Submitted:
25 April 2025
Posted:
28 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. rHFSCs-Derived Secretome and Exosomes Isolation
2.2. rHFSCs-Derived Exosomes Analysis
2.3. Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
2.2.1. RNA Isolation and cDNA Synthesis
2.2.2. Quantitative RT-PCR Assay
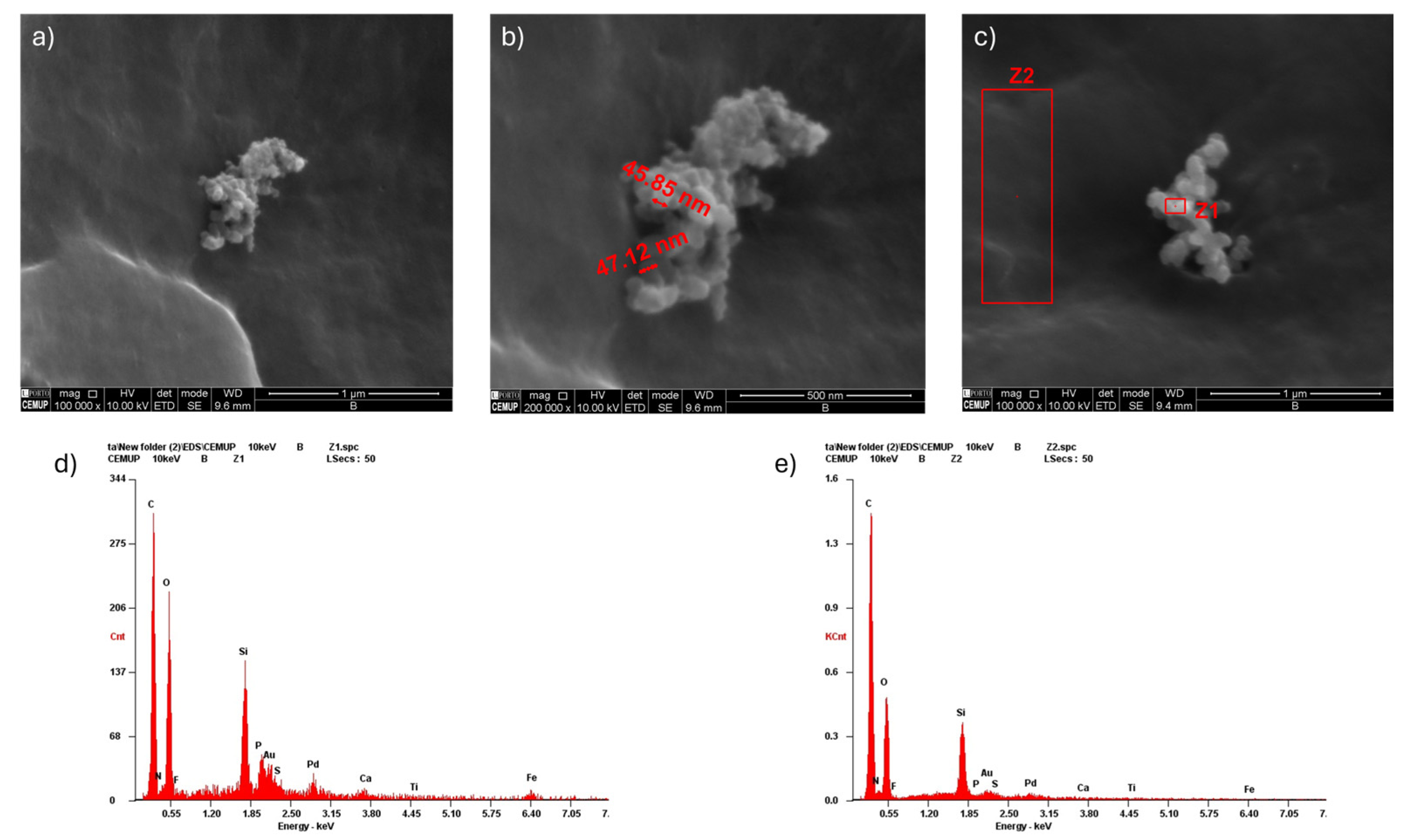
2.4. Scanning Electron Microscopy (SEM) and Energy Dispersive Spectroscopy (EDS)
2.5. Total Protein Quantification
2.6. Prestoblue™ Assay
2.7. Scratch Assay
2.8. Statistical Analysis
3. Results
3.1. rHFSCs-Derived Exosomes Analysis and Comparison to Secretome
3.2. RT-PCR
3.3. Scanning Electron Microscopy (SEM) with Energy-Dispersive X-Ray Spectroscopy (EDS)
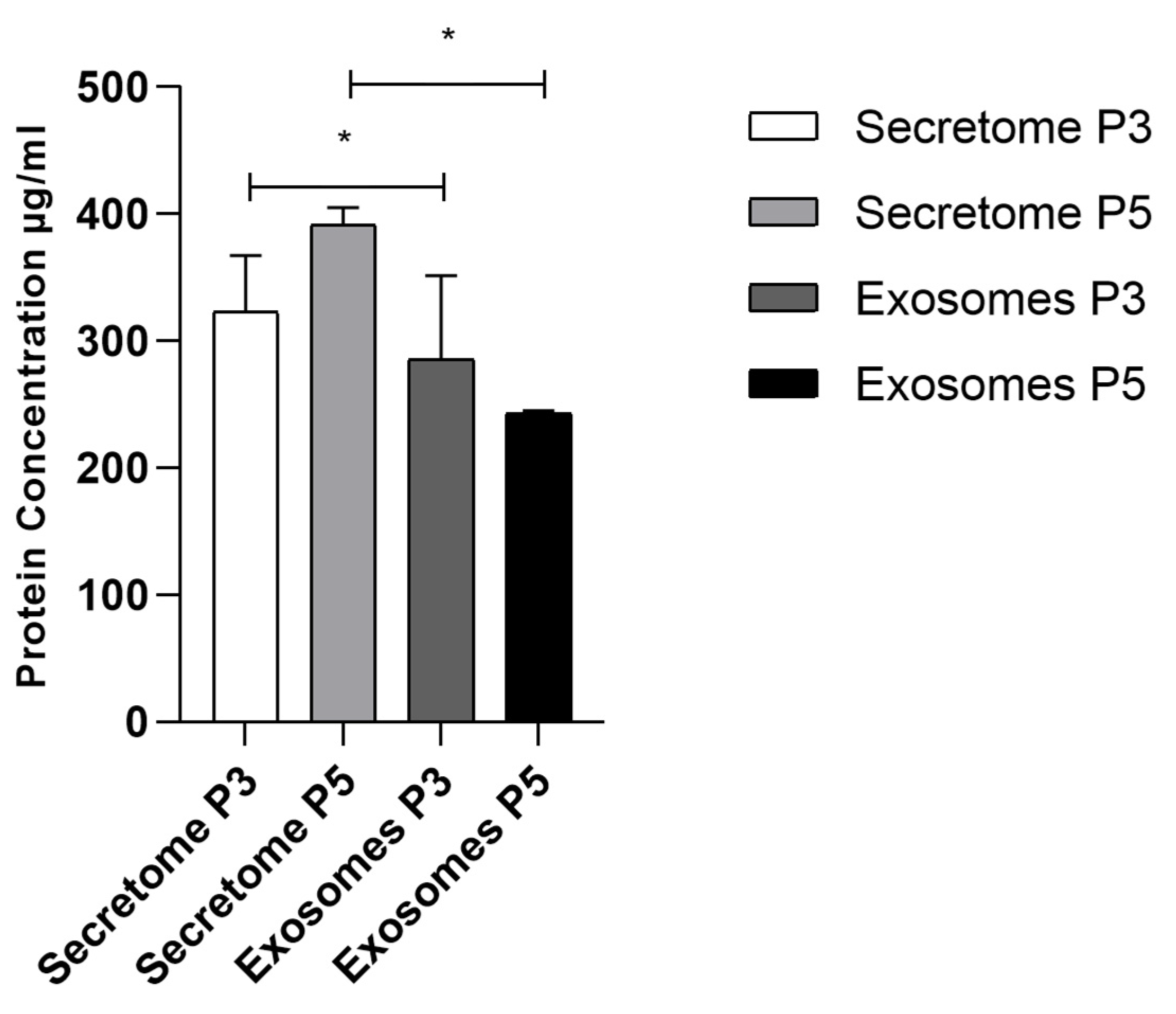
3.4. Total Protein Quantification
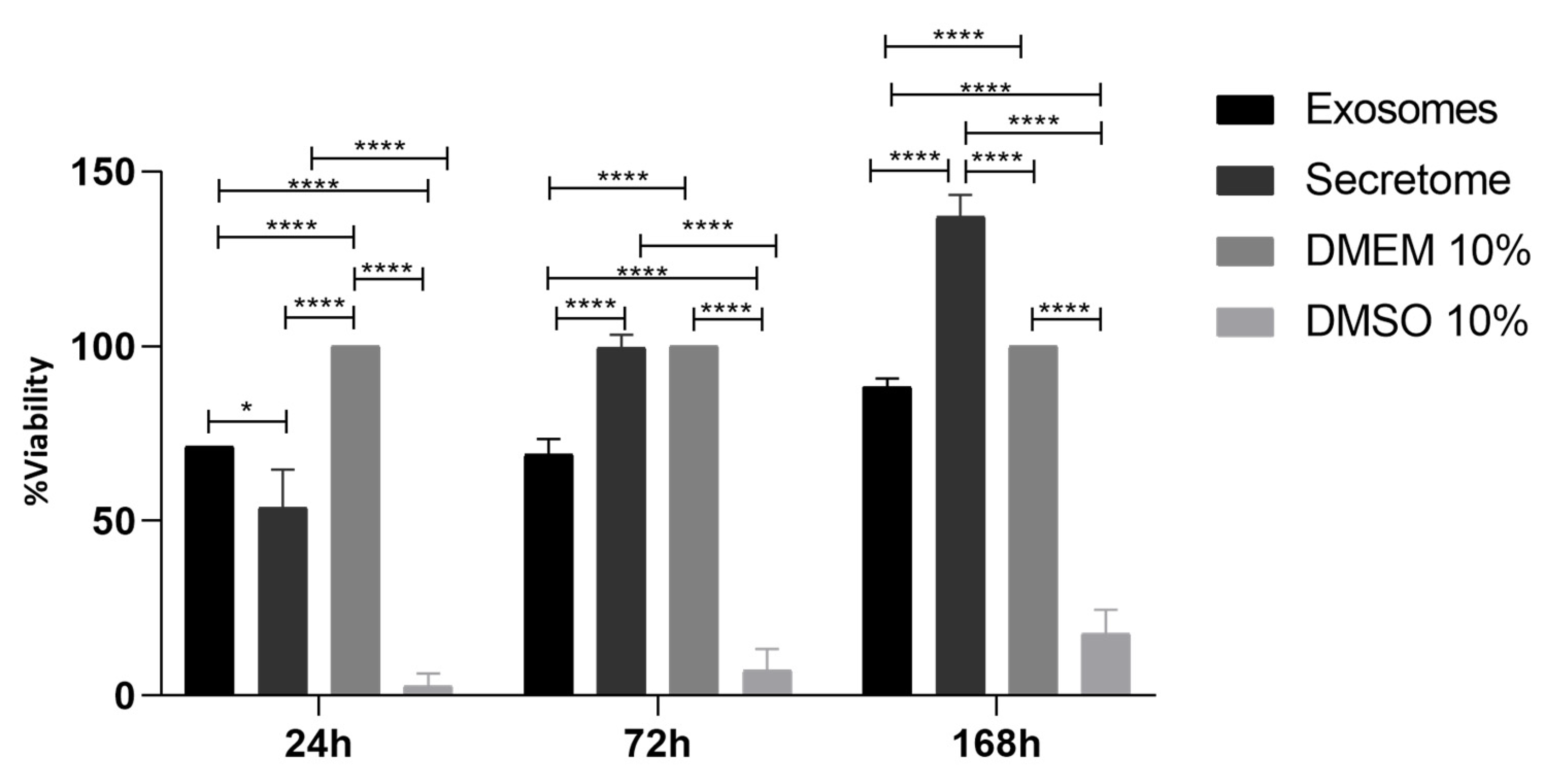
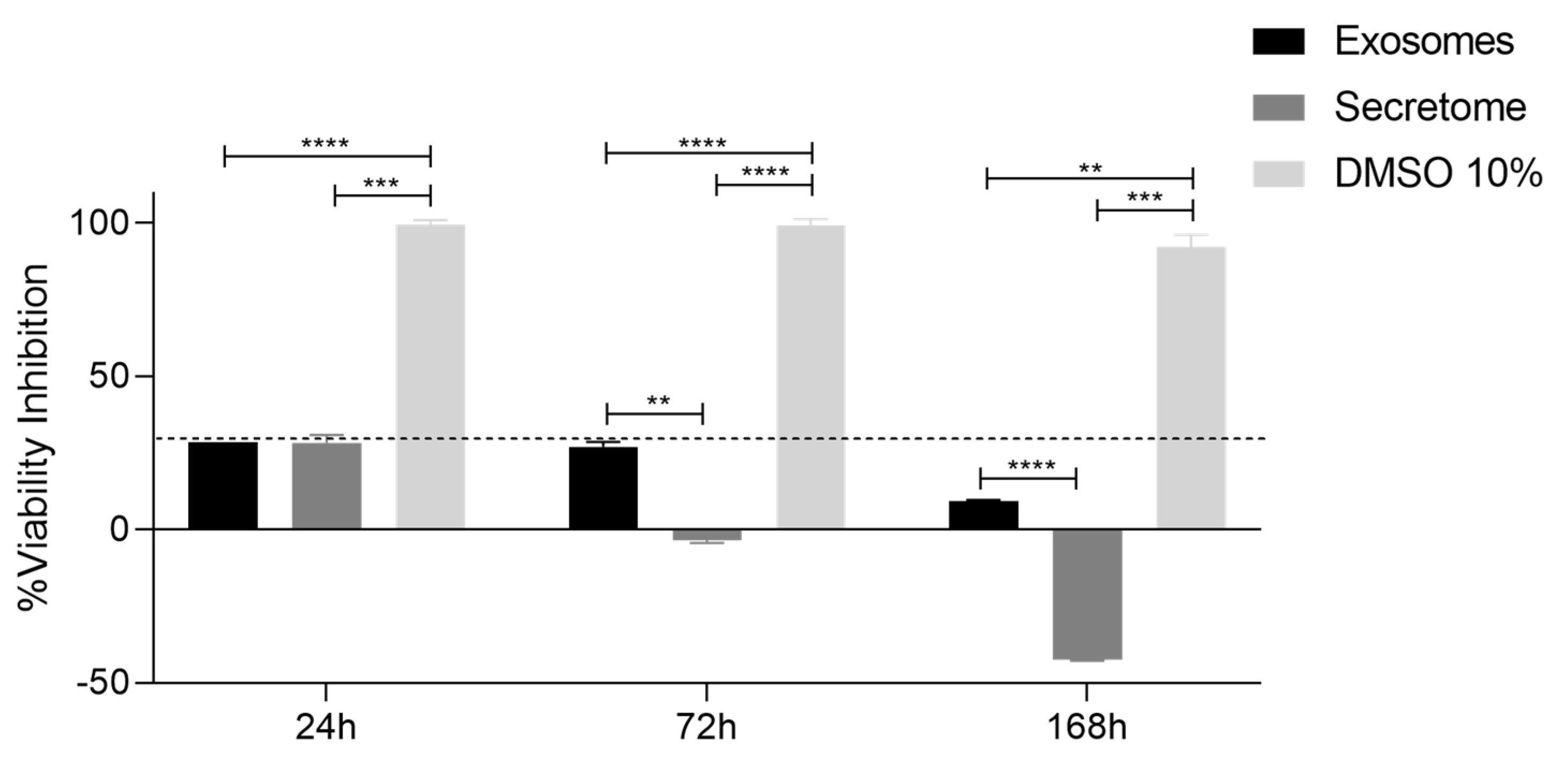
3.5. Prestoblue™ Assay
