Submitted:
28 April 2025
Posted:
29 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Amyloid Beta 42 Structure
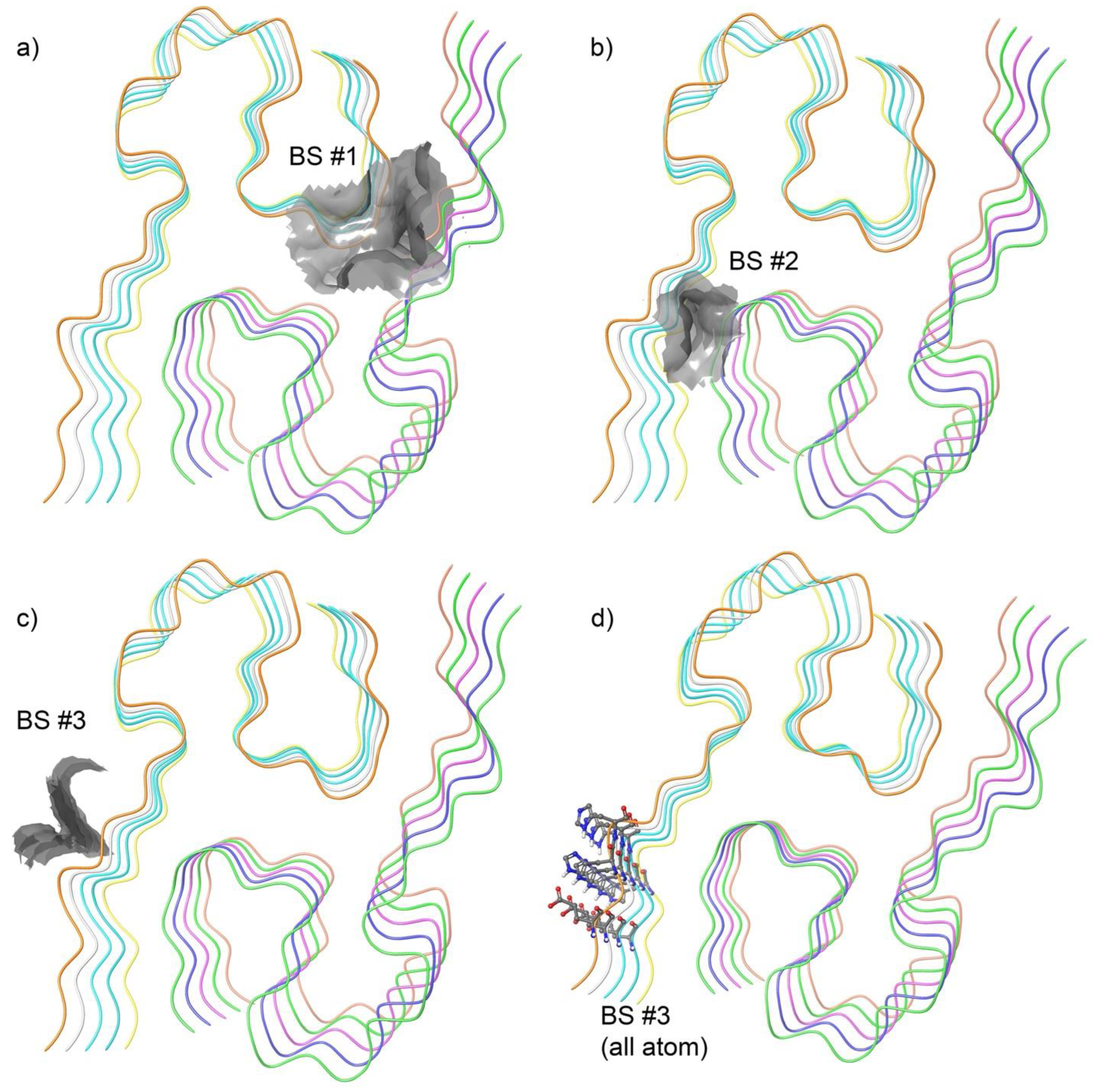
2.2. Binding Site Identified in Amyloid Beta42
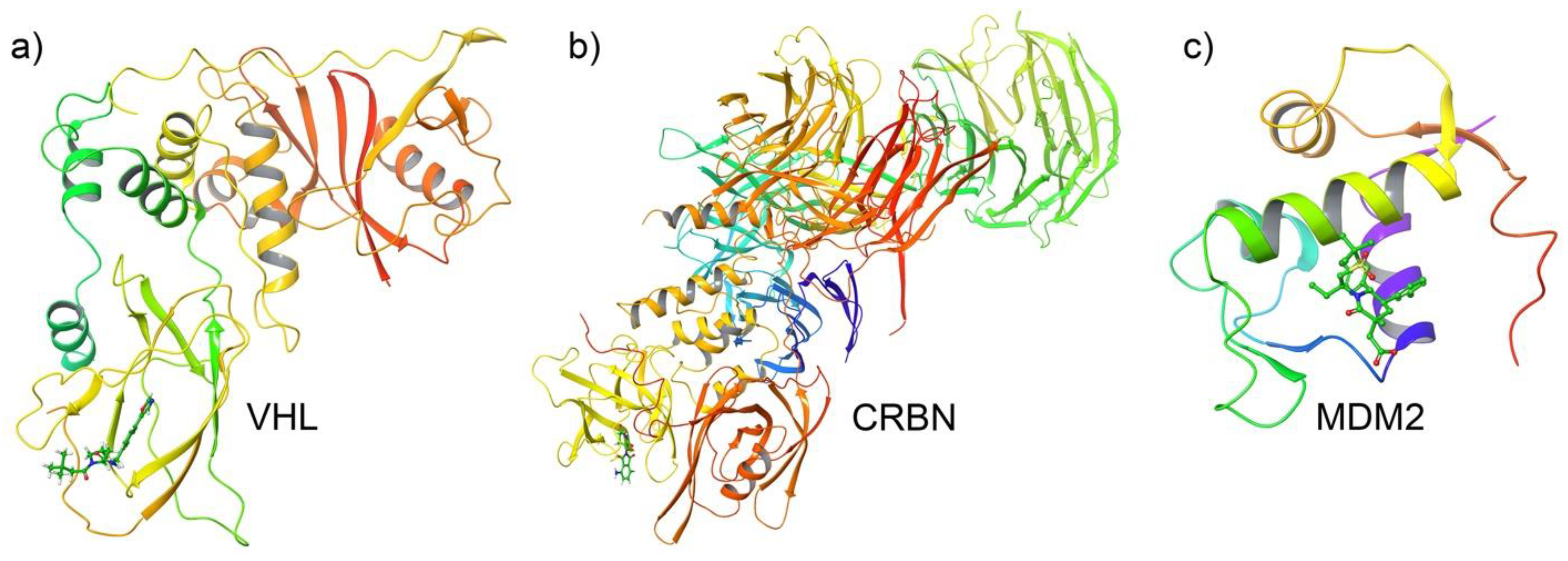
2.3. E3 Ligase Structure Selection and Preparation
2.4. ADMET Based Filtering and Docking
2.5. Dataset Construction and Feature Engineering for Ligase-Conditioned Molecular Glue Design
2.6. Development of Ligase-Conditioned JT-VAE Incorporating Torsional Features for Molecular Glue Design
2.7. Encoding E3 Ligase Binding Site Sequences
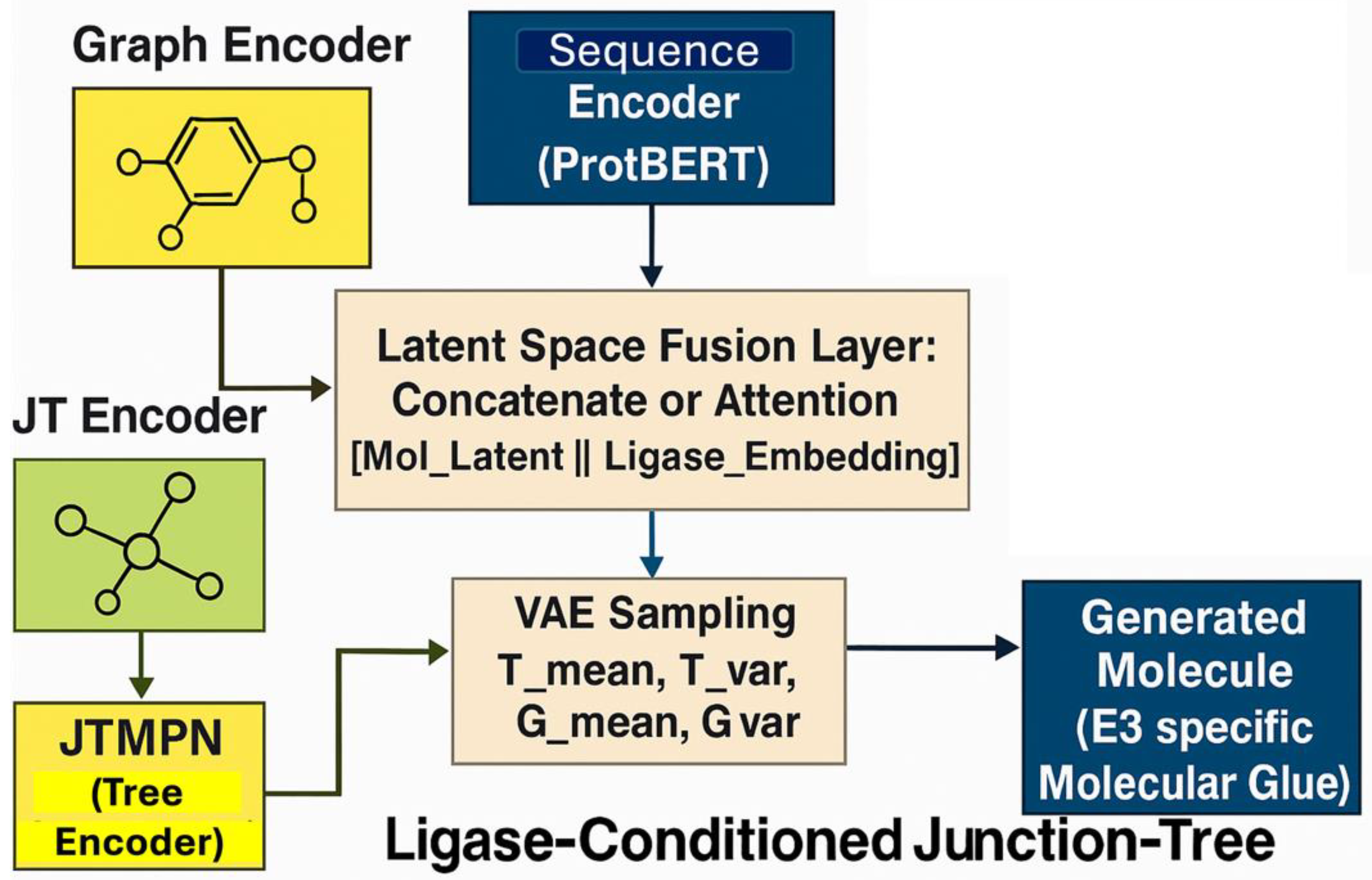
2.8. LC-JT-VAE Architecture and Latent Space Fusion
2.9. Detailed Architectural Breakdown and Conditioning Mechanism
- Molecular Graph and Junction Tree Encoding: Molecules were processed as graphs and junction trees using MPN and JTMPN. Bond features were enhanced by incorporating torsional angles computed via RDKit, improving the model's ability to capture conformational flexibility crucial for protein–ligand interactions.
- Ligase Sequence Encoding: Binding site sequences were embedded using ProtBERT or BiLSTM-based encoders and projected to match the molecular latent space dimensions.
-
Latent Space Fusion Strategies:
- ○
-
Concatenation and Linear Projection:z_fused = ReLU(W[z_mol; z_seq] + b)
- ○
-
Cross-Attention Mechanism:z_mol' = MultiHeadAttention(z_mol, z_seq, z_seq)
-
Conditional Decoding:
- ○
- Tree Decoder: Reconstructs the junction tree scaffold.
- ○
- Graph Decoder: Rebuilds the full molecular graph conditioned on the fused latent vector.
2.10. Model Training, Optimization, and Implementation Details
3. Results
3.1. Identification of a Ligandable Interface on Amyloid-β42 Fibrils
3.2. ADMET-Based Filtering of Compound Libraries
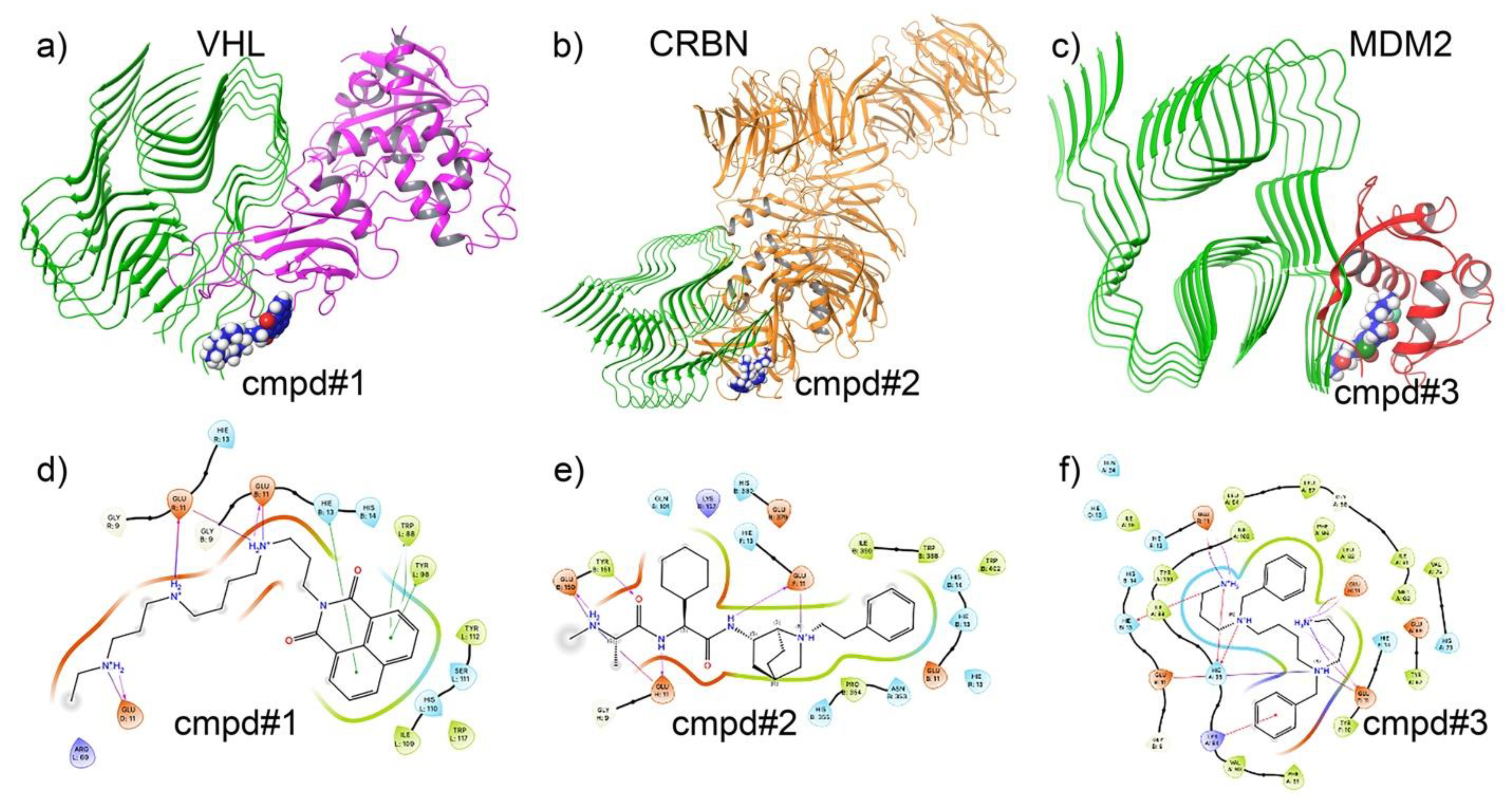
3.3. Docking Analysis of Ternary Complexes Formed by E3 Ligases and Amyloid Beta-42 via Molecular Glue Compounds
3.4. Training Data Compounds Characterization to Generate the Ligase-Conditioned Junction Tree Variational Autoencoder (LCJTVAE)
3.5. Graph and Junction Tree Representations of Molecules for Model Training
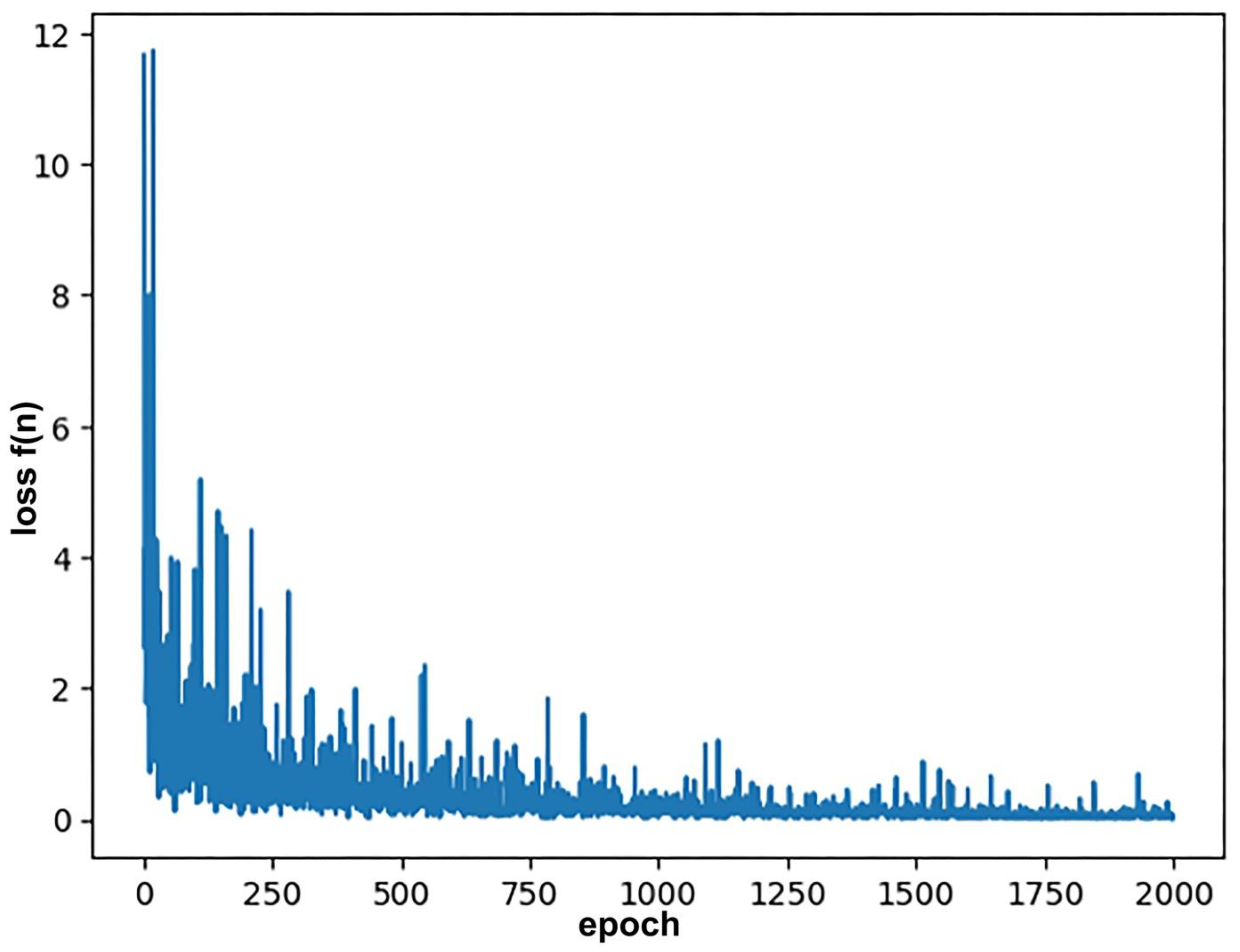
3.6. The Loss Function
3.7. AI-Generated Compounds and Model Performance
3.7.1. Molecular Diversity Visualization via t-SNE
3.7.2. Model Performance Across E3 Ligases
3.7.3. E3 Ligase Protein-Specific Structural Insights into AI-Generated Compounds
3.8. Target-Specific Docking Validates the Precision of AI-Generated Compounds
