High-Risk Features. Diagnostics (Basel) 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Antonsen, S.L.; et al. HE4 and CA125 levels in the preoperative assessment of endometrial cancer patients: a prospective multicenter study (ENDOMET). Acta Obstetricia et Gynecologica Scandinavica 2013, 92, 1313–1322. [Google Scholar] [CrossRef]
- Gong, S.; et al. The value of serum HE4 and CA125 levels for monitoring the recurrence and risk stratification of endometrial endometrioid carcinoma. Heliyon 2023, 9, e18016. [Google Scholar] [CrossRef]
- O'Toole, S.A.; et al. HE4 and CA125 as preoperative risk stratifiers for lymph node metastasis in endometrioid carcinoma of the endometrium: A retrospective study in a cohort with histological proof of lymph node status. Gynecologic Oncology 2021, 160, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Brennan, D.J.; et al. Serum HE4 as a prognostic marker in endometrial cancer — A population based study. Gynecologic Oncology 2014, 132, 159–165. [Google Scholar] [CrossRef]
- Behrouzi, R.; Barr, C.E.; Crosbie, E.J. HE4 as a Biomarker for Endometrial Cancer. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Degez, M.; et al. Endometrial cancer: A systematic review of HE4, REM and REM-B. Clinica Chimica Acta 2021, 515, 27–36. [Google Scholar] [CrossRef]
- He, Y.; et al. Role of Human Epididymis Protein 4 (HE4) in Determining Survival of Patients With Endometrial Cancer: A Meta-Analysis. Technol Cancer Res Treat 2020, 19, 1533033820971660. [Google Scholar] [CrossRef]
- Tangen, I.L.; et al. Blood steroids are associated with prognosis and fat distribution in endometrial cancer. Gynecol Oncol 2019, 152, 46–52. [Google Scholar] [CrossRef]
- Forsse, D.; et al. Blood steroid levels predict survival in endometrial cancer and reflect tumor estrogen signaling. Gynecologic oncology 2019. [Google Scholar] [CrossRef] [PubMed]
- Dahmani, C.; et al. Circulating adrenal 11-oxygenated androgens are associated with clinical outcome in endometrial cancer. Frontiers in Endocrinology 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; et al. Endometrial cancer diagnostic and prognostic algorithms based on proteomics, metabolomics, and clinical data: a systematic review. Front Oncol 2023, 13, 1120178. [Google Scholar] [CrossRef] [PubMed]
- Tea Lanišnik Rižner, A.R. The discovery of biomarkers for endometrial cancer: update over the last years. Expert Review of Molecular Diagnostics 2025. [Google Scholar]
- Ura, B.; et al. A Targeted Proteomics Approach for Screening Serum Biomarkers Observed in the Early Stage of Type I Endometrial Cancer. Biomedicines 2022, 10. [Google Scholar] [CrossRef]
- Ura, B.; et al. Two Dimensional-Difference in Gel Electrophoresis (2D-DIGE) Proteomic Approach for the Identification of Biomarkers in Endometrial Cancer Serum. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Enroth, S.; et al. A two-step strategy for identification of plasma protein biomarkers for endometrial and ovarian cancer. Clin Proteomics 2018, 15, 38. [Google Scholar] [CrossRef]
