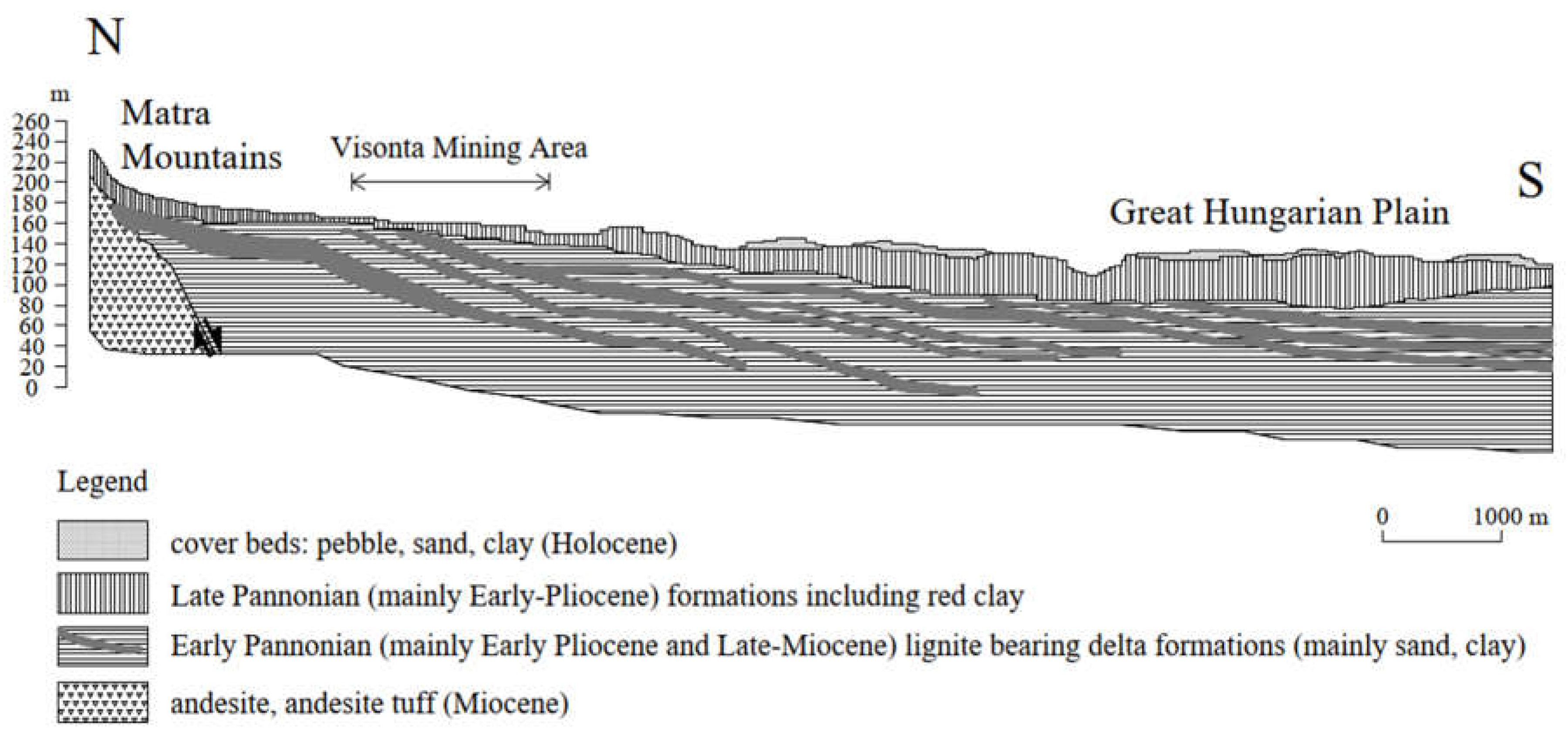
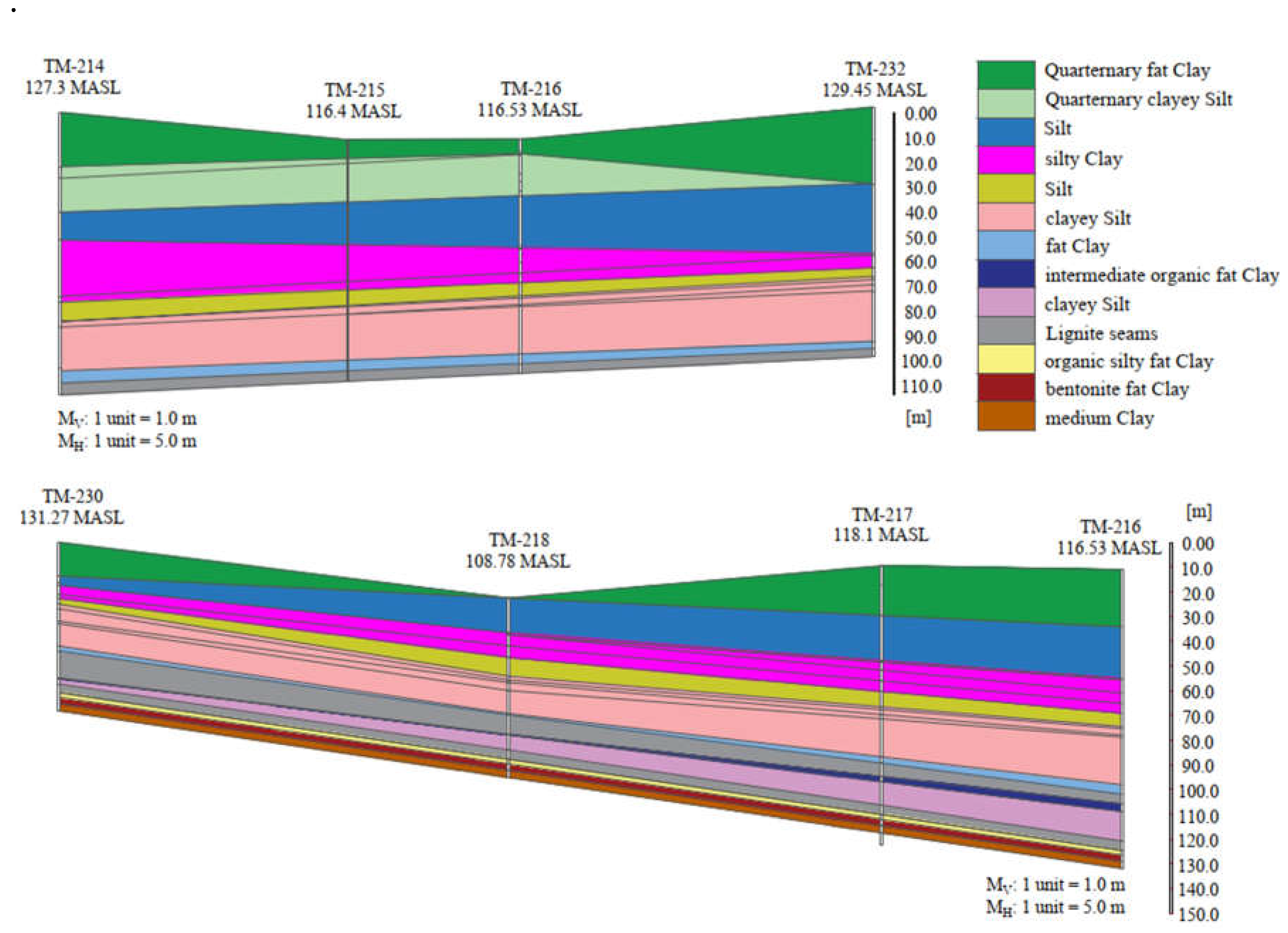
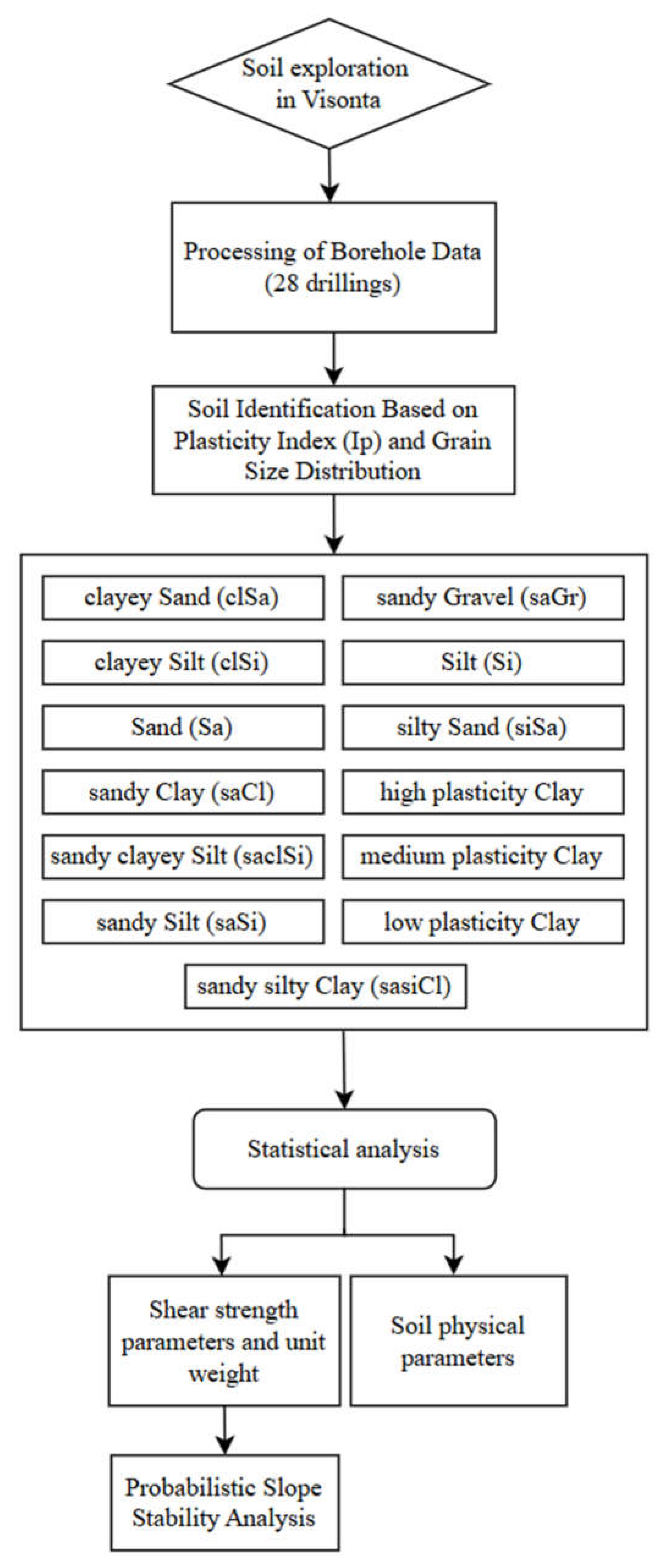
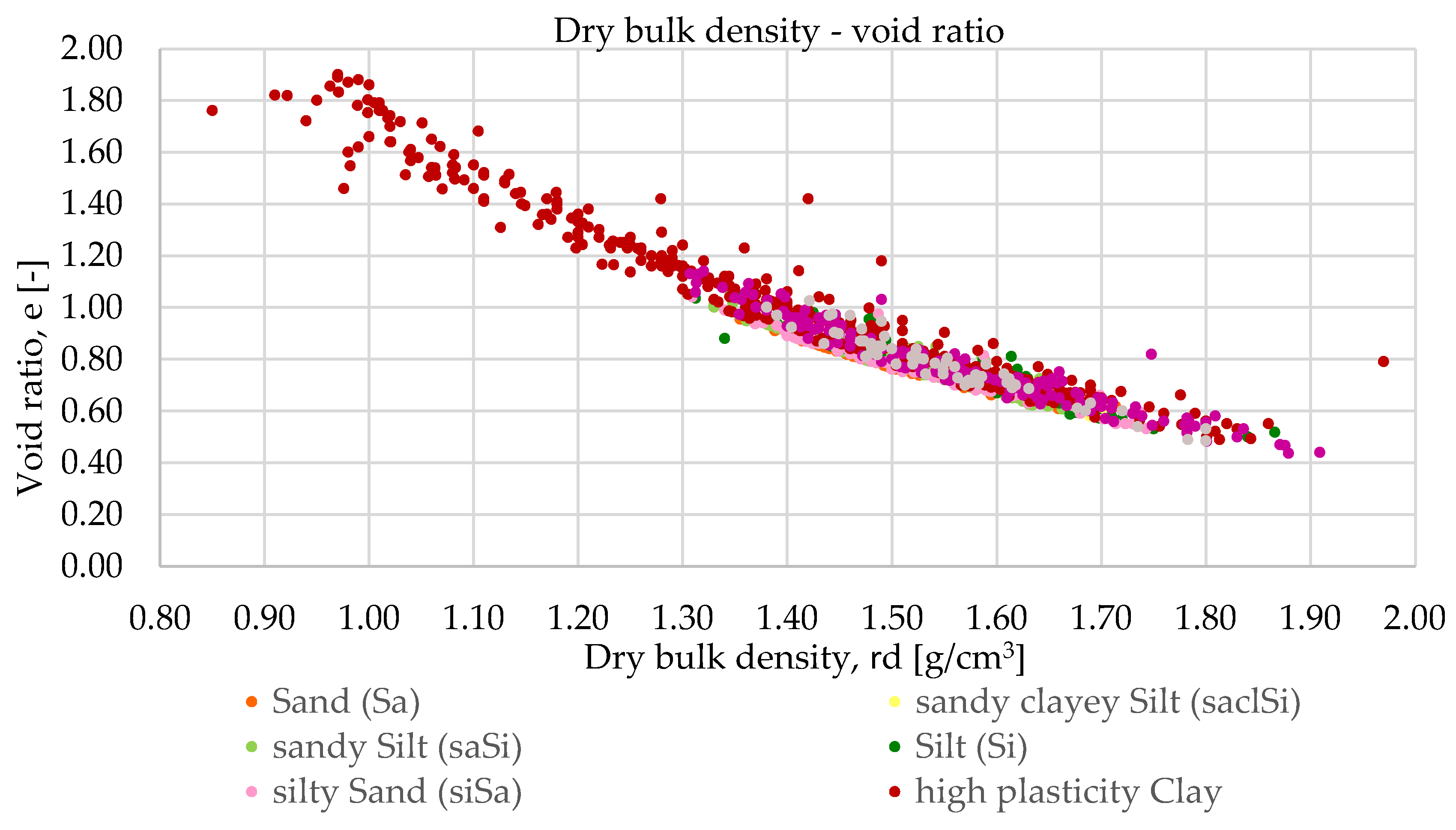
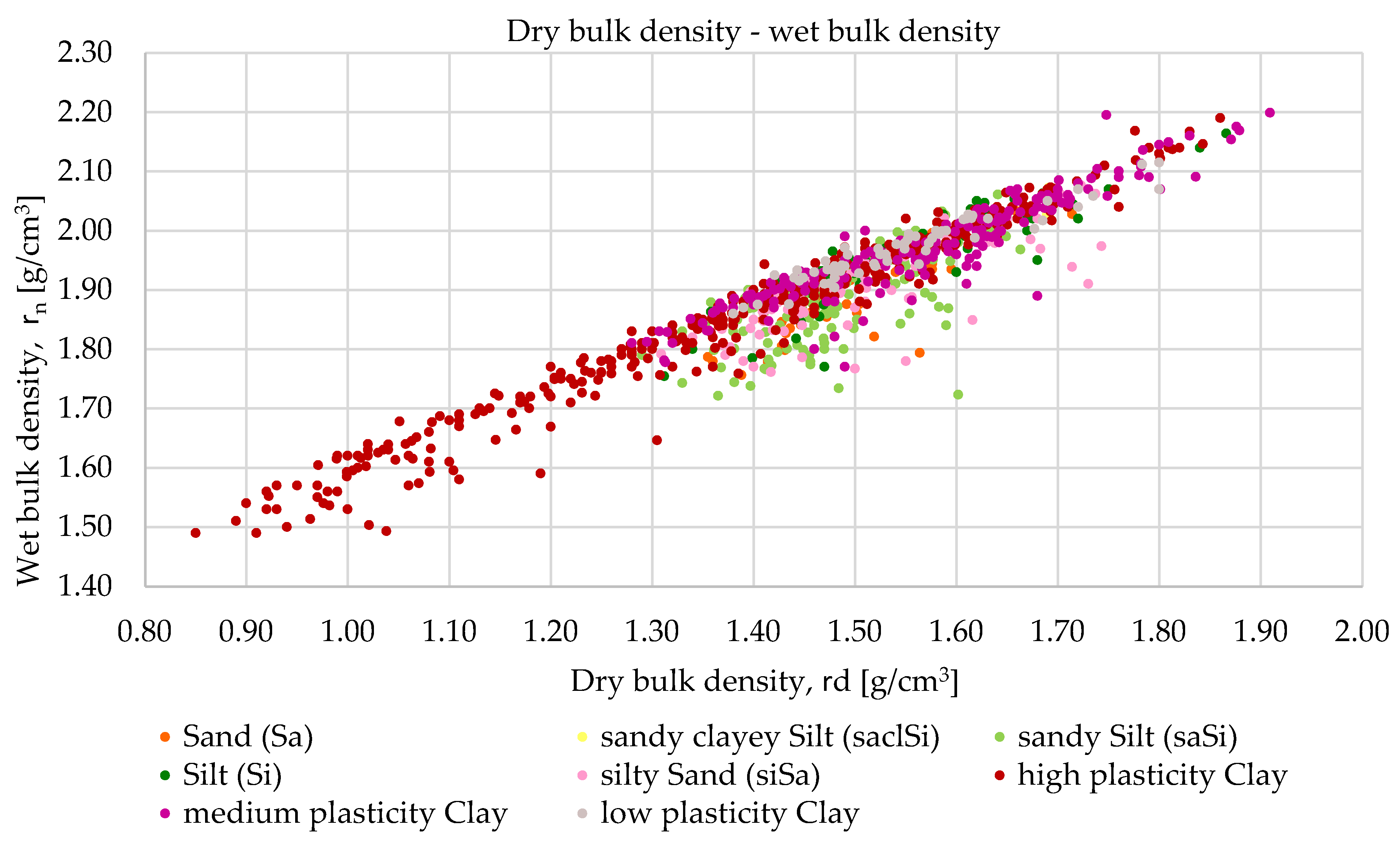
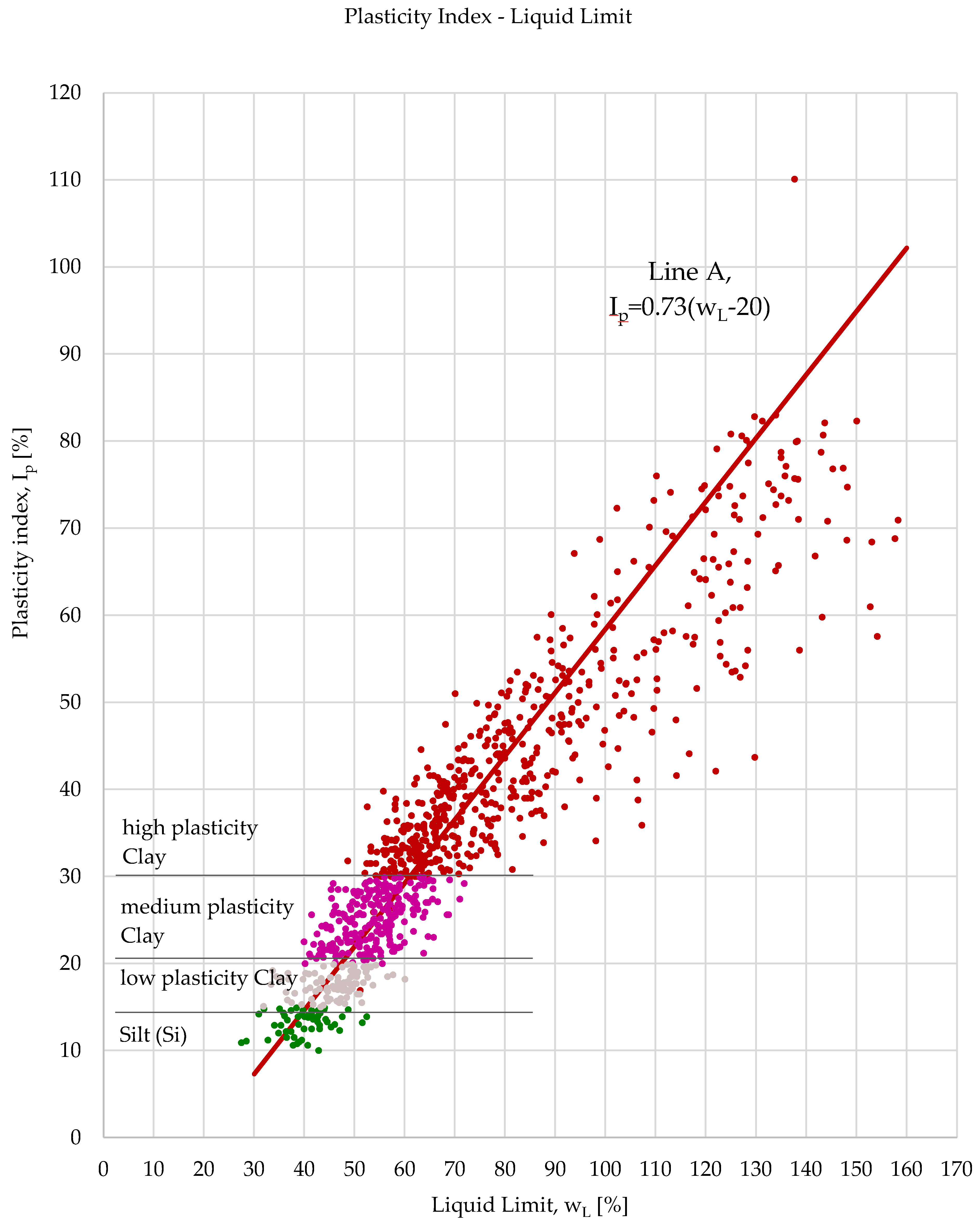
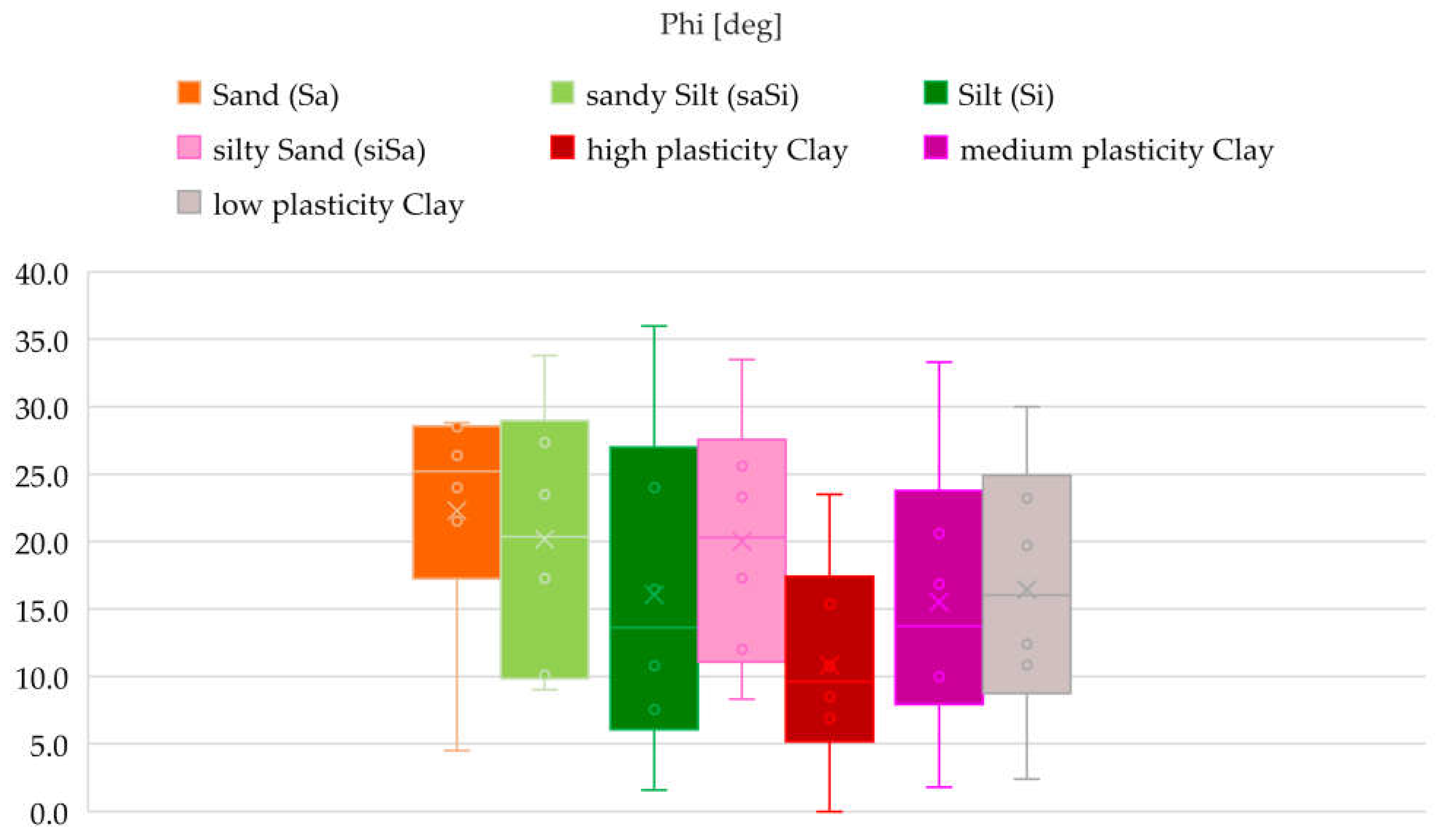