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Selkoe, D.J.; Hardy, J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol Med 2016, 8, 595-608. [CrossRef]
- Hoe, H.S.; Lee, H.K.; Pak, D.T. The upside of APP at synapses. CNS Neurosci Ther 2012, 18, 47-56. [CrossRef]
- Haass, C.; Selkoe, D.J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol 2007, 8, 101-112. [CrossRef]
- Koch, L. Non-coding RNA: A protective role for TERRA at telomeres. Nat Rev Genet 2017, 18, 453. [CrossRef]
- Li, Y.; Schindler, S.E.; Bollinger, J.G.; Ovod, V.; Mawuenyega, K.G.; Weiner, M.W.; Shaw, L.M.; Masters, C.L.; Fowler, C.J.; Trojanowski, J.Q., et al. Validation of Plasma Amyloid-beta 42/40 for Detecting Alzheimer Disease Amyloid Plaques. Neurology 2022, 98, e688-e699. [CrossRef]
- Hampel, H.; Hardy, J.; Blennow, K.; Chen, C.; Perry, G.; Kim, S.H.; Villemagne, V.L.; Aisen, P.; Vendruscolo, M.; Iwatsubo, T., et al. The Amyloid-β Pathway in Alzheimer’s Disease. Molecular Psychiatry 2021, 26, 5481-5503. [CrossRef]
- Lu, M.; Williamson, N.; Mishra, A.; Michel, C.H.; Kaminski, C.F.; Tunnacliffe, A.; Kaminski Schierle, G.S. Structural progression of amyloid-beta Arctic mutant aggregation in cells revealed by multiparametric imaging. The Journal of biological chemistry 2019, 294, 1478-1487. [CrossRef]
- Lin, J.Y.; Wang, W.A.; Zhang, X.; Liu, H.Y.; Zhao, X.L.; Huang, F.D. Intraneuronal accumulation of Abeta42 induces age-dependent slowing of neuronal transmission in Drosophila. Neurosci Bull 2014, 30, 185-190. [CrossRef]
- Zeidel, M.L.; Hammond, T.; Botelho, B.; Harris, H.W., Jr. Functional and structural characterization of endosomes from toad bladder epithelial cells. Am J Physiol 1992, 263, F62-76. [CrossRef]
- Hardy, J.A.; Higgins, G.A. Alzheimer's disease: the amyloid cascade hypothesis. Science 1992, 256, 184-185. [CrossRef]
- Imbimbo, B.P.; Panza, F.; Frisardi, V.; Solfrizzi, V.; D'Onofrio, G.; Logroscino, G.; Seripa, D.; Pilotto, A. Therapeutic intervention for Alzheimer's disease with gamma-secretase inhibitors: still a viable option? Expert Opin Investig Drugs 2011, 20, 325-341. [CrossRef]
- Sevigny, J.; Suhy, J.; Chiao, P.; Chen, T.; Klein, G.; Purcell, D.; Oh, J.; Verma, A.; Sampat, M.; Barakos, J. Amyloid PET Screening for Enrichment of Early-Stage Alzheimer Disease Clinical Trials: Experience in a Phase 1b Clinical Trial. Alzheimer Dis Assoc Disord 2016, 30, 1-7. [CrossRef]
- van Dyck, C.H.; Swanson, C.J.; Aisen, P.; Bateman, R.J.; Chen, C.; Gee, M.; Kanekiyo, M.; Li, D.; Reyderman, L.; Cohen, S., et al. Lecanemab in Early Alzheimer's Disease. N Engl J Med 2023, 388, 9-21. [CrossRef]