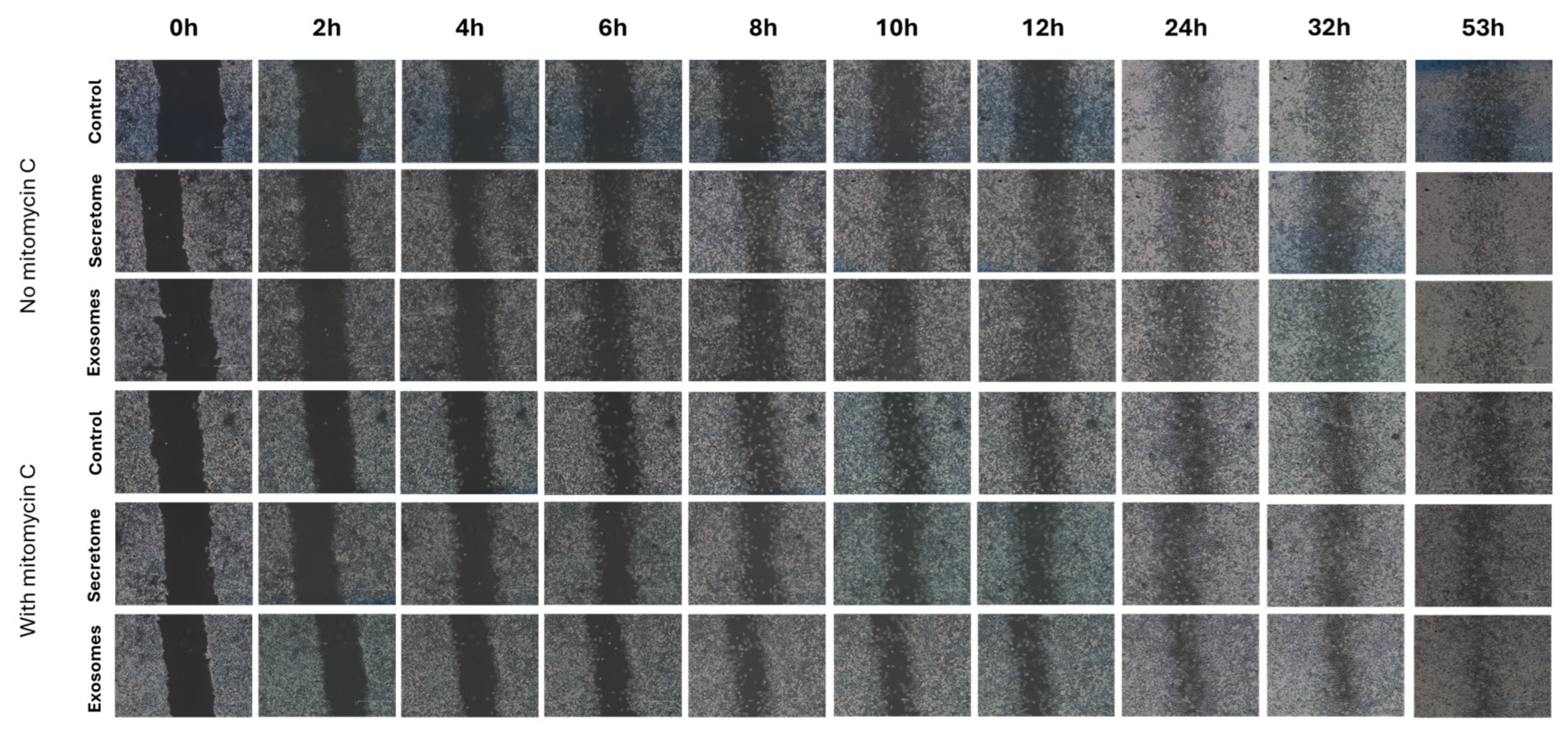
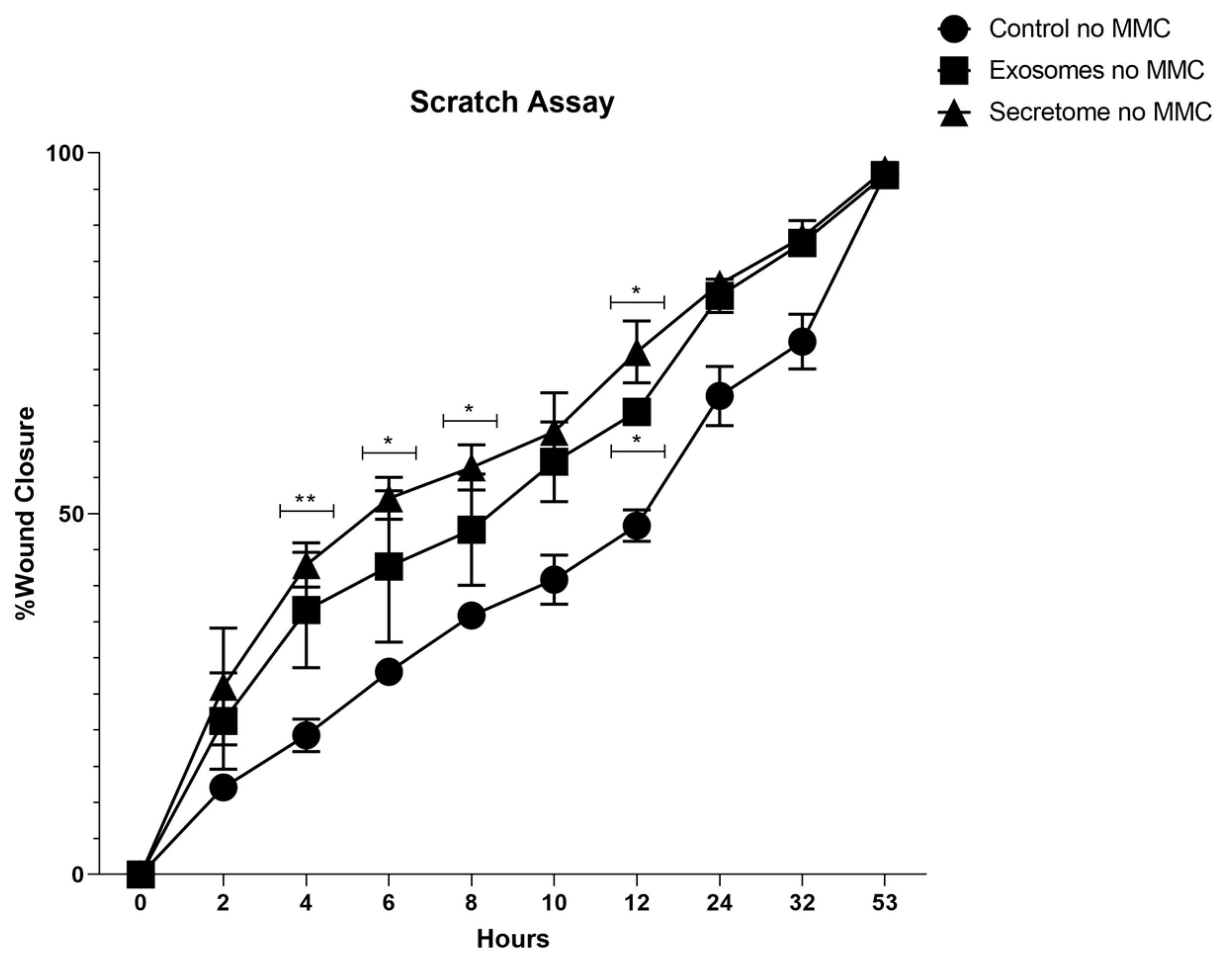
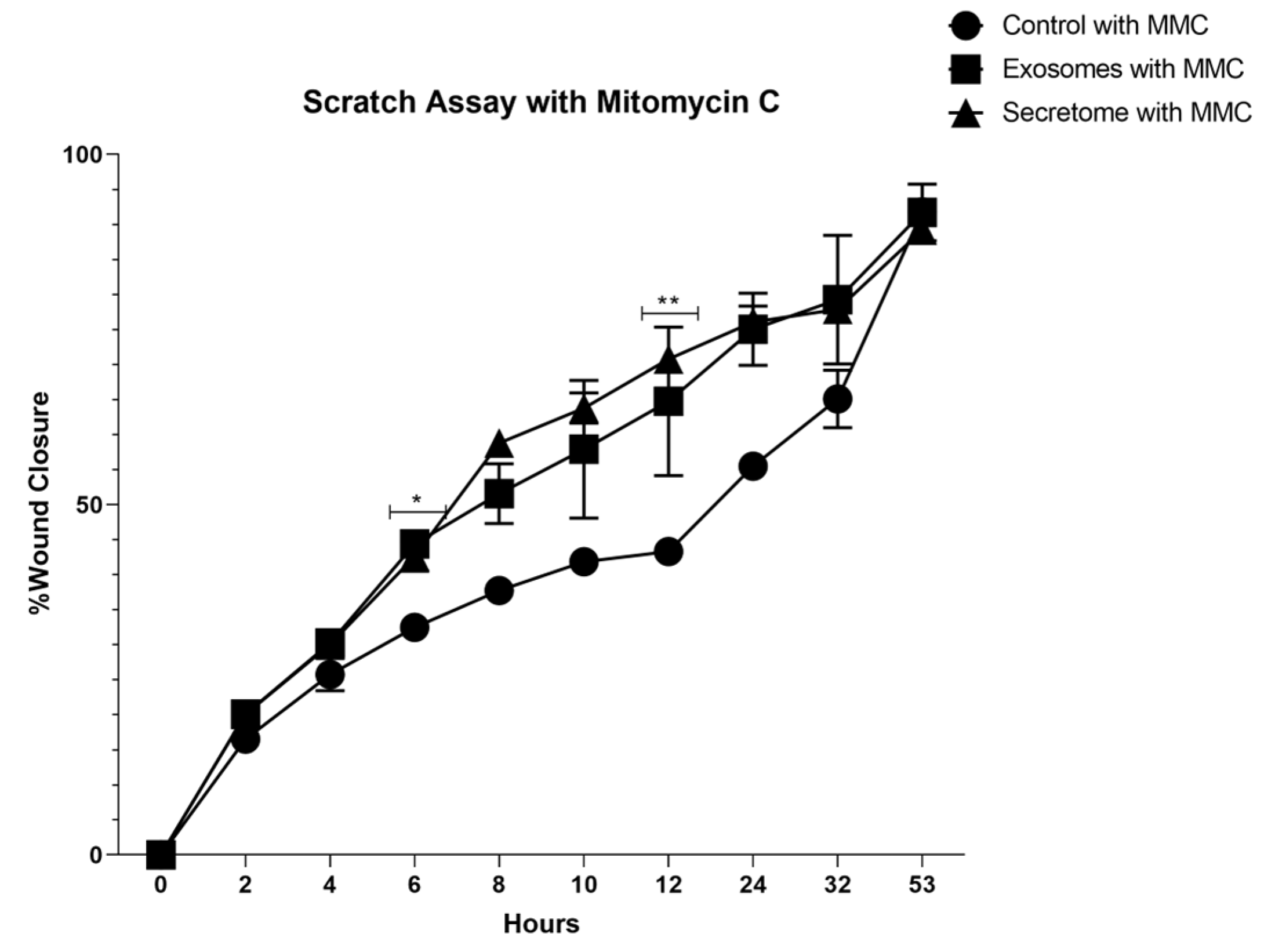
3.6. Scratch Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACTB | Beta-Actin |
| Au | Gold |
| BCA | Bicinchoninic Acid |
| C | Carbon |
| Ca | Calcium |
| CM2D | Conditioned Medium 2D |
| Ct | Threshold cycle |
| DMSO | Dimethyl Sulfoxide |
| EBSD | Electron Backscattered Diffraction |
| ECM | Extracellular Matrix |
| EDS | Energy-Dispersive X-ray Spectroscopy |
| EGF | Epidermal Growth Factor Recombinant Protein |
| EVs | Extracellular Vesicles |
| FBS | Bovine Fetal Serum |
| Fe | Iron |
| GAPDH | Glyceraldehyde 3-phosphate Dehydrogenase |
| G-CSF | Granulocyte Colony-Stimulating Factor |
| GM-CSF | Granulocyte-Macrophage Colony-Stimulating Factor |
| GRO/KC/CINC-1 | Human Growth-Regulated Oncogene/Keratinocyte Chemoattractant/ Cytokine-Induced Neutrophil Chemoattractant-1 |