- Tarney, C.M.; et al. Biomarker panel for early detection of endometrial cancer in the Prostate, Lung, Colorectal, and Ovarian cancer screening trial. Am J Obstet Gynecol 2019, 221, 472.e1–472e10. [Google Scholar] [CrossRef]
- Celsi, F.; et al. Gel-Based Proteomic Identification of Suprabasin as a Potential New Candidate Biomarker in Endometrial Cancer. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef]
- Ceylan, Y.; et al. Proteomic analysis in endometrial cancer and endometrial hyperplasia tissues by 2D-DIGE technique. J Gynecol Obstet Hum Reprod 2020, 49, 101652. [Google Scholar] [CrossRef]
- Akkour, K.; et al. Tissue-Based Proteomic Profiling in Patients with Hyperplasia and Endometrial Cancer. Cells 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Hishinuma, E.; et al. Identification of predictive biomarkers for endometrial cancer diagnosis and treatment response monitoring using plasma metabolome profiling. Cancer & Metabolism 2023, 11, 16. [Google Scholar]
- Ihata, Y.; et al. Amino acid profile index for early detection of endometrial cancer: verification as a novel diagnostic marker. Int J Clin Oncol 2014, 19, 364–72. [Google Scholar] [CrossRef]
- Njoku, K.; et al. Metabolomic Biomarkers for the Detection of Obesity-Driven Endometrial Cancer. Cancers 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Audet-Delage, Y.; et al. Identification of Metabolomic Biomarkers for Endometrial Cancer and Its Recurrence after Surgery in Postmenopausal Women. Frontiers in Endocrinology 2018, 9. [Google Scholar] [CrossRef]
- Troisi, J.; et al. Metabolomic Signature of Endometrial Cancer. J Proteome Res 2018, 17, 804–812. [Google Scholar] [CrossRef]
- Bahado-Singh, R.O.; et al. Metabolomic prediction of endometrial cancer. Metabolomics 2017, 14, 6. [Google Scholar] [CrossRef]
- Gu, M.; et al. A metabolomic signature of FIGO stage I and II endometrial cancer. Neoplasma 2021, 68, 1283–1291. [Google Scholar] [CrossRef]
- Yan, X.; et al. A serum lipidomics study for the identification of specific biomarkers for endometrial polyps to distinguish them from endometrial cancer or hyperplasia. Int J Cancer 2022, 150, 1549–1559. [Google Scholar] [CrossRef]
- Schuhn, A.; et al. Potential of blood-based biomarker approaches in endometrium and breast cancer: a case-control comparison study. Arch Gynecol Obstet 2022, 306, 1623–1632. [Google Scholar] [CrossRef]
- Cheng, S.-C.; et al. Metabolomic biomarkers in cervicovaginal fluid for detecting endometrial cancer through nuclear magnetic resonance spectroscopy. Metabolomics 2019, 15, 146. [Google Scholar] [CrossRef]
- Yi, R.; et al. Multi-Omic Profiling of Multi-Biosamples Reveals the Role of Amino Acid and Nucleotide Metabolism in Endometrial Cancer. Front Oncol 2022, 12, 861142. [Google Scholar] [CrossRef] [PubMed]