- Menzies, F.M.; Fleming, A.; Caricasole, A.; Bento, C.F.; Andrews, S.P.; Ashkenazi, A.; Fullgrabe, J.; Jackson, A.; Jimenez Sanchez, M.; Karabiyik, C., et al. Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron 2017, 93, 1015-1034. [CrossRef]
- Elsworthy, R.J.; Crowe, J.A.; King, M.C.; Dunleavy, C.; Fisher, E.; Ludlam, A.; Parri, H.R.; Hill, E.J.; Aldred, S. The effect of citalopram treatment on amyloid-beta precursor protein processing and oxidative stress in human hNSC-derived neurons. Transl Psychiatry 2022, 12, 285. [CrossRef]
- Chung, C.W.; Stephens, A.D.; Konno, T.; Ward, E.; Avezov, E.; Kaminski, C.F.; Hassanali, A.A.; Kaminski Schierle, G.S. Intracellular Abeta42 Aggregation Leads to Cellular Thermogenesis. J Am Chem Soc 2022, 144, 10034-10041. [CrossRef]
- Ohyagi, Y.; Tsuruta, Y.; Motomura, K.; Miyoshi, K.; Kikuchi, H.; Iwaki, T.; Taniwaki, T.; Kira, J. Intraneuronal amyloid beta42 enhanced by heating but counteracted by formic acid. J Neurosci Methods 2007, 159, 134-138. [CrossRef]
- Kitajima, Y.; Yoshioka, K.; Suzuki, N. The ubiquitin–proteasome system in regulation of the skeletal muscle homeostasis and atrophy: from basic science to disorders. The Journal of Physiological Sciences 2020, 70, 40. [CrossRef]
- Zhang, Y.; Chen, H.; Li, R.; Sterling, K.; Song, W. Amyloid β-based therapy for Alzheimer’s disease: challenges, successes and future. Signal Transduction and Targeted Therapy 2023, 8, 248. [CrossRef]
- Wertz, I.E.; Wang, X. From Discovery to Bedside: Targeting the Ubiquitin System. Cell Chem Biol 2019, 26, 156-177. [CrossRef]
- Thibaudeau, T.A.; Anderson, R.T.; Smith, D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat Commun 2018, 9, 1097. [CrossRef]
- King, E.A.; Cho, Y.; Hsu, N.S.; Dovala, D.; McKenna, J.M.; Tallarico, J.A.; Schirle, M.; Nomura, D.K. Chemoproteomics-enabled discovery of a covalent molecular glue degrader targeting NF-kappaB. Cell Chem Biol 2023, 30, 394-402 e399. [CrossRef]
- Hanzl, A.; Winter, G.E. Targeted protein degradation: current and future challenges. Curr Opin Chem Biol 2020, 56, 35-41. [CrossRef]
- Potjewyd, F.M.; Axtman, A.D. Exploration of Aberrant E3 Ligases Implicated in Alzheimer's Disease and Development of Chemical Tools to Modulate Their Function. Front Cell Neurosci 2021, 15, 768655. [CrossRef]
- Ivanenkov, Y.A.; Polykovskiy, D.; Bezrukov, D.; Zagribelnyy, B.; Aladinskiy, V.; Kamya, P.; Aliper, A.; Ren, F.; Zhavoronkov, A. Chemistry42: An AI-Driven Platform for Molecular Design and Optimization. J Chem Inf Model 2023, 63, 695-701. [CrossRef]