| HMDS | Hexamethyldisilazane |
| IFNγ | Interferon Gamma |
| IL | Interleukin |
| IP-10 | Interferon-Gamma Inducible Protein |
| MCP-1 | Monocyte Chemoattractant Protein-1 |
| MIP | Macrophage Inflammatory Protein |
| MMC | Mitomycin C |
| N | Nitrogen |
| O | Oxygen |
| P | Phosphorus |
| PCR | Polymerase Chain Reaction |
| Pd | Palladium |
| RANTES | Regulated upon Activation, Normal T Cell Expressed and Presumably Secreted |
| rHFSCs | Rat Hair Follicle Stem Cells |
| RT-PCR | Reverse Transcriptase Polymerase Chain Reaction |
| S | Sulfur |
| SEM | Standard Error of the Mean |
| Si | Silicon |
| TGFβ | Transforming Growth Factor Beta |
| Ti | Titanium |
| TNFα | Tumor Necrosis Factor-Alpha |
| UP | University of Porto |
| VEGF | Vascular Endothelial Growth Factor |
| Z | Zone |
References
- Sousa, P., et al., Advancements and Insights in Exosome-Based Therapies for Wound Healing: A Comprehensive Systematic Review (2018–June 2023). Biomedicines, 2023. 11(8): p. 2099.
- Sousa, P., et al., Isolation, Expansion, and Characterization of Rat Hair Follicle Stem Cells and Their Secretome: Insights into Wound Healing Potential. Biomedicines, 2024. 12(12): p. 2854.
- Lopes, B., et al., The Application of Mesenchymal Stem Cells on Wound Repair and Regeneration. Applied Sciences, 2021. 11(7): p. 3000.
- Zhou, C., et al., Stem cell-derived exosomes: emerging therapeutic opportunities for wound healing. Stem Cell Res Ther, 2023. 14(1): p. 107.
- Cerqueira, M.T.e.a., Using Stem Cells in Skin Regeneration: Possibilities and Reality. Stem Cells and Development, 2012. 21(8): p. 1201-1214.
- Rezaie, F., M. Momeni-Moghaddam, and H. Naderi-Meshkin, Regeneration and Repair of Skin Wounds: Various Strategies for Treatment. The International Journal of Lower Extremity Wounds, 2019. 18(3): p. 247-261.
- Dean, J., et al., Advancements in bioengineered and autologous skin grafting techniques for skin reconstruction: a comprehensive review. Front Bioeng Biotechnol, 2024. 12: p. 1461328.
- do Amaral, R.J.F.C., et al., Functionalising Collagen-Based Scaffolds With Platelet-Rich Plasma for Enhanced Skin Wound Healing Potential. Frontiers in Bioengineering and Biotechnology, 2019. Volume 7 - 2019.
- Chocarro-Wrona, C., et al., Therapeutic strategies for skin regeneration based on biomedical substitutes. Journal of the European Academy of Dermatology and Venereology, 2019. 33(3): p. 484-496.
- Zhao, M., et al., Advances on Graphene-Based Nanomaterials and Mesenchymal Stem Cell-Derived Exosomes Applied in Cutaneous Wound Healing. Int J Nanomedicine, 2021. 16: p. 2647-2665.