- Troisi, J.; et al. The Metabolomic Approach for the Screening of Endometrial Cancer: Validation from a Large Cohort of Women Scheduled for Gynecological Surgery. Biomolecules 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; et al. Three plasma-based microRNAs as potent diagnostic biomarkers for endometrial cancer. Cancer Biomark 2021, 31, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; et al. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int J Cancer 2013, 132, 1633–45. [Google Scholar] [CrossRef]
- Liu, J.; et al. Early detection of uterine corpus endometrial carcinoma utilizing plasma cfDNA fragmentomics. BMC Medicine 2024, 22, 310. [Google Scholar] [CrossRef]
- Zou, J.; Wang, E. eTumorType, An Algorithm of Discriminating Cancer Types for Circulating Tumor Cells or Cell-Free DNAs in Blood. Genomics, Proteomics & Bioinformatics 2017, 15, 130–140. [Google Scholar]
- Ashley, C.W.; et al. High-Sensitivity Mutation Analysis of Cell-Free DNA for Disease Monitoring in Endometrial Cancer. Clin Cancer Res 2023, 29, 410–421. [Google Scholar] [CrossRef]
- Yayla Abide, C.; et al. Evaluation of complete blood count parameters to predict endometrial cancer. J Clin Lab Anal 2018, 32, e22438. [Google Scholar] [CrossRef]
- Marin, A.-G.; et al. Clinical Correlations between Serological Markers and Endometrial Cancer. Cancers 2024, 16. [Google Scholar] [CrossRef]
- Ronsini, C.; et al. SIR-EN-New Biomarker for Identifying Patients at Risk of Endometrial Carcinoma in Abnormal Uterine Bleeding at Menopause. Cancers (Basel) 2024, 16. [Google Scholar] [CrossRef] [PubMed]
| Variable [median (IQR) or n (%)] |
Controls (n=70; 100%) |
Cases (n=62; 100%) |
P value ǂ |
|---|---|---|---|
| Age (years) | 64.0 (55.0-71.0) | 64.5 (59.3-71.0) | 0.624 |
| BMI (kg/m2) | 27.4 (23.8-30.5) | 30.4 (27.2-35.2) | 0.005 |
| BMI category | |||
| Normal (<25 kg/m2) | 25 (35.7%) | 14 (22.6%) | 0.055 |
| Overweight (25-30 kg/m2) | 26 (37.2%) | 19 (30.6%) | |
| Obese (≥ 30 kg/m2) | 19 (27.1%) | 29 (46.8%) | |
| Menopausal status | |||
| Premenopausal | 8 (11.4%) | 2 (3.2%) | 0.148 |
| Postmenopausal | 62 (88.6%) | 60 (96.8%) | |
| Diabetes type 2 | |||
| Yes | 10 (14.3%) | 9 (14.5%)) | 0.900 |
| No | 60 (85.7%)) | 52 (83.8%) | |
| Missing data | 0 (0%) | 1 (1.6%) | |
| Arterial hypertension | |||
| Yes | 40 (57.1%) | 38 (61.3%) | 0.473 |
| No | 30 (42.9) | 23 (37.1%) | |
| Missing data | 0 (0%) | 1 1.6%) | |
| Hormonal therapy in the past | |||
| Yes | 2 (2.9%) | 1 (1.6%) | 0.100 |
| No | 67 (95.7%) | 59 (95.2%) | |
| Missing data | 1 (1.4%) | 2 (3.2%) | |
| Medication intake | |||
| Yes | 46 (65.7%) | 45 (72.6%) | 0.508 |
| No | 24 (34.3%) | 17 (27.4%) | |
| Smoking status | |||
| Nonsmoker | 45 (64.3%) | 45 (72.6%) | 0.170 |
| Ever-smoker | 20 (28.6%) | 11 (17.7%) | |
| Missing data | 5 (7.1%) | 6 (9.7%) | |
| Clinical biomarkers | |||
| CA125 (kU/L) | 13.0 (9.1-19.0) | 21.7 (13.4-34.5) | <0.001 |
| HE4 (pmol/L) |
55.6 (45.6-69.7) | 86.0 (64.7-130.4) | <0.001 |
| Controls (n=70; 100%) |