- McNair, D. Artificial Intelligence and Machine Learning for Lead-to-Candidate Decision-Making and Beyond. Annu Rev Pharmacol Toxicol 2023, 63, 77-97. [CrossRef]
- Singh, S.; Gupta, H.; Sharma, P.; Sahi, S. Advances in Artificial Intelligence (AI)-assisted approaches in drug screening. Artificial Intelligence Chemistry 2024, 2, 100039. [CrossRef]
- Paul, D.; Sanap, G.; Shenoy, S.; Kalyane, D.; Kalia, K.; Tekade, R.K. Artificial intelligence in drug discovery and development. Drug Discov Today 2021, 26, 80-93. [CrossRef]
- Perron, Q.; Mirguet, O.; Tajmouati, H.; Skiredj, A.; Rojas, A.; Gohier, A.; Ducrot, P.; Bourguignon, M.P.; Sansilvestri-Morel, P.; Do Huu, N., et al. Deep generative models for ligand-based de novo design applied to multi-parametric optimization. J Comput Chem 2022, 43, 692-703. [CrossRef]
- Luukkonen, S.; van den Maagdenberg, H.W.; Emmerich, M.T.M.; van Westen, G.J.P. Artificial intelligence in multi-objective drug design. Current Opinion in Structural Biology 2023, 79, 102537. [CrossRef]
- Ramirez-Mora, T.; Retana-Lobo, C.; Valle-Bourrouet, G. Biochemical characterization of extracellular polymeric substances from endodontic biofilms. PLoS One 2018, 13, e0204081. [CrossRef]
- Jin, W.; Barzilay, R.; Jaakkola, T. Junction Tree Variational Autoencoder for Molecular Graph Generation. arXiv [cs.LG] 2019.
- Truong, G.-B.; Pham, T.-A.; To, V.-T.; Le, H.-S.L.; Van Nguyen, P.-C.; Trinh, T.-C.; Phan, T.-L.; Truong, T.N. Discovery of Vascular Endothelial Growth Factor Receptor 2 Inhibitors Employing Junction Tree Variational Autoencoder with Bayesian Optimization and Gradient Ascent. ACS Omega 2024, 9, 47180-47193. [CrossRef]
- Kondratyev, V.; Dryzhakov, M.; Gimadiev, T.; Slutskiy, D. Generative model based on junction tree variational autoencoder for HOMO value prediction and molecular optimization. Journal of Cheminformatics 2023, 15, 11. [CrossRef]
- Arus-Pous, J.; Johansson, S.V.; Prykhodko, O.; Bjerrum, E.J.; Tyrchan, C.; Reymond, J.L.; Chen, H.; Engkvist, O. Randomized SMILES strings improve the quality of molecular generative models. J Cheminform 2019, 11, 71. [CrossRef]
- Reiser, P.; Neubert, M.; Eberhard, A.; Torresi, L.; Zhou, C.; Shao, C.; Metni, H.; van Hoesel, C.; Schopmans, H.; Sommer, T., et al. Graph neural networks for materials science and chemistry. Communications Materials 2022, 3, 93. [CrossRef]
- Meyers, J.; Fabian, B.; Brown, N. De novo molecular design and generative models. Drug Discovery Today 2021, 26, 2707-2715. [CrossRef]
- Hamidizadeh, A.; Shen, T.; Ester, M. Semi-Supervised Junction Tree Variational Autoencoder for Molecular Property Prediction. arXiv [cs.LG] 2023.