- Vizoso, F.J., et al., Mesenchymal Stem Cell Secretome: Toward Cell-Free Therapeutic Strategies in Regenerative Medicine. International Journal of Molecular Sciences, 2017. 18(9): p. 1852.
- Teixeira, F.G. and A.J. Salgado, Mesenchymal stem cells secretome: current trends and future challenges. Neural Regen Res, 2020. 15(1): p. 75-77.
- Bormann, D., et al., Therapeutic Application of Cell Secretomes in Cutaneous Wound Healing. Journal of Investigative Dermatology, 2023. 143(6): p. 893-912.
- Md Fadilah, N.I., et al., Cell secretomes for wound healing and tissue regeneration: Next generation acellular based tissue engineered products. J Tissue Eng, 2022. 13: p. 20417314221114273.
- Suhandi, C., et al., The Effect of Stem Cell Secretome on the Improvement of Diabetic Wound Recovery: A Systematic Review and Meta-Analysis of In Vivo Studies. Current Therapeutic Research, 2025: p. 100778.
- Suhandi, C., et al., Effectiveness of Mesenchymal Stem Cell Secretome on Wound Healing: A Systematic Review and Meta-analysis. Tissue Engineering and Regenerative Medicine, 2023. 20(7): p. 1053-1062.
- Marques da Silva, M., et al., Mesenchymal Stromal Cell Secretome for Therapeutic Application in Skin Wound Healing: A Systematic Review of Preclinical Studies. Cells Tissues Organs, 2022. 212(6): p. 567-582.
- Kim, J.H., et al., Identification and characterization of stem cell secretome-based recombinant proteins for wound healing applications. Frontiers in Bioengineering and Biotechnology, 2022. 10.
- Prasai, A., et al., Role of Exosomes in Dermal Wound Healing: A Systematic Review. Journal of Investigative Dermatology, 2022. 142(3, Part A): p. 662-678.e8.
- Ye, H., et al., Advancements in engineered exosomes for wound repair: current research and future perspectives. Frontiers in Bioengineering and Biotechnology, 2023. 11.
- Lv, H., et al., Exosome derived from stem cell: A promising therapeutics for wound healing. Front Pharmacol, 2022. 13: p. 957771.
- Armstrong, J.P., M.N. Holme, and M.M. Stevens, Re-Engineering Extracellular Vesicles as Smart Nanoscale Therapeutics. ACS Nano, 2017. 11(1): p. 69-83.
- Zhu, Z., et al., Exosomes Derived From Umbilical Cord Mesenchymal Stem Cells Treat Cutaneous Nerve Damage and Promote Wound Healing. Front Cell Neurosci, 2022. 16: p. 913009.
- Zhu, J. and H. Quan, Adipose-derived stem cells-derived exosomes facilitate cutaneous wound healing by delivering XIST and restoring discoidin domain receptor 2. Cytokine, 2022. 158: p. 155981.
- Zhou, Z., et al., Exosomes derived from dental pulp stem cells accelerate cutaneous wound healing by enhancing angiogenesis via the Cdc42/p38 MAPK pathway. Int J Mol Med, 2022. 50(6).
- Tienda-Vázquez, M.A., et al., Exosomes: A Promising Strategy for Repair, Regeneration and Treatment of Skin Disorders. Cells, 2023. 12(12).
- Golchin, A., et al., Combination Therapy of Stem Cell-derived Exosomes and Biomaterials in the Wound Healing. Stem Cell Reviews and Reports, 2022. 18(6): p. 1892-1911.
- Han, X., et al., Exosome-coated oxygen nanobubble-laden hydrogel augments intracellular delivery of exosomes for enhanced wound healing. Nature Communications, 2024. 15(1): p. 3435.
- Narauskaitė, D., et al., Extracellular Vesicles in Skin Wound Healing. Pharmaceuticals, 2021. 14(8): p. 811.
- Sun, Y., et al., Therapeutic application of mesenchymal stem cell-derived exosomes in skin wound healing. Frontiers in Bioengineering and Biotechnology, 2024. 12.
- Qiao, Z., et al., The effectiveness of cell-derived exosome therapy for diabetic wound: A systematic review and meta-analysis. Ageing Research Reviews, 2023. 85: p. 101858.
- Zhang, W., et al., Cell-free therapy based on adipose tissue stem cell-derived exosomes promotes wound healing via the PI3K/Akt signaling pathway. Exp Cell Res, 2018. 370(2): p. 333-342.
- Ahmadpour, F., et al., Effects of exosomes derived from fibroblast cells on skin wound healing in Wistar rats. Burns, 2023.
- Shakhakarmi, K., et al., EGF, a veteran of wound healing: highlights on its mode of action, clinical applications with focus on wound treatment, and recent drug delivery strategies. Archives of Pharmacal Research, 2023. 46(4): p. 299-322.
- Choi, S.M., et al., Effects of structurally stabilized EGF and bFGF on wound healing in type I and type II diabetic mice. Acta Biomaterialia, 2018. 66: p. 325-334.
- Huang, H., et al., Granulocyte-Colony Stimulating Factor (G-CSF) Accelerates Wound Healing in Hemorrhagic Shock Rats by Enhancing Angiogenesis and Attenuating Apoptosis. Med Sci Monit, 2017. 23: p. 2644-2653.
- Shen, G.-Y., et al., Local injection of granulocyte-colony stimulating factor accelerates wound healing in a rat excisional wound model. Tissue Engineering and Regenerative Medicine, 2016. 13(3): p. 297-303.
- Goswami, A.G., et al., An appraisal of vascular endothelial growth factor (VEGF): the dynamic molecule of wound healing and its current clinical applications. Growth Factors, 2022. 40(3-4): p. 73-88.
- Wilgus, T.A. and L.A. DiPietro, Complex roles for VEGF in dermal wound healing. J Invest Dermatol, 2012. 132(2): p. 493-4.
- Wong, R.S.-Y., et al., The role of cytokines in wound healing: from mechanistic insights to therapeutic applications. Exploration of Immunology, 2025. 5: p. 1003183.
- Aslan, C., et al., Development of Interleukin-2 Loaded Chitosan-Based Nanogels Using Artificial Neural Networks and Investigating the Effects on Wound Healing in Rats. AAPS PharmSciTech, 2017. 18(4): p. 1019-1030.
- Biglari, S., et al., Simulating Inflammation in a Wound Microenvironment Using a Dermal Wound-on-a-Chip Model. Advanced Healthcare Materials, 2019. 8(1): p. 1801307.