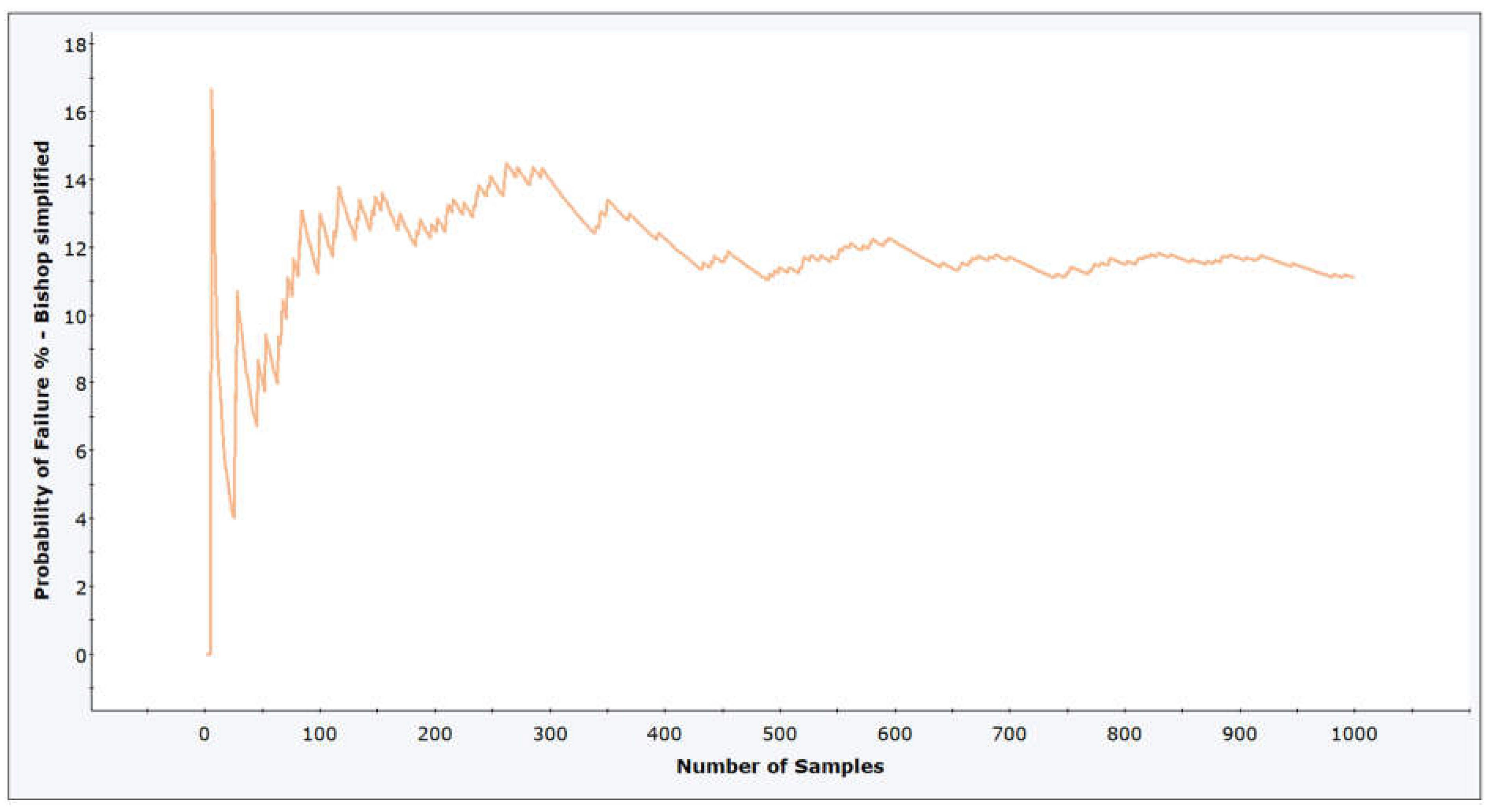
3.1. Methodology of Correlation Analysis
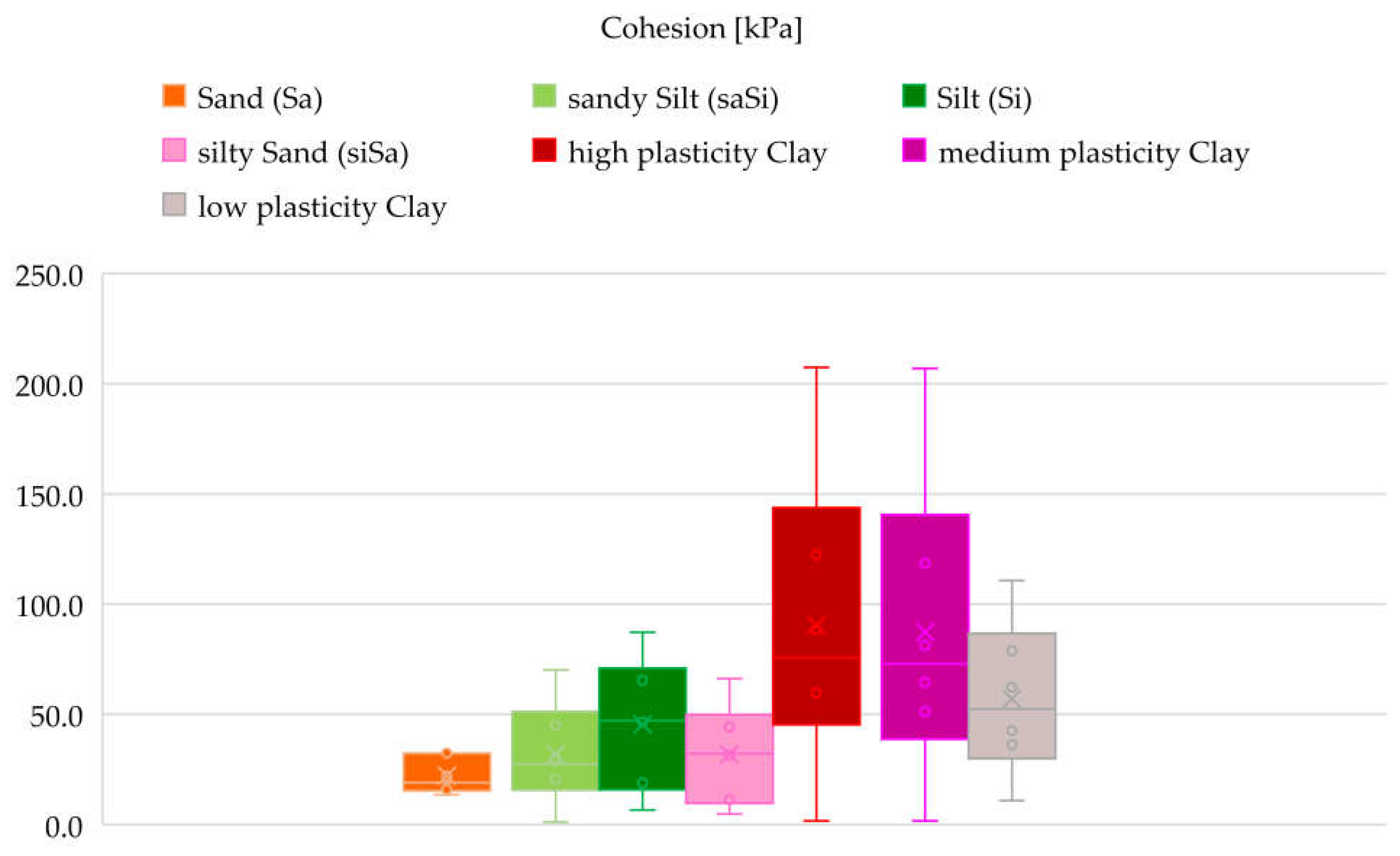
A total of thirteen soil types were analysed: clayey Sand (clSa), clayey Silt (clSi), Sand (Sa), sandy Clay (saCl), sandy clayey Silt (saclSi), sandy Silt (saSi), sandy silty Clay (saclSi), sandy Gravel (saGr), Silt (Si), silty Sand (siSa), high plasticity Clay, medium plasticity Clay, and low plasticity Clay.
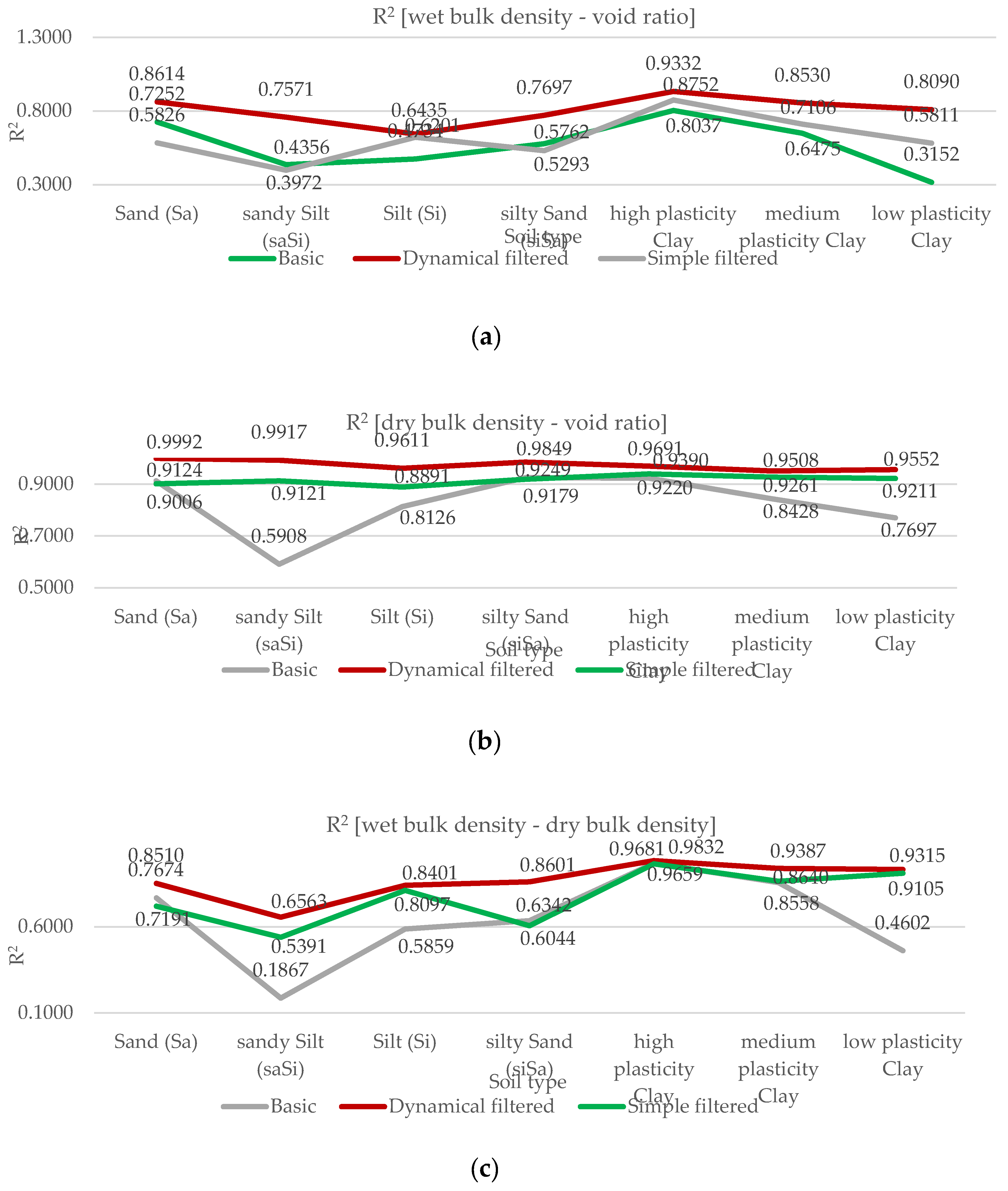
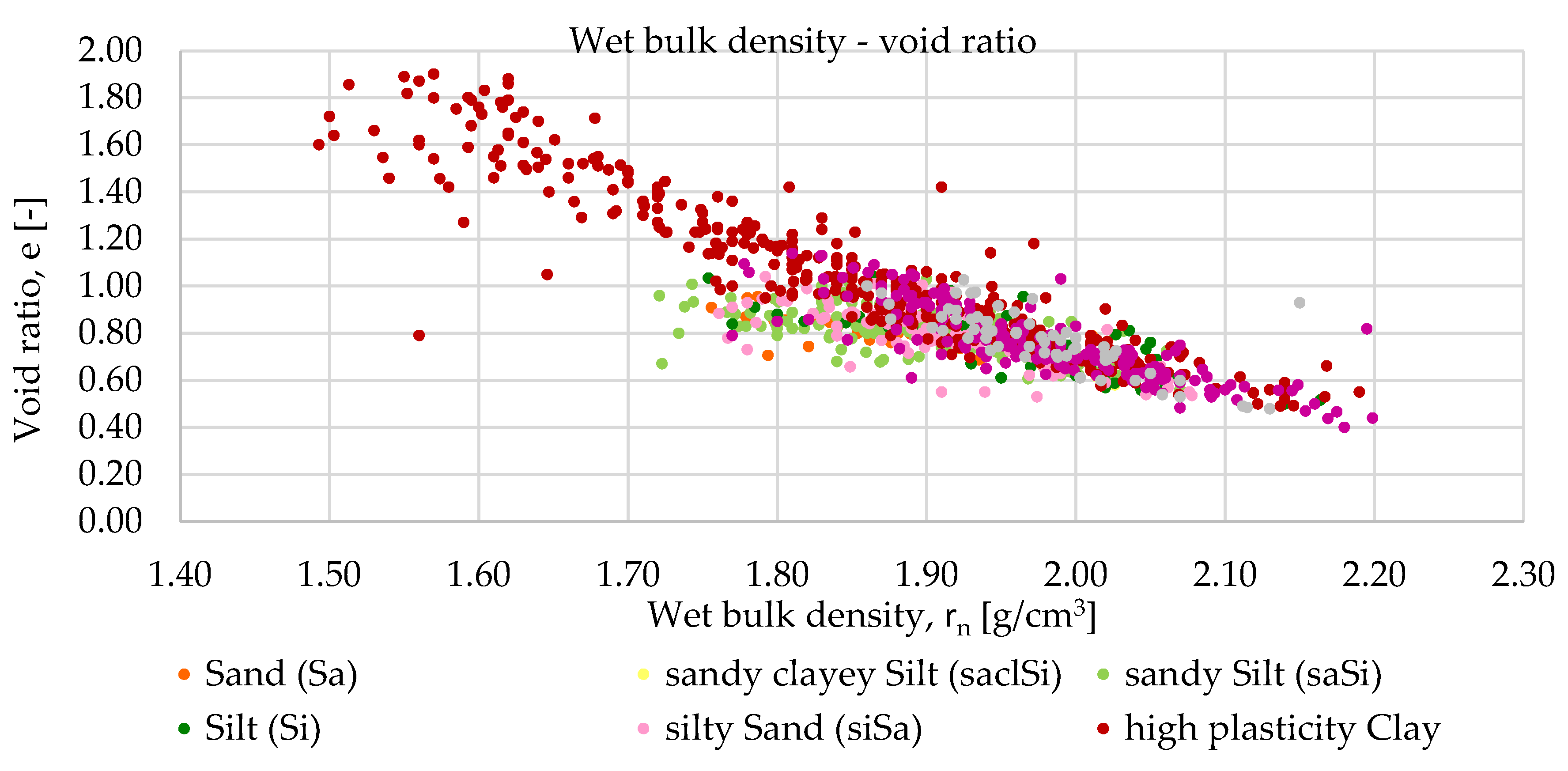
Relationships were examined between the following parameters: wet bulk density and void ratio, dry bulk density and void ratio, wet bulk density and dry bulk density and plasticity index and liquid limit. The study was conducted in three phases. The first phase served as a baseline, where all available laboratory results for each parameter related to each soil type were included.