- Yang, Z.; Hu, Z.; Salakhutdinov, R.; Berg-Kirkpatrick, T. Improved Variational Autoencoders for Text Modeling using Dilated Convolutions. arXiv [cs.NE] 2017.
- Rigoni, D.; Navarin, N.; Sperduti, A. RGCVAE: relational graph conditioned variational autoencoder for molecule design. Machine Learning 2025, 114, 47. [CrossRef]
- McLoughlin, K.S.; Shi, D.; Mast, J.E.; Bucci, J.; Williams, J.P.; Jones, W.D.; Miyao, D.; Nam, L.; Osswald, H.L.; Zegelman, L., et al. Generative Molecular Design and Experimental Validation of Selective Histamine H1 Inhibitors. bioRxiv 2023. [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res 2000, 28, 235-242. [CrossRef]
- Lee, M.; Yau, W.-M.; Louis, J.M.; Tycko, R. Structures of brain-derived 42-residue amyloid-β fibril polymorphs with unusual molecular conformations and intermolecular interactions. Proceedings of the National Academy of Sciences 2023, 120, e2218831120. [CrossRef]
- Dey, A.; Verma, A.; Bhaskar, U.; Sarkar, B.; Kallianpur, M.; Vishvakarma, V.; Das, A.K.; Garai, K.; Mukherjee, O.; Ishii, K., et al. A Toxicogenic Interaction between Intracellular Amyloid-beta and Apolipoprotein-E. ACS Chem Neurosci 2024, 15, 1265-1275. [CrossRef]