- Nguyen, J.K., et al., The IL-4/IL-13 axis in skin fibrosis and scarring: mechanistic concepts and therapeutic targets. Archives of Dermatological Research, 2020. 312(2): p. 81-92.
- Serezani, A.P.M., et al., IL-4 impairs wound healing potential in the skin by repressing fibronectin expression. Journal of Allergy and Clinical Immunology, 2017. 139(1): p. 142-151.e5.
- Coden, M.E. and S. Berdnikovs, Eosinophils in wound healing and epithelial remodeling: Is coagulation a missing link? Journal of Leukocyte Biology, 2020. 108(1): p. 93-103.
- Singampalli, K.L., et al., The Role of an IL-10/Hyaluronan Axis in Dermal Wound Healing. Frontiers in Cell and Developmental Biology, 2020. Volume 8 - 2020.
- Short, W.D., et al., IL-10 promotes endothelial progenitor cell infiltration and wound healing via STAT3. The FASEB Journal, 2022. 36(7): p. e22298.
- Takagi, N., et al., IL-17A promotes neutrophilic inflammation and disturbs acute wound healing in skin. Experimental Dermatology, 2017. 26(2): p. 137-144.
- Ahmed, M. and J.R. Huh, Cutting edge: interleukin-17a prompts HIF1α for wound healing. Trends in Immunology, 2022. 43(11): p. 861-863.
- Wood, S., et al., Pro-Inflammatory Chemokine CCL2 (MCP-1) Promotes Healing in Diabetic Wounds by Restoring the Macrophage Response. PLOS ONE, 2014. 9(3): p. e91574.
- Badr, G., et al., Treatment of diabetic mice with undenatured whey protein accelerates the wound healing process by enhancing the expression of MIP-1α, MIP-2, KC, CX3CL1 and TGF-β in wounded tissue. BMC Immunology, 2012. 13(1): p. 32.
- Ritsu, M., et al., Critical role of tumor necrosis factor-α in the early process of wound healing in skin. Journal of Dermatology & Dermatologic Surgery, 2017. 21(1): p. 14-19.
- Ashcroft, G.S., et al., Tumor necrosis factor-alpha (TNF-α) is a therapeutic target for impaired cutaneous wound healing. Wound Repair and Regeneration, 2012. 20(1): p. 38-49.
- Skoda, M., A. Stangret, and D. Szukiewicz, Fractalkine and placental growth factor: A duet of inflammation and angiogenesis in cardiovascular disorders. Cytokine & Growth Factor Reviews, 2018. 39: p. 116-123.
- Szukiewicz, D., CX3CL1 (Fractalkine)-CX3CR1 Axis in Inflammation-Induced Angiogenesis and Tumorigenesis. International Journal of Molecular Sciences, 2024. 25(9): p. 4679.
- Yuan, C., et al., Current knowledge of leptin in wound healing: A collaborative review. Frontiers in Pharmacology, 2022. Volume 13 - 2022.
- Tadokoro, S., et al., Leptin Promotes Wound Healing in the Skin. PLOS ONE, 2015. 10(3): p. e0121242.
- Shen, H., et al., Interferon-gamma inhibits healing post scald burn injury. Wound Repair and Regeneration, 2012. 20(4): p. 580-591.
- Yates-Binder, C.C., et al., An IP-10 (CXCL10)-Derived Peptide Inhibits Angiogenesis. PLOS ONE, 2012. 7(7): p. e40812.
- Korbecki, J., et al., The Potential Importance of CXCL1 in the Physiological State and in Noncancer Diseases of the Cardiovascular System, Respiratory System and Skin. International Journal of Molecular Sciences, 2023. 24(1): p. 205.
- Zhang, J., et al., The Stimulation of Macrophages by Systematical Administration of GM-CSF Can Accelerate Adult Wound Healing Process. International Journal of Molecular Sciences, 2022. 23(19): p. 11287.
- Lichtman, M.K., M. Otero-Vinas, and V. Falanga, Transforming growth factor beta (TGF-β) isoforms in wound healing and fibrosis. Wound Repair and Regeneration, 2016. 24(2): p. 215-222.
- Kiritsi, D. and A. Nyström, The role of TGFβ in wound healing pathologies. Mechanisms of Ageing and Development, 2018. 172: p. 51-58.
- Théry, C., et al., Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles, 2018. 7(1): p. 1535750.
- Li, Y., et al., p63: a crucial player in epithelial stemness regulation. Oncogene, 2023. 42(46): p. 3371-3384.
- Melino, G., et al., Maintaining epithelial stemness with p63. Science Signaling, 2015. 8(387): p. re9-re9.
- Sidney, L.E., et al., Concise Review: Evidence for CD34 as a Common Marker for Diverse Progenitors. Stem Cells, 2014. 32(6): p. 1380-1389.
- Joulai Veijouye, S., et al., Bulge Region as a Putative Hair Follicle Stem Cells Niche: A Brief Review. Iran J Public Health, 2017. 46(9): p. 1167-1175.
- Kloepper, J.E., et al., Immunophenotyping of the human bulge region: the quest to define useful in situ markers for human epithelial hair follicle stem cells and their niche. Experimental Dermatology, 2008. 17(7): p. 592-609.
- Xu, J., et al., Potential mechanisms underlying the Runx2 induced osteogenesis of bone marrow mesenchymal stem cells. Am J Transl Res, 2015. 7(12): p. 2527-35.
- Rumiński, S., I. Kalaszczyńska, and M. Lewandowska-Szumieł, Effect of cAMP Signaling Regulation in Osteogenic Differentiation of Adipose-Derived Mesenchymal Stem Cells. Cells, 2020. 9(7): p. 1587.
- Nirenjen, S., et al., Exploring the contribution of pro-inflammatory cytokines to impaired wound healing in diabetes. Frontiers in Immunology, 2023. 14.
- Nosenko, M.A., S.G. Ambaryan, and M.S. Drutskaya, Proinflammatory Cytokines and Skin Wound Healing in Mice. Molecular Biology, 2019. 53(5): p. 653-664.
- Nurkesh, A., et al., Recent Advances in the Controlled Release of Growth Factors and Cytokines for Improving Cutaneous Wound Healing. Frontiers in Cell and Developmental Biology, 2020. 8.
- Schaffrick, L., et al., The dynamic changes of monocytes and cytokines during wound healing post-burn injury. Cytokine, 2023. 168: p. 156231.