Subsequently, outlier values were filtered for each soil type and for each studied soil parameter using manual, basic filtering based on the Interquartile Range (IQR) method. The IQR filtering method is used to remove data points that significantly differ from the rest, known as outliers. The central part of the data is identified, and the range between the lower and upper quartiles is measured. Based on this range, boundaries are determined, and any values falling outside these limits are excluded, as they are considered unusual or potentially incorrect. This procedure can be performed in Python, and from Microsoft Excel 2016 onwards, the quartile function is also supported for this purpose.
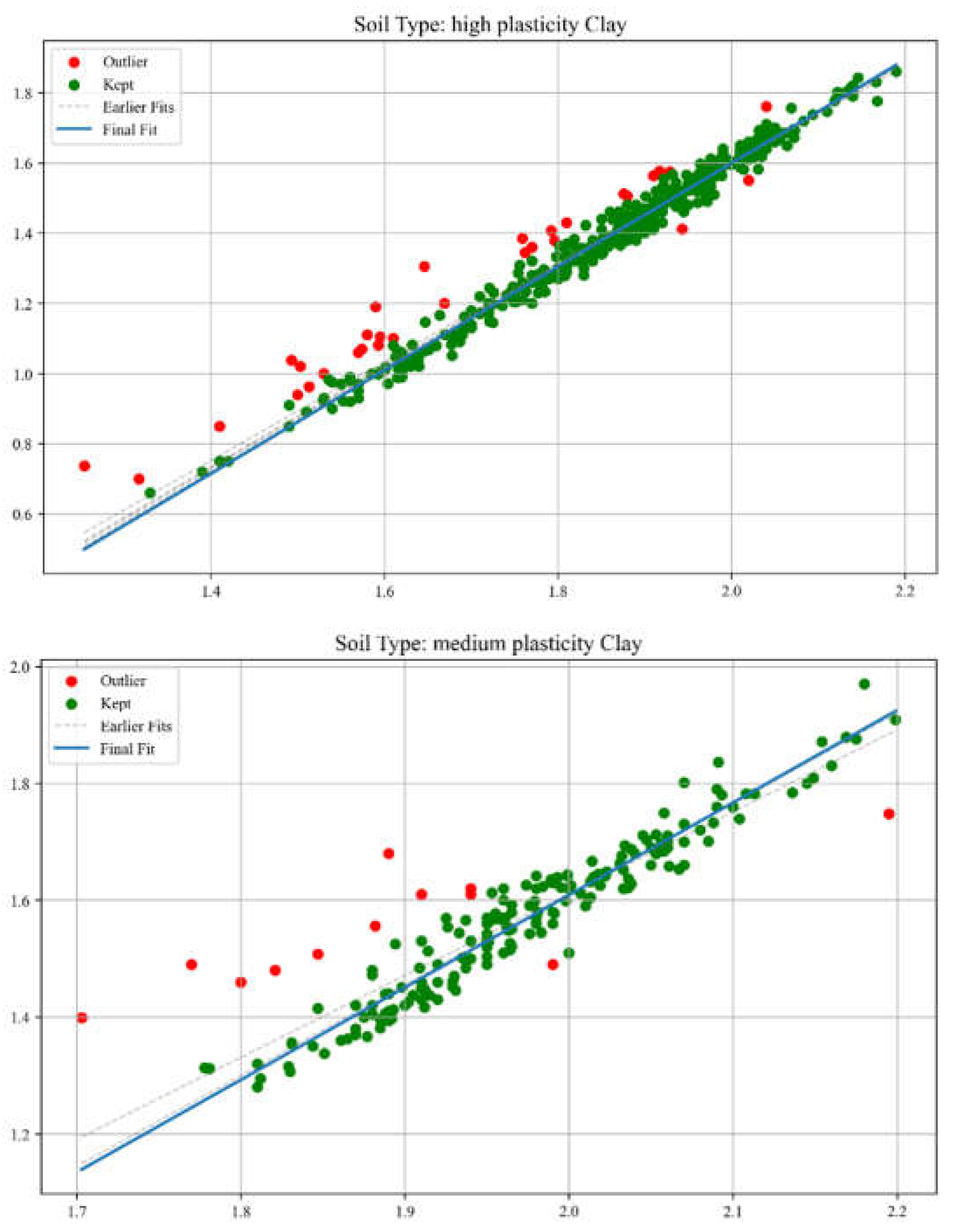
The same procedure was later carried out using a more professional and efficient approach—dynamical filtering—implemented through Python code. The Python program performs iterative outlier detection and visualization on soil-related experimental data. It applies linear regression within each soil type group and iteratively removes statistical outliers based on interquartile range (IQR) analysis of residuals. Each iteration’s regression line is recorded: all intermediate fits are shown as dashed grey lines, while the final fit is highlighted in bold. The tool facilitates reproducible and transparent data cleaning for soil parameter analyses. Dashed grey lines represent earlier regression fits, while the final regression model is shown as a solid blue line. This method allows for very accurate data filtering but carries the risk of excessive data loss. The amount of data lost is shown in