- Bhachoo, J.; Beuming, T. Investigating Protein-Peptide Interactions Using the Schrodinger Computational Suite. Methods Mol Biol 2017, 1561, 235-254. [CrossRef]
- Hsu, P.W.; Lin, L.Z.; Hsu, S.D.; Hsu, J.B.; Huang, H.D. ViTa: prediction of host microRNAs targets on viruses. Nucleic Acids Res 2007, 35, D381-385. [CrossRef]
- Zdrazil, B.; Felix, E.; Hunter, F.; Manners, E.J.; Blackshaw, J.; Corbett, S.; de Veij, M.; Ioannidis, H.; Lopez, D.M.; Mosquera, J.F., et al. The ChEMBL Database in 2023: a drug discovery platform spanning multiple bioactivity data types and time periods. Nucleic Acids Res 2024, 52, D1180-D1192. [CrossRef]
- Shaikh, F.; Siu, S.W. Identification of novel natural compound inhibitors for human complement component 5a receptor by homology modeling and virtual screening. Med Chem Res 2016, 25, 1564-1573. [CrossRef]
- Jin, W.; Barzilay, R.; Jaakkola, T.S. Junction Tree Variational Autoencoder for Molecular Graph Generation. CoRR 2018, abs/1802.04364.
- Jin, W.; Barzilay, R.; Jaakkola, T. Junction Tree Variational Autoencoder for Molecular Graph Generation. In Proceedings of Proceedings of the 35th International Conference on Machine Learning, 2018; pp. 2323-2332.
- Karauda, T.; Kornicki, K.; Jarri, A.; Antczak, A.; Milkowska-Dymanowska, J.; Piotrowski, W.J.; Majewski, S.; Gorski, P.; Bialas, A.J. Eosinopenia and neutrophil-to-lymphocyte count ratio as prognostic factors in exacerbation of COPD. Sci Rep 2021, 11, 4804. [CrossRef]
- Thireou, T.; Reczko, M. Bidirectional Long Short-Term Memory Networks for predicting the subcellular localization of eukaryotic proteins. IEEE/ACM Trans Comput Biol Bioinform 2007, 4, 441-446. [CrossRef]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. arXiv [stat.ML] 2022.
- Csiszar, I. I-Divergence Geometry of Probability Distributions and Minimization Problems. Ann. Probab 1975, 3, 146-158. [CrossRef]
- Colaboratory, G. Jupyter Notebook. Availabe online: https://colab.research.google.com (accessed on 2024).
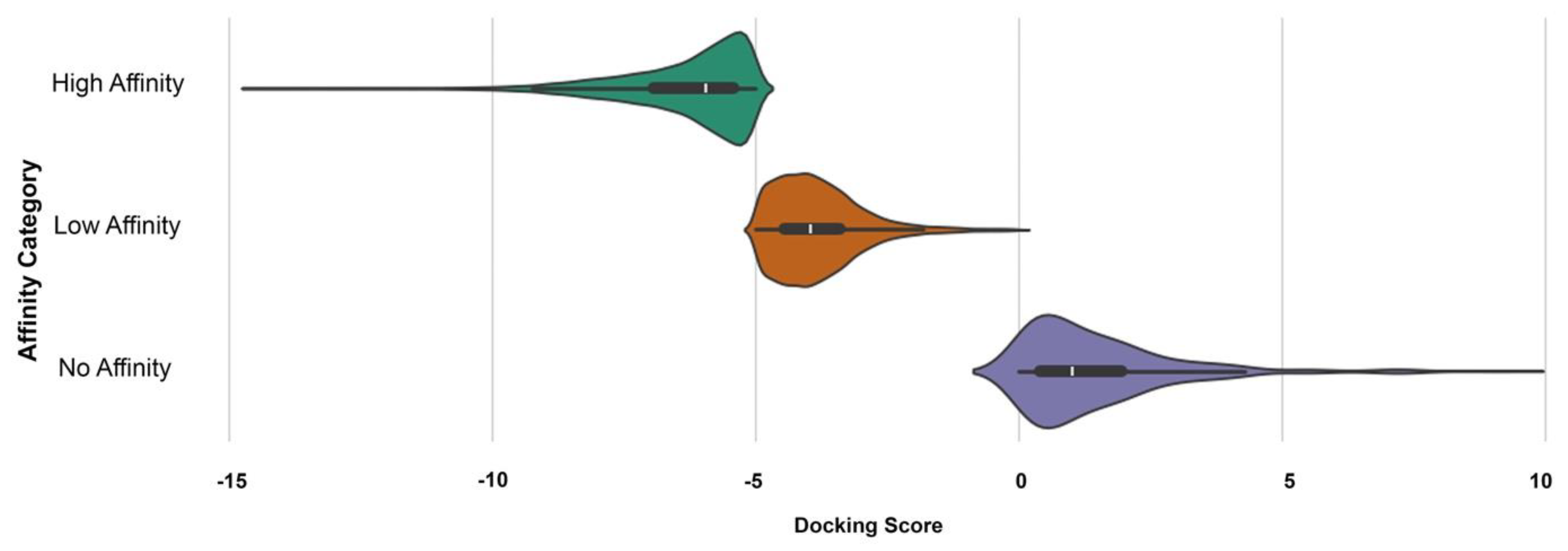
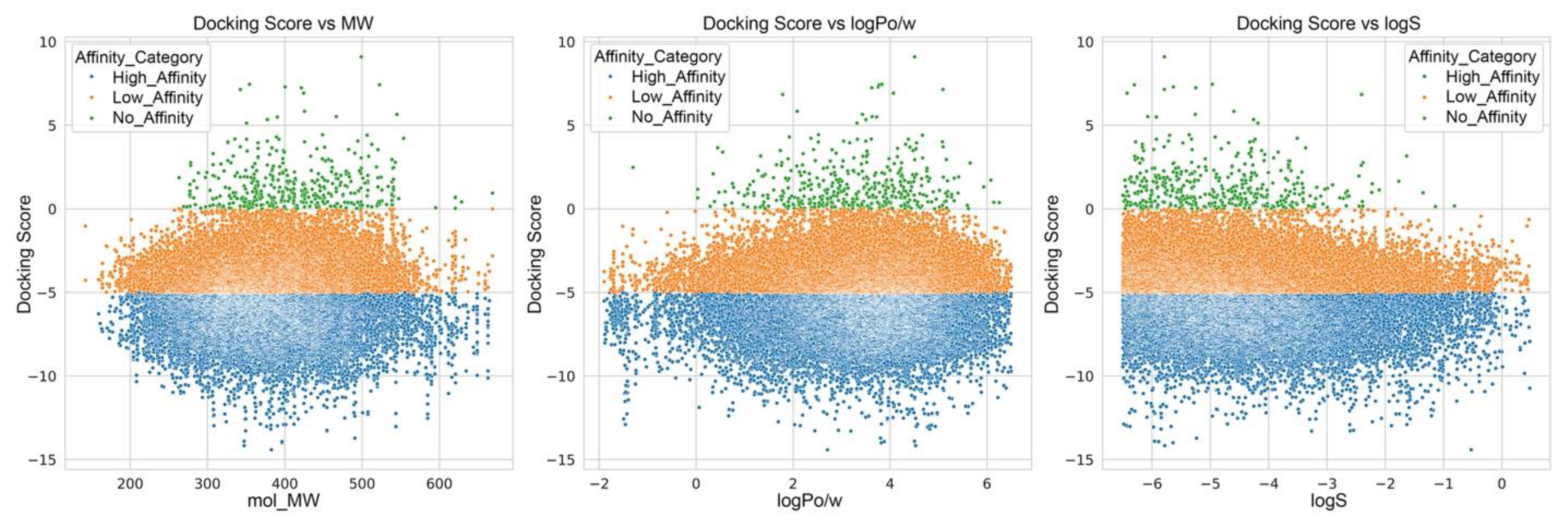