Cases (n=62; 100%) |
P value | |||
|---|---|---|---|---|---|
| Analyte | Median | IQR | Median | IQR | |
| 11-oxyandrogens (nM) | |||||
| 11OHA4 | 13.29 | 10.30-17.56 | 16.62 | 11.88-21.14 | 0.010 |
| 11KA4 | 5.84 | 4.31-6.99 | 5.40 | 3.78-7.95 | 0.810 |
| 11OHT | 0.75 | 0.49-1.06 | 1.00 | 0.66-1.48 | 0.006 |
| 11KT | 1.64 | 1.11-2.02 | 1.73 | 1.10-2.20 | 0.564 |
| Classic androgens (nM) | |||||
| DHEA | 9.29 | 6.96-14.39 | 10.98 | 7.53-18.14 | 0.123 |
| A4 | 2.99 | 2.28-4.06 | 3.67 | 2.70-4.68 | 0.029 |
| T | 0.77 | 0.58-1.30 | 1.18 | 0.81-1.65 | 0.010 |
| Glucocorticoids (nM) | |||||
| 17α-hydroxy-progesterone | 0.95 | 0.66-1.42 | 1.28 | 0.83-1.85 | 0.041 |
| 11-deoxycortisol | 1.07 | 0.73-1.60 | 1.66 | 0.93-2.37 | 0.012 |
| Cortisol | 370.3 | 257.70-495.30 | 420.7 | 298.60-602.10 | 0.055 |
| Cortisone | 64.62 | 53.27-75.86 | 64.98 | 55.73-72.94 | 0.995 |
| Mineralocorticoids (nM) | |||||
| Corticosterone | 9.45 | 5.69-16.96 | 11.54 | 6.16-22.54 | 0.127 |
| LVSI | Deep MI a | |||||
|---|---|---|---|---|---|---|
| Analyte [median (IQR)]) |
Negative (n=50; 81%) |
Positive (n=12; 19%) | P value | No (n=44, 73%) |
Yes (n=16, 27%) |
P value |
| 11-oxyandrogens (nM) | ||||||
| 11OHA4 | 16.40 (11.75-20.61) | 19.51 (14.84-29.40) | 0.187 | 16.77 (10.80-21.71) | 16.89 (15.26-21.88) | 0.483 |
| 11KA4 | 5.07 (3.76-7.82) | 5.95 (5.19-8.33) | 0.269 | 4.85 (3.72-7.98) | 6.04 (4.72-8.05) | 0.367 |
| 11OHT | 0.99 (0.65-1.42) | 1.18 (0.74-1.48) | 0.782 | 1.00 (0.63-1.53) | 0.97 (0.76-1.44) | 0.848 |
| 11KT | 1.67 (1.10-2.20) | 1.88 (1.10-2.15) | 0.838 | 1.78 (0.92-2.31) | 1.73 (1.23-1.97) | 0.821 |
| Classic androgens (nM) | ||||||
| DHEA | 11.25 (7.72-18.17) | 9.82 (5.98-15.31) | 0.364 | 11.73 (7.91-20.01) | 9.16 (7.25-12.27) | 0.116 |
| A4 | 3.61 (2.79-4.68) | 4.31 (2.49-4.68) | 0.972 | 3.67 (2.79-5.12) | 3.53 (2.64-4.40) | 0.559 |
| T | 1.19 (0.84-1.65) | 1.05 (0.65-1.28) | 0.402 | 1.15 (0.79-1.65) | 1.14 (0.77-1.27) | 0.353 |
| Glucocorticoids (nM) | ||||||
| 17α-hydroxy-progesterone | 1.28 (0.82-1.72) | 1.24 (0.87-1.99) | 0.762 | 1.25 (0.82-1.88) | 1.41 (0.84-1.80) | 0.763 |
| 11-deoxycortisol | 1.64 (0.83-2.37) | 1.69 (1.26-2.06) | 0.831 | 1.64 (0.89-2.43) | 1.75 (1.15-2.33) | 0.780 |
| Cortisol | 396.9 (275.3-589.8) | 497.7 (422.5-646.1) | 0.125 | 401.5 (281.1-620.7) | 438.0 (362.2-594.9) | 0.688 |
| Cortisone | 64.45 (54.37-72.94) | 68.96 (60.73-72.81) | 0.465 | 65.86 (52.18-73.92) | 63.43 (60.76-68.34) | 0.973 |
| Mineralocorticoids (nM) | ||||||
| Corticosterone | 10.66 (5.41-23.55) | 14.76 (11.62-21.75) | 0.144 | 10.83 (5.90-24.80) | 13.61 (9.86-20.21) | 0.493 |
| Clinical biomarkers | ||||||
| CA125 (kU/L) | 20.26 (13.10-30.75) | 39.66 (21.37-57.32) | 0.018 | 21.82 (12.37-34.85) | 23.99 (19.69-35.77) | 0.285 |
| HE4 (pmol/L) | 79.46 (61.84-103.92) | 131.51 (114.60-366.27) | 0.001 | 79.46 (61.35-114.95) | 118.95 (87.42-188.53) | 0.004 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