Table 2. The simple filter called IQR filtering with MS Excel was used during the research, since too much data loss can have negative consequences because it reduces the representativeness of the data.
Figure 6.
Regression Fit and Outlier Filtering (examples of dynamical filtering method).
Figure 6.
Regression Fit and Outlier Filtering (examples of dynamical filtering method).
The essential part of the Python script code has been highlighted below.
def iterative_outlier_filtering(group):
group = group.copy()
all_outliers = []
regression_lines = []
num_iterations = 0
while True:
if len(group) < 2:
break
slope, intercept, _, _, _ = linregress(group['Value1'], group['Value2'])
regression_lines.append((slope, intercept))
num_iterations += 1
group['Predicted_Value2'] = group['Value1'] * slope + intercept
group['Residual'] = group[' Value2'] - group['Predicted_ Value2']
q1 = group['Residual'].quantile(0.25)
q3 = group['Residual'].quantile(0.75)
iqr = q3 - q1
lower_bound = q1 - 1.5 * iqr
upper_bound = q3 + 1.5 * iqr
group['Outlier'] = ~group['Residual'].between(lower_bound, upper_bound)
outliers = group[group['Outlier']]
if outliers.empty:
break
all_outliers.append(outliers)
group = group[~group['Outlier']]
group['Outlier'] = False
if all_outliers:
all_outliers_df = pd.concat(all_outliers)
all_outliers_df['Outlier'] = True
final_group = pd.concat([group, all_outliers_df])
else:
final_group = group
final_group = final_group.sort_index()
final_slope, final_intercept = regression_lines[-1] if regression_lines else (0, 0)
return final_group, final_slope, final_intercept, regression_lines, num_iterations
3.1. Methodology of the Statistical Analysis
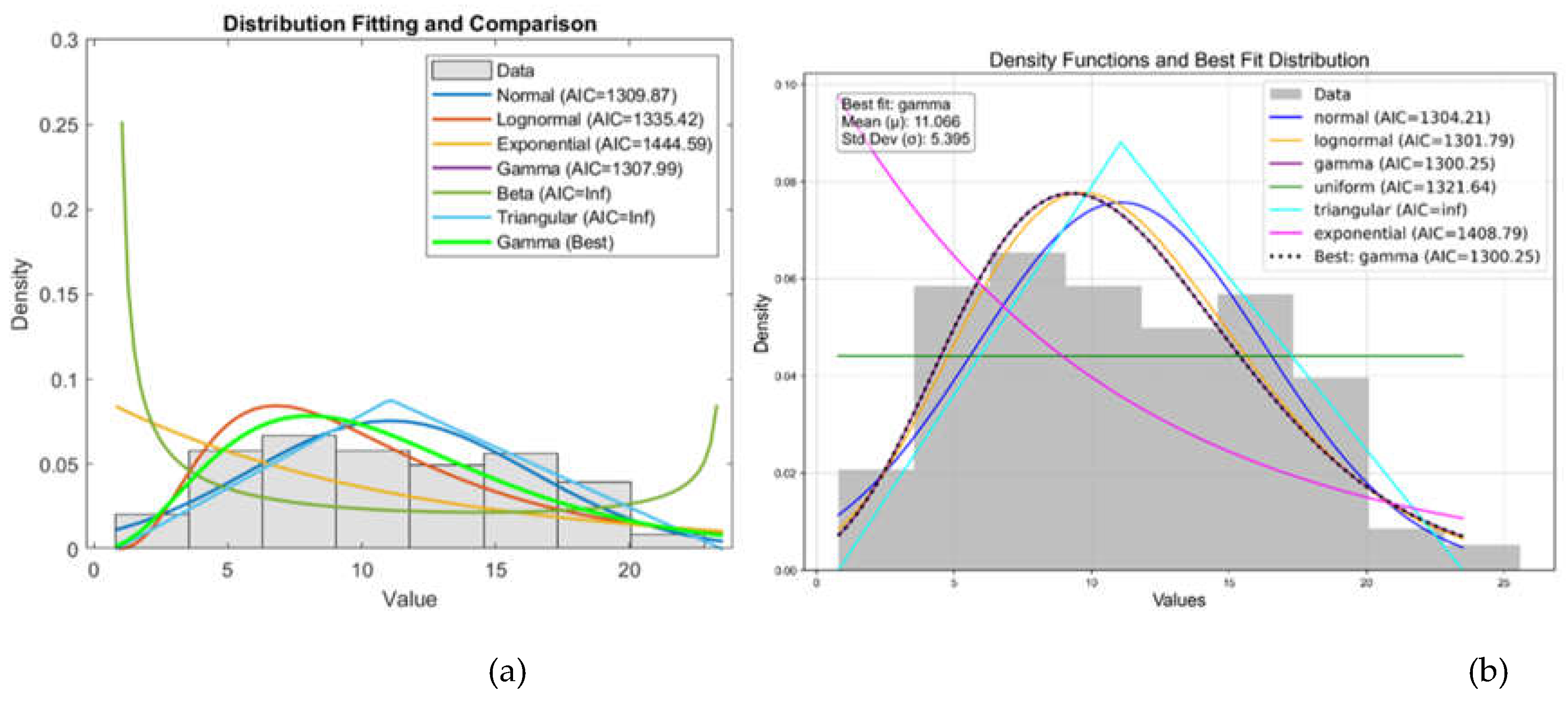