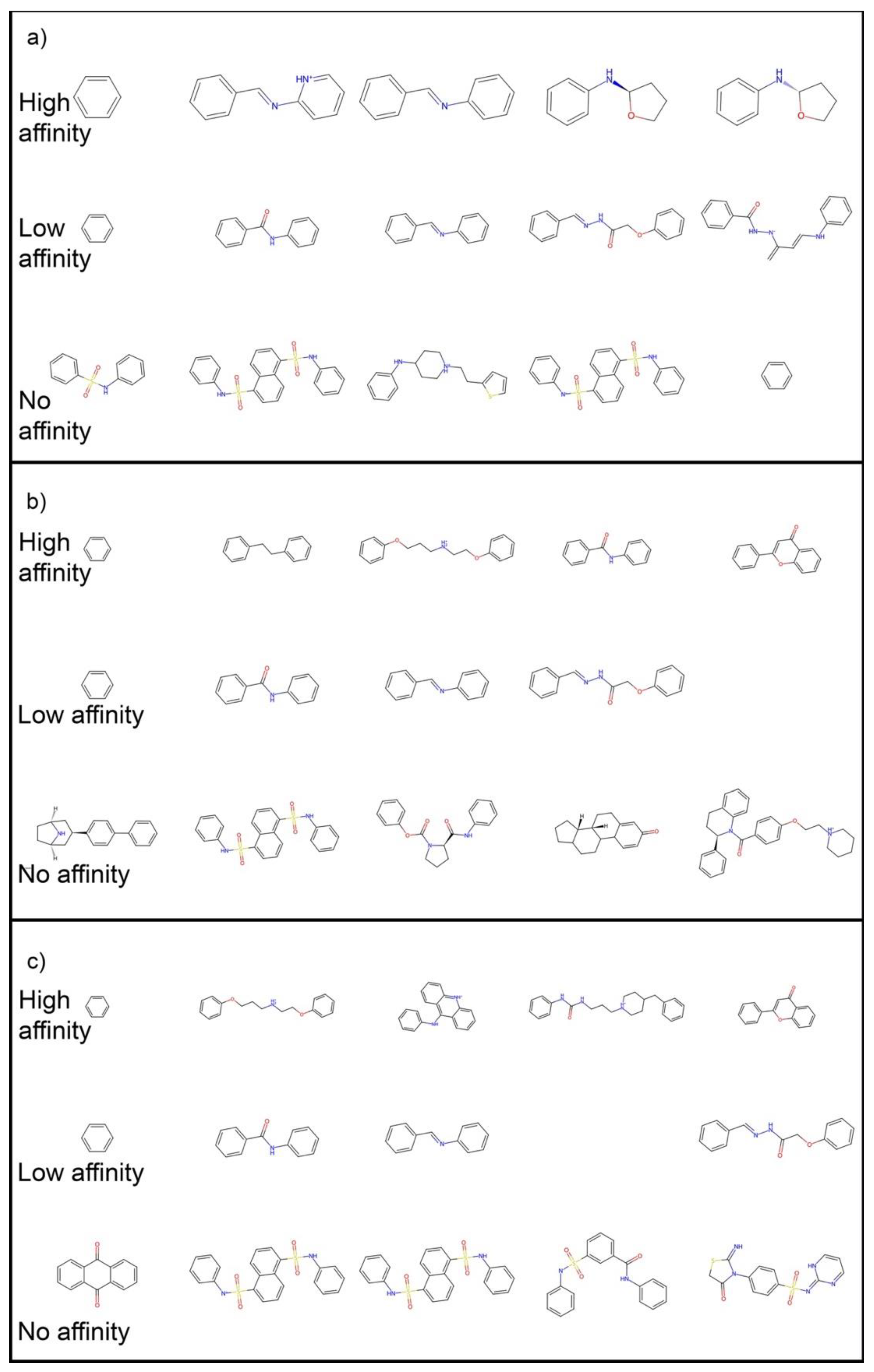
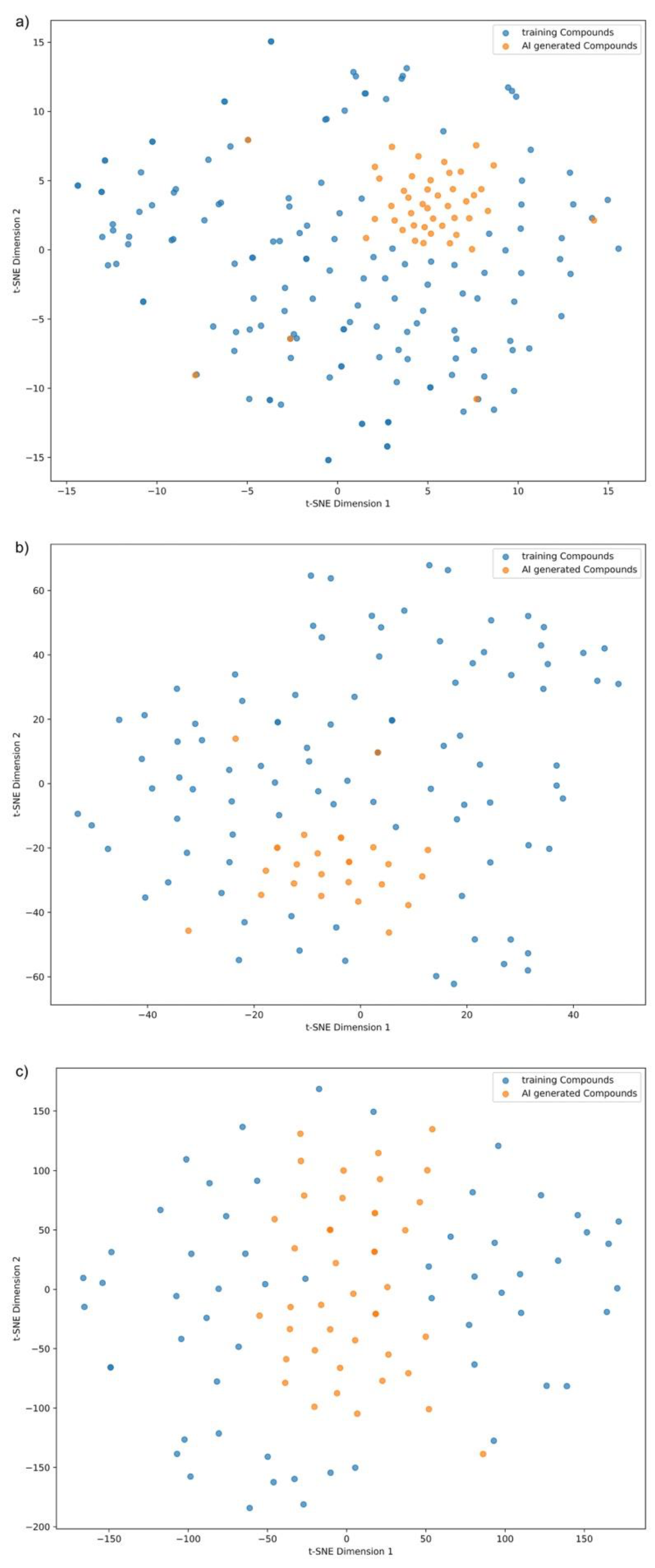
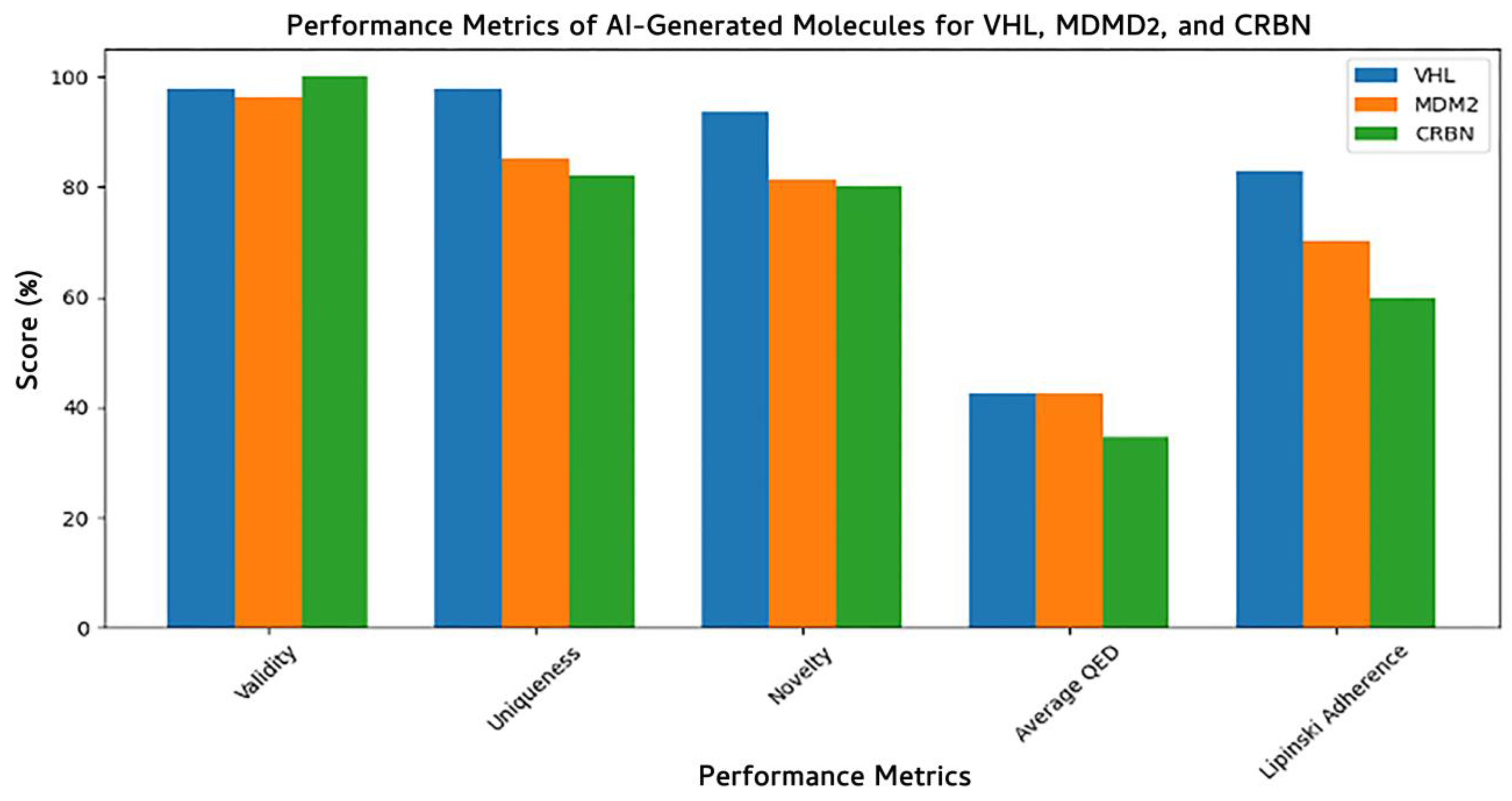
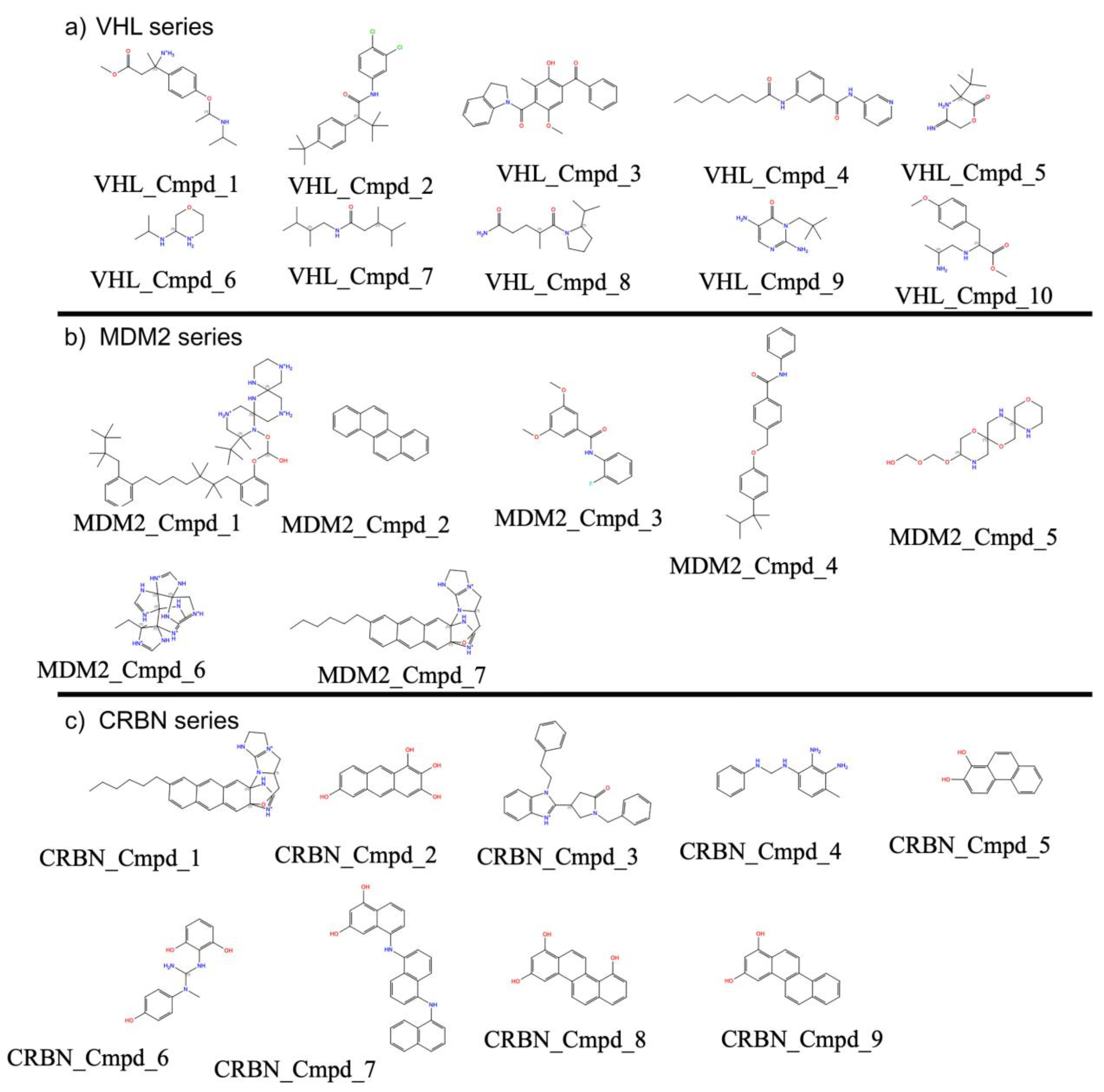
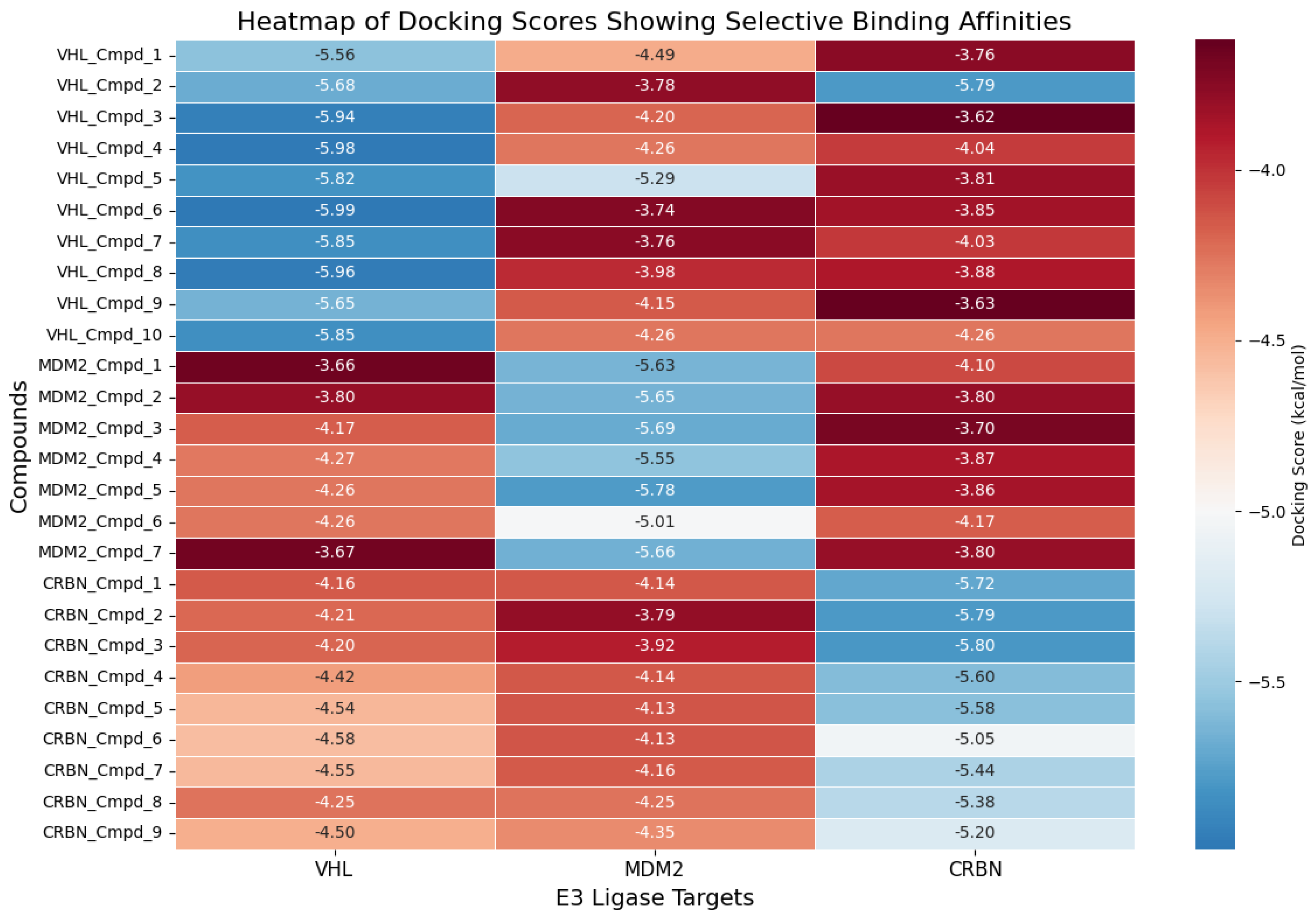
| Property | Count | Mean | Std Dev | Min | 25% | 50% | 75% | Max |
|---|---|---|---|---|---|---|---|---|
| MW | 65998 | 366.68 | 70.31 | 142.24 | 316.42 | 361.35 | 411.45 | 668.72 |
| logPo/w | 65998 | 3.39 | 1.29 | -1.89 | 2.60 | 3.54 | 4.31 | 6.49 |
| logS | 65998 | -4.58 | 1.29 | -6.5 | -5.59 | -4.77 | -3.83 | 0.47 |
| #metab | 65998 | 3.64 | 1.76 | 1 | 2 | 3 | 5 | 8 |
| Rule-Of-Five | 65998 | 0.14 | 0.35 | 0 | 0 | 0 | 0 | 1 |
| Ligase | Library | High_Affinity | Low_Affinity | No_Affinity | Total |
|---|---|---|---|---|---|
| CRBN | Chemble | 4834 | 7448 | 36 | 12318 |
| Vitas | 1748 | 11949 | 73 | 13770 | |
| MDM2 | Chemble | 131 | 51 | 0 | 182 |
| Vitas | 2302 | 11361 | 86 | 13749 | |
| VHL | Chemble | 8792 | 3414 | 80 | 12286 |
| Vitas | 6607 | 6949 | 137 | 13693 | |
| Total | 65998 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