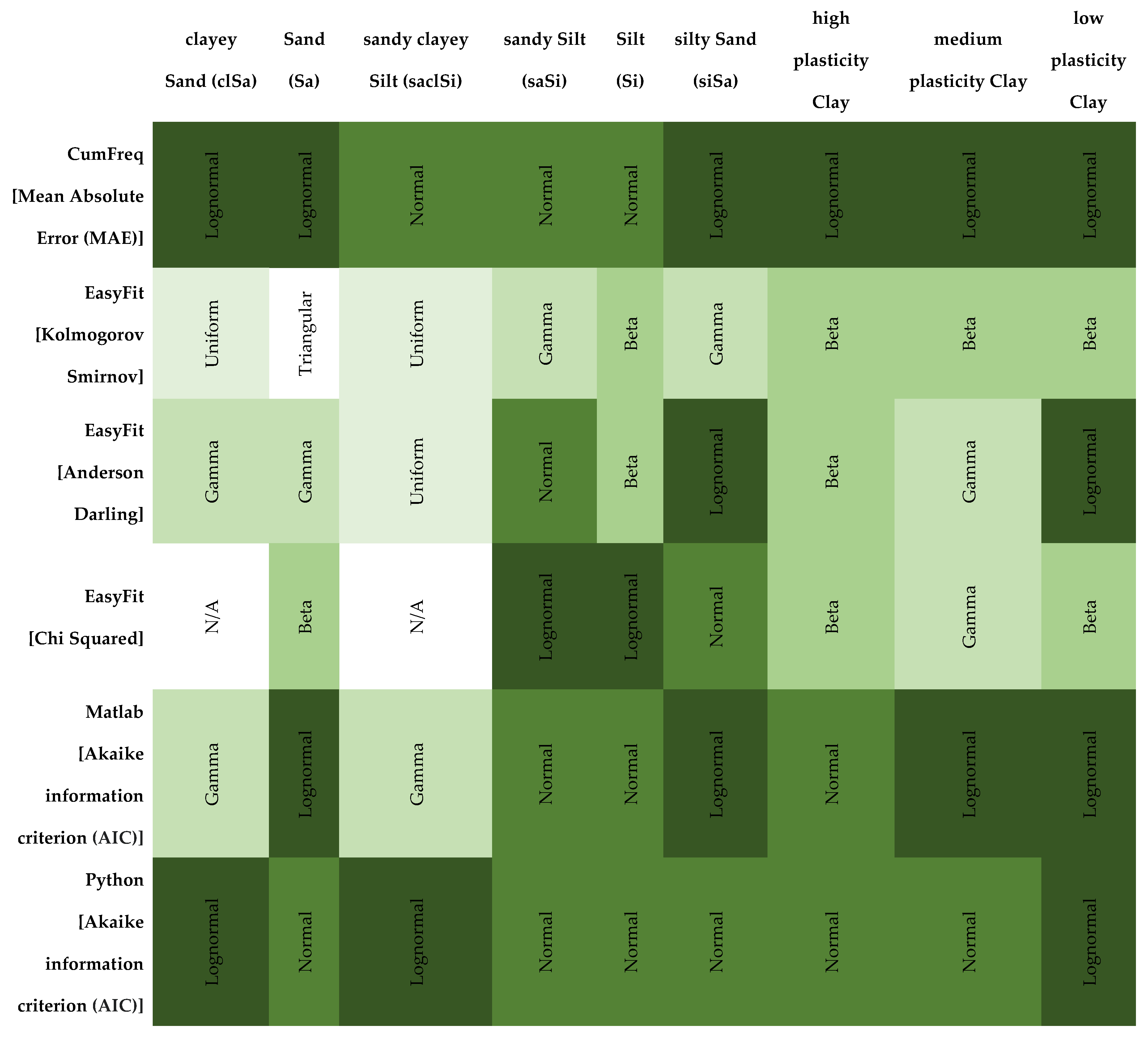
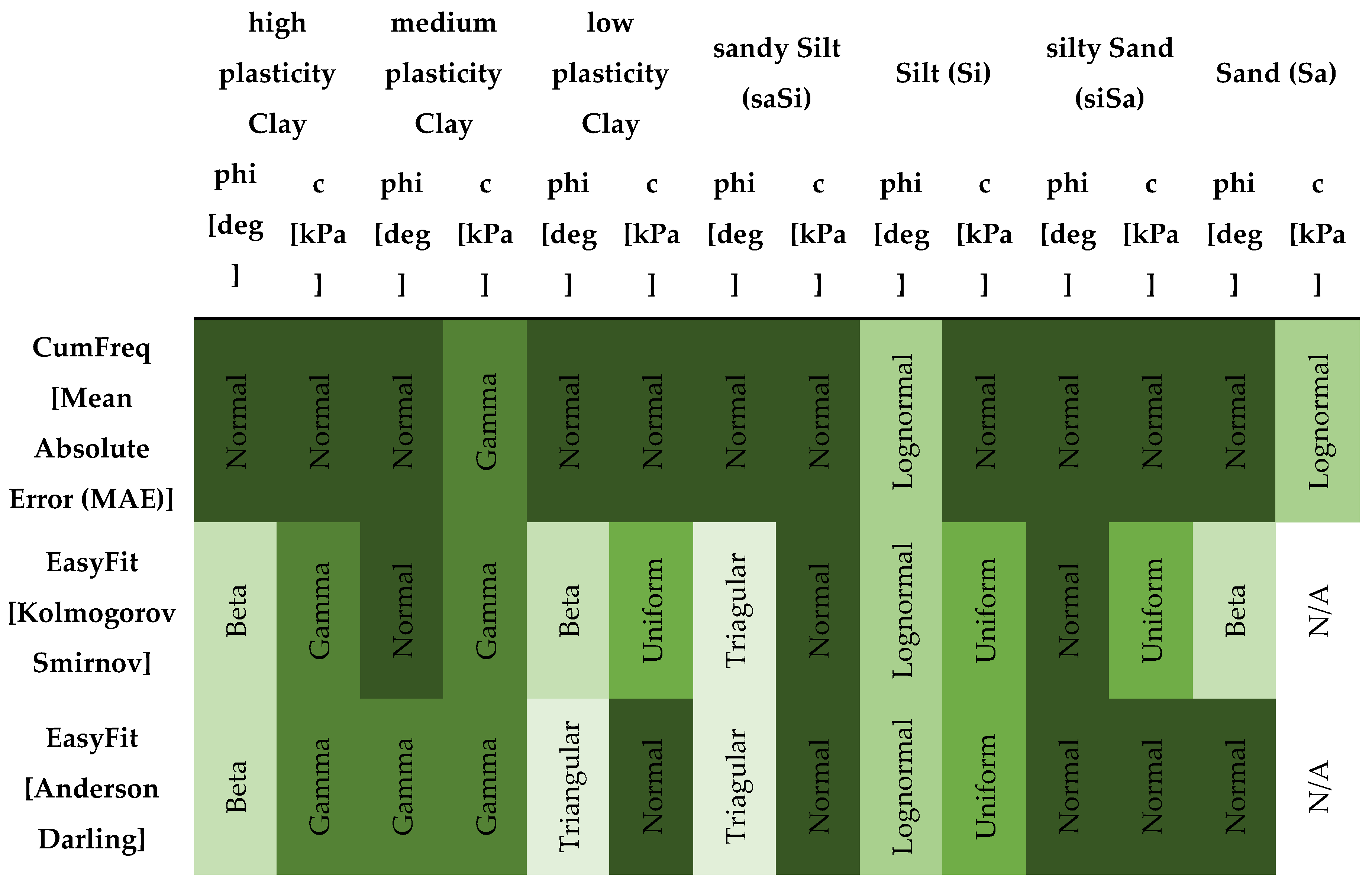
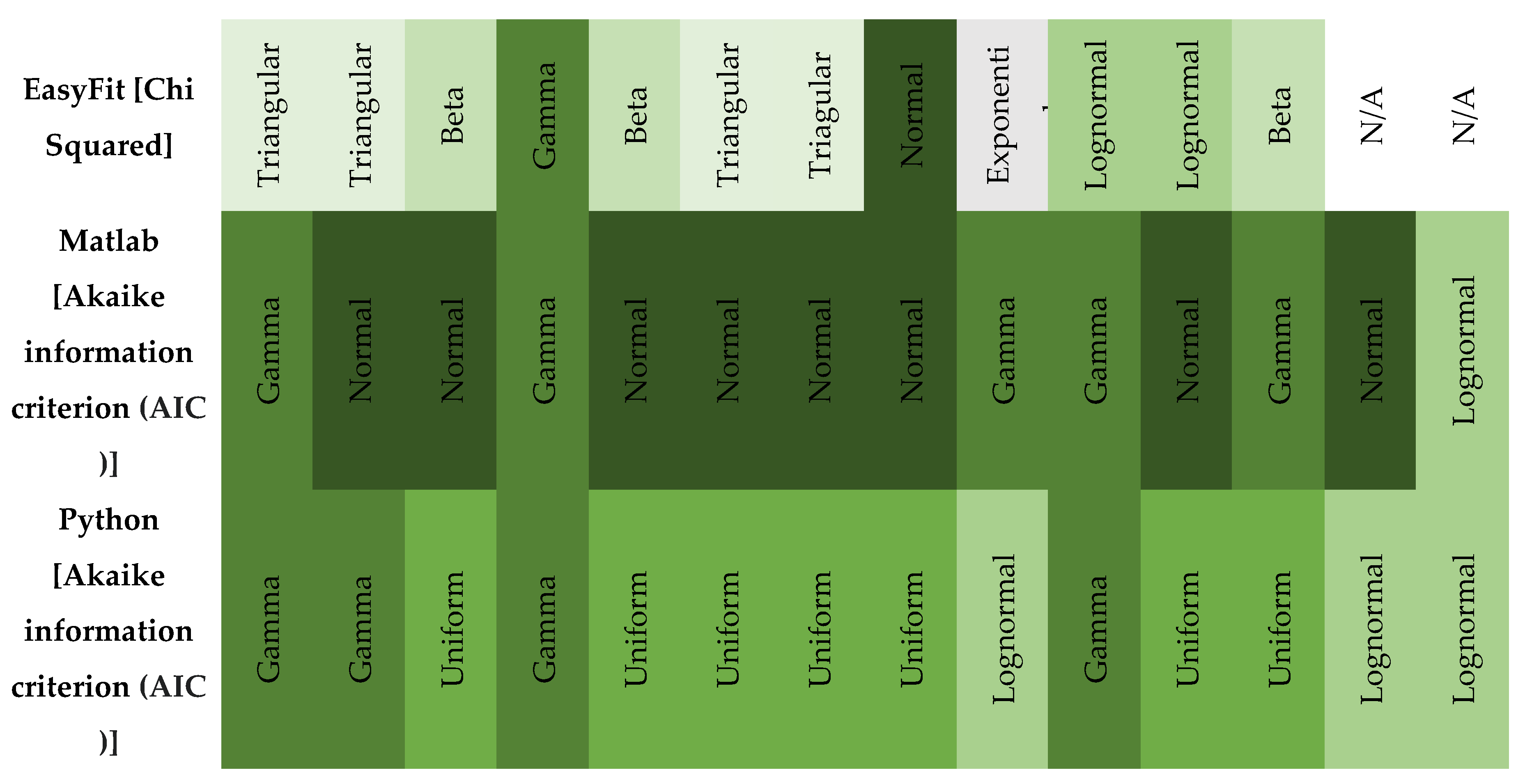
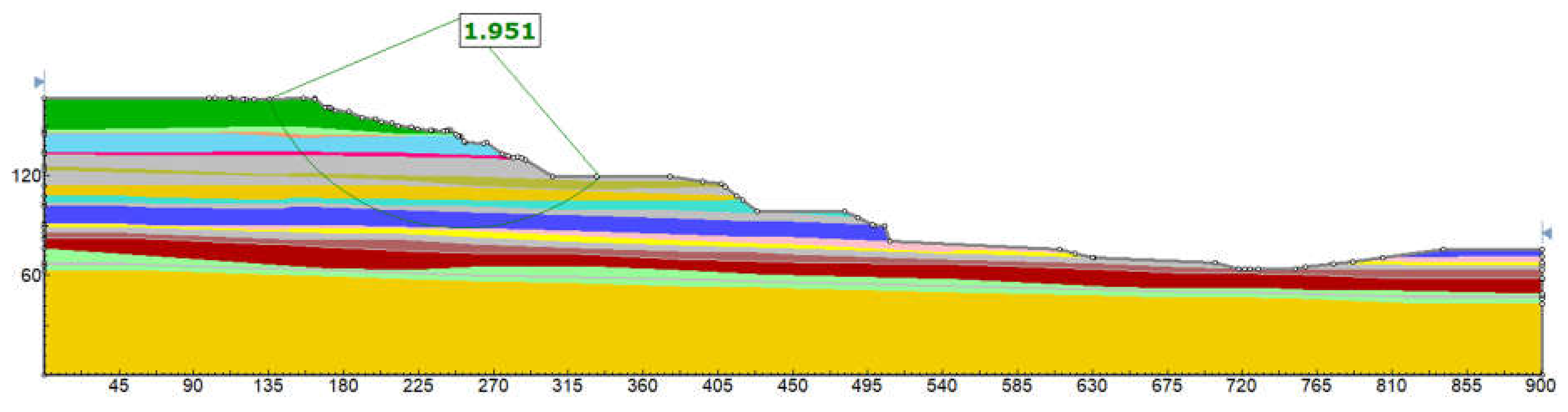
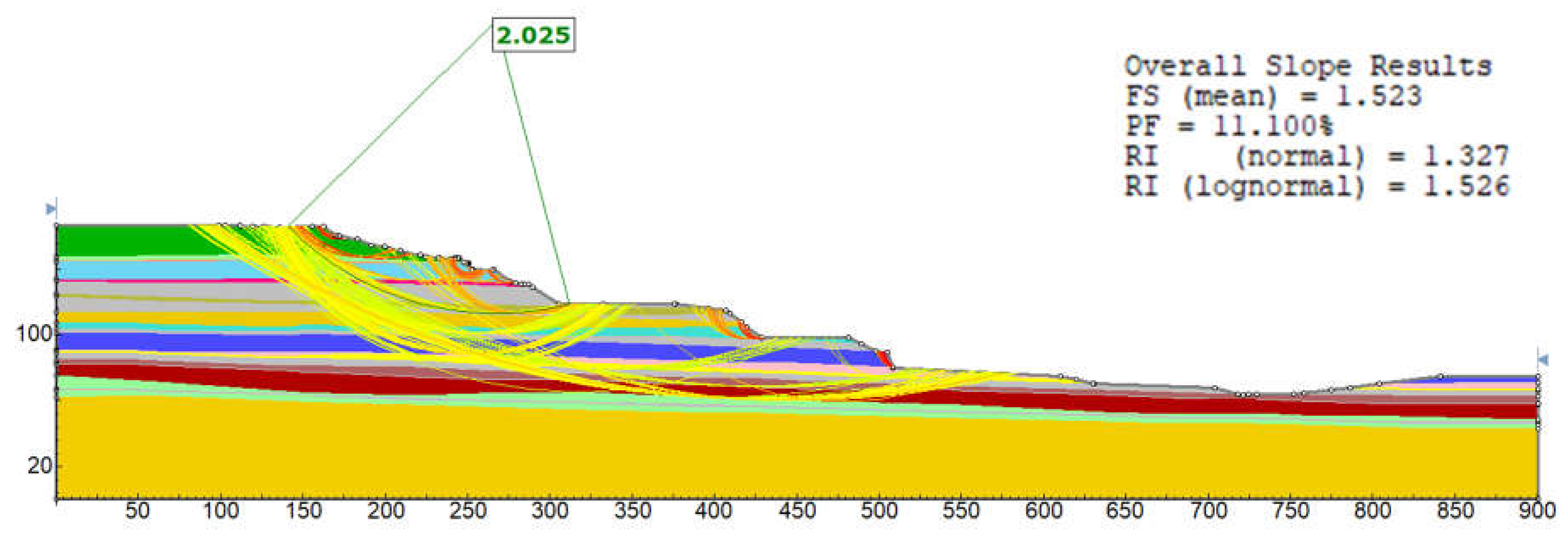
In probabilistic slope stability analysis, accurately defining the statistical properties of input parameters is essential. This includes determining the appropriate probability distribution, mean, standard deviation, and range. These parameters ensure that variability is realistically captured in the model. The analysis in this study uses Rocscience software, which limits inputs to seven distribution types: Normal, Lognormal, Triangular, Gamma, Beta, Uniform, and Exponential. Hence, selecting the most representative distribution for each parameter is critical. A custom MATLAB script was developed specifically for this study. It focuses on the seven distributions supported by Rocscience. By estimating parameters, the script fits multiple distributions: normal, lognormal, exponential, gamma, beta, uniform, and triangular. Each model is evaluated using log-likelihood and the Akaike Information Criterion (AIC), which balances model fit with complexity. The best-fitting model is identified based on the lowest AIC value. Visualization includes a histogram with overlaid probability density functions (PDFs) for each distribution, each plotted in a distinct color.
The best-fitting model is emphasized with a thicker line and annotated with the distribution name, AIC, mean, and standard deviation. For improved usability and validation, a parallel Python implementation was created. It mirrors the MATLAB version but includes more robust error handling and input validation.
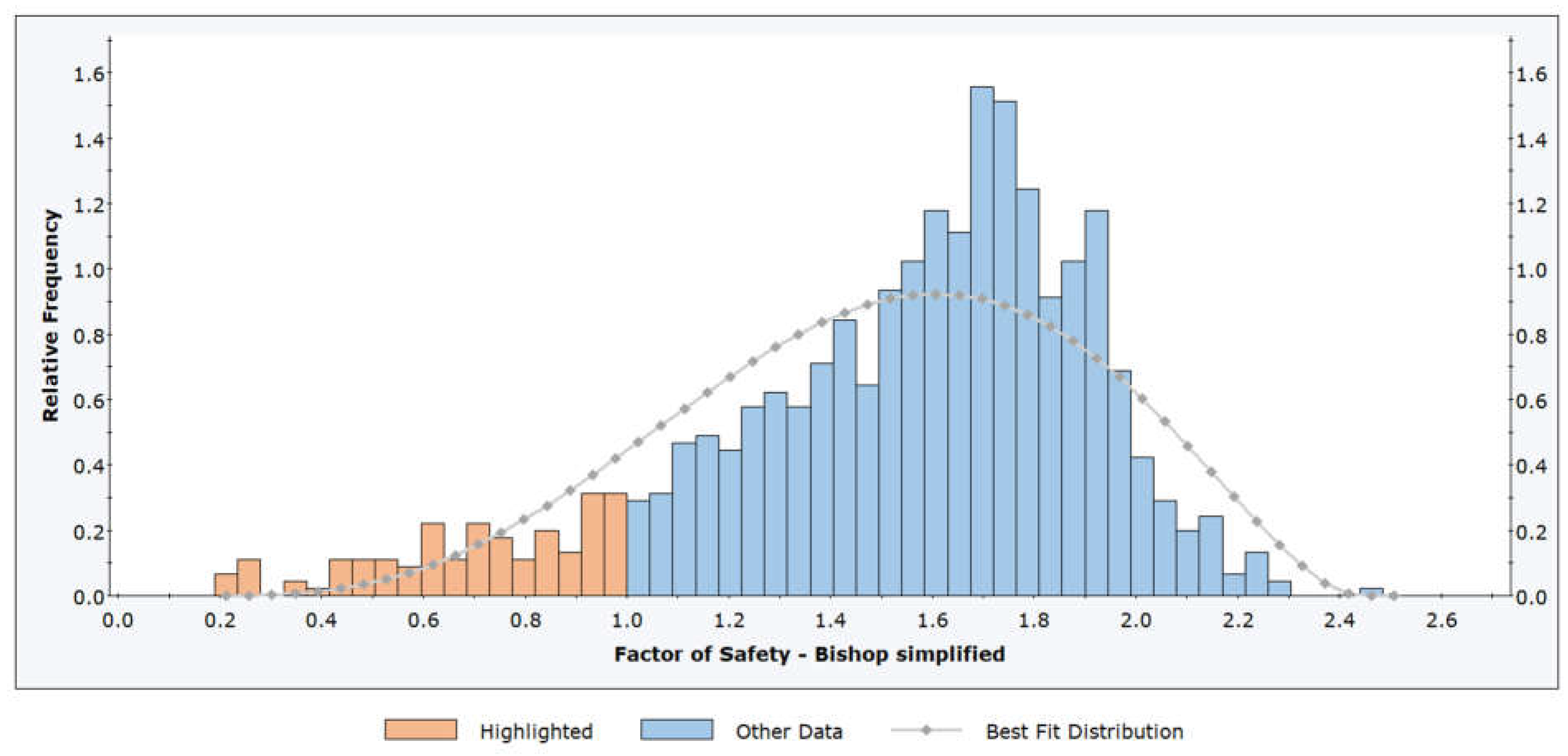
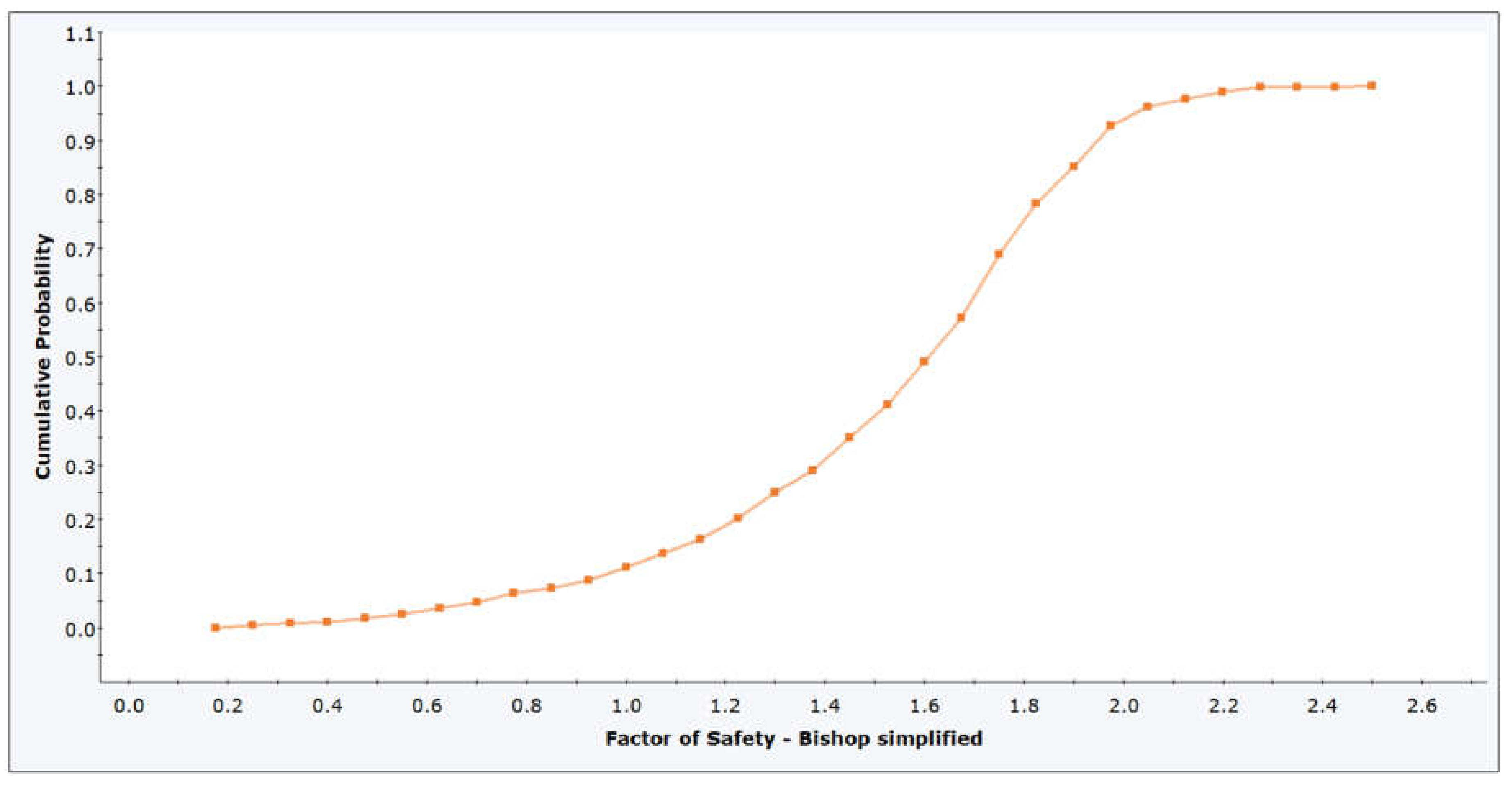
Figure 7.
Comparison of Probability Distribution Fits Using Matlab (a) and Python (b).
Figure 7.
Comparison of Probability Distribution Fits Using Matlab (a) and Python (b).
The left figure shows a Matlab result, while the right figure shows the same result calculated using Python code. The two plots show the results of a distribution fitting of a dataset, comparing several probability distributions based on the quality of the fit. In both plots, several distributions (normal, lognormal, gamma, uniform, triangular, and exponential) are fitted to the histogram of the data. The gamma distribution is the best fit, with the lowest AIC value. The gamma distribution closely follows the shape of the histogram, indicating a good fit. In both plots, AIC (Akaike Information Criterion) values were used to determine the fit: lower values indicate a better fit.
The MATLAB and Python code snippets both aim to identify the best-fitting statistical distribution based on the Akaike Information Criterion (AIC), yet exhibit notable differences. The MATLAB script employs a straightforward loop structure, normalizes data specifically for the Beta distribution without parameter constraints, computes log-likelihood directly from fitted distributions, and stores only the best-fitting model parameters and their AIC. In contrast, the Python implementation utilizes explicit normalization parameters (floc, fscale) for Beta distributions, separately manages the triangular distribution as a special case, computes additional statistics (mean, standard deviation) for each fit, systematically captures fitting errors via structured warnings, and returns comprehensive results for all distributions tested, including detailed statistical summaries. Overall, the Python approach provides greater analytical depth, explicit error handling, and enhanced result organization compared to the concise, more simplistic MATLAB methodology.
The essential parts of the codes have been highlighted below.
Matlab code for selecting best fit distribution:
% Fit each distribution and compute AIC
for i = 1:size(distributions, 1)
distName = distributions{i, 1};
distFunc = distributions{i, 2};
try
if strcmp(distName, 'Beta')
normalized_data = (data - min(data)) / (max(data) - min(data));
pd = distFunc(normalized_data);
else
pd = distFunc(data);
end
logL = sum(log(pdf(pd, data)));
k = length(pd.ParameterValues);
AIC = 2 * k - 2 * logL;
AIC_values(i) = AIC;
if AIC < bestAIC
bestAIC = AIC;
bestDist = distName;
bestParam = pd.ParameterValues;
bestPD = pd;
end
catch ME
fprintf('Error fitting %s distribution: %s\n', distName, ME.message);
end
end
Python code or selecting best fit distribution:
def fit_and_compare_distributions(data):
results = []
for name, dist in distributions.items():
try:
if name == "beta":
data_min, data_max = np.min(data), np.max(data)
normalized_data = (data - data_min) / (data_max - data_min)
params = dist.fit(normalized_data, floc=0, fscale=1)
ll = np.sum(dist.logpdf(normalized_data, *params))
mean, var = dist.stats(*params, moments="mv")
mean = mean * (data_max - data_min) + data_min
std = np.sqrt(var) * (data_max - data_min)
elif name == "triangular":
a, c = np.min(data), np.max(data)
b = np.mean(data)
loc = a
scale = c - a
params = ((b - a) / scale, loc, scale)
ll = np.sum(dist.logpdf(data, *params))
mean, var = dist.stats(*params, moments="mv")
std = np.sqrt(var)
else:
params = dist.fit(data)
ll = np.sum(dist.logpdf(data, *params))
mean, var = dist.stats(*params, moments="mv")
std = np.sqrt(var)
k = len(params)
aic = 2 * k - 2 * ll
results.append({
"distribution": name,
"params": params,
"log_likelihood": ll,
"aic": aic,
"mean": mean,
"std": std,
})
except Exception as e:
warnings.warn(f"Error fitting {name} distribution: {e}")
if results:
return results, min(results, key=lambda x: x["aic"])
else:
warnings.warn("No distributions successfully fitted to the data.")
return [], None