- Papait, A., et al., Comparison of EV-free fraction, EVs, and total secretome of amniotic mesenchymal stromal cells for their immunomodulatory potential: a translational perspective. Front Immunol, 2022. 13: p. 960909.
- Claudinot, S., et al., Tp63-expressing adult epithelial stem cells cross lineages boundaries revealing latent hairy skin competence. Nature Communications, 2020. 11(1): p. 5645.
- Lee, B.-W., et al., Expression of p63 and its association with cell proliferation at different stages of murine hair follicle cycle. Journal of Biomedical Translational Research, 2018. 19(1): p. 10-15.
- Ohyama, M., et al., Characterization and isolation of stem cell–enriched human hair follicle bulge cells. The Journal of Clinical Investigation, 2006. 116(1): p. 249-260.
- Hoogduijn, M.J., E. Gorjup, and P.G. Genever, Comparative characterization of hair follicle dermal stem cells and bone marrow mesenchymal stem cells. Stem Cells Dev, 2006. 15(1): p. 49-60.
- Komori, T., Whole Aspect of Runx2 Functions in Skeletal Development. International Journal of Molecular Sciences, 2022. 23(10): p. 5776.
- Komori, T., Regulation of Skeletal Development and Maintenance by Runx2 and Sp7. International Journal of Molecular Sciences, 2024. 25(18): p. 10102.
- Zhu, M., et al., Osteogenically-induced exosomes stimulate osteogenesis of human adipose-derived stem cells. Cell Tissue Bank, 2021. 22(1): p. 77-91.
- Ievlev, V., et al., Krt14 and Krt15 differentially regulate regenerative properties and differentiation potential of airway basal cells. JCI Insight, 2023. 8(2).
- Quan, R., et al., Culture and characterization of rat hair follicle stem cells. Cytotechnology, 2016. 68(4): p. 621-628.
- Guda, P.R., et al., Nanoscopic and Functional Characterization of Keratinocyte-Originating Exosomes in the Wound Fluid of Non-Diabetic and Diabetic Chronic Wound Patients. Nano Today, 2023. 52.
- Garcia-Martin, R., et al., Tissue differences in the exosomal/small extracellular vesicle proteome and their potential as indicators of altered tissue metabolism. Cell Rep, 2022. 38(3): p. 110277.
- Zhang, Y., et al., Hair follicle stem cells promote epidermal regeneration under expanded condition. Frontiers in Physiology, 2024. 15.
- Samundeshwari, E.L., et al., Prominent Expression of COL2A1, ACAN and IHH Genes are Observed in the Differentiation of Human Hematopoietic Stem Cells into Articular Type of Chondrocytes. Stem Cell Rev Rep, 2024. 20(5): p. 1370-1373.
- Levis, H., et al., Multiplex gene editing to promote cell survival using low-pH clustered regularly interspaced short palindromic repeats activation (CRISPRa) gene perturbation. Cytotherapy, 2023. 25(10): p. 1069-1079.
- Luo, S., et al., ITGB1 promotes the chondrogenic differentiation of human adipose-derived mesenchymal stem cells by activating the ERK signaling. J Mol Histol, 2020. 51(6): p. 729-739.
- Rompolas, P. and V. Greco, Stem cell dynamics in the hair follicle niche. Semin Cell Dev Biol, 2014. 25-26: p. 34-42.
- Çankirili, N.K., O. Altundag, and B. Çelebi-Saltik, Skin Stem Cells, Their Niche and Tissue Engineering Approach for Skin Regeneration, in Cell Biology and Translational Medicine, Volume 6: Stem Cells: Their Heterogeneity, Niche and Regenerative Potential, K. Turksen, Editor. 2020, Springer International Publishing: Cham. p. 107-126.
- Thiagarajan, L., H.A.M. Abu-Awwad, and J.E. Dixon, Osteogenic Programming of Human Mesenchymal Stem Cells with Highly Efficient Intracellular Delivery of RUNX2. Stem Cells Transl Med, 2017. 6(12): p. 2146-2159.
- Wang, S., et al., Single cell transcriptomics of human epidermis identifies basal stem cell transition states. Nature Communications, 2020. 11(1): p. 4239.
- Doyle, L.M. and M.Z. Wang, Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells, 2019. 8(7).
- Gurunathan, S., et al., Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells, 2019. 8(4).
- Soo, C.Y., et al., Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology, 2012. 136(2): p. 192-7.
- Franquesa, M., et al., Update on controls for isolation and quantification methodology of extracellular vesicles derived from adipose tissue mesenchymal stem cells. Front Immunol, 2014. 5: p. 525.
- Teng, L., et al., Exosomes Derived from Human Umbilical Cord Mesenchymal Stem Cells Accelerate Diabetic Wound Healing via Promoting M2 Macrophage Polarization, Angiogenesis, and Collagen Deposition. Int J Mol Sci, 2022. 23(18).
- Narayanan, K.B., et al., Polydopamine-Functionalized Bacterial Cellulose as Hydrogel Scaffolds for Skin Tissue Engineering. Gels, 2023. 9(8).
- Rasti, M., et al., Enhancing the wound healing process through local injection of exosomes derived from blood serum: An in vitro and in vivo assessment. Regenerative Therapy, 2024. 26: p. 281-289.
- Choudhary, V., M. Choudhary, and W.B. Bollag, Exploring Skin Wound Healing Models and the Impact of Natural Lipids on the Healing Process. International Journal of Molecular Sciences, 2024. 25(7): p. 3790.
- Zhao, B., et al., Exosomes derived from human amniotic epithelial cells accelerate wound healing and inhibit scar formation. J Mol Histol, 2017. 48(2): p. 121-132.
- Cooper, D.R., et al., Human Adipose-Derived Stem Cell Conditioned Media and Exosomes Containing MALAT1 Promote Human Dermal Fibroblast Migration and Ischemic Wound Healing. Adv Wound Care (New Rochelle), 2018. 7(9): p. 299-308.
- Villatoro, A.J., et al., Comparative analysis and characterization of soluble factors and exosomes from cultured adipose tissue and bone marrow mesenchymal stem cells in canine species. Veterinary Immunology and Immunopathology, 2019. 208: p. 6-15.
- Villatoro, A.J., et al., Proteomic Analysis of the Secretome and Exosomes of Feline Adipose-Derived Mesenchymal Stem Cells. Animals, 2021. 11(2): p. 295.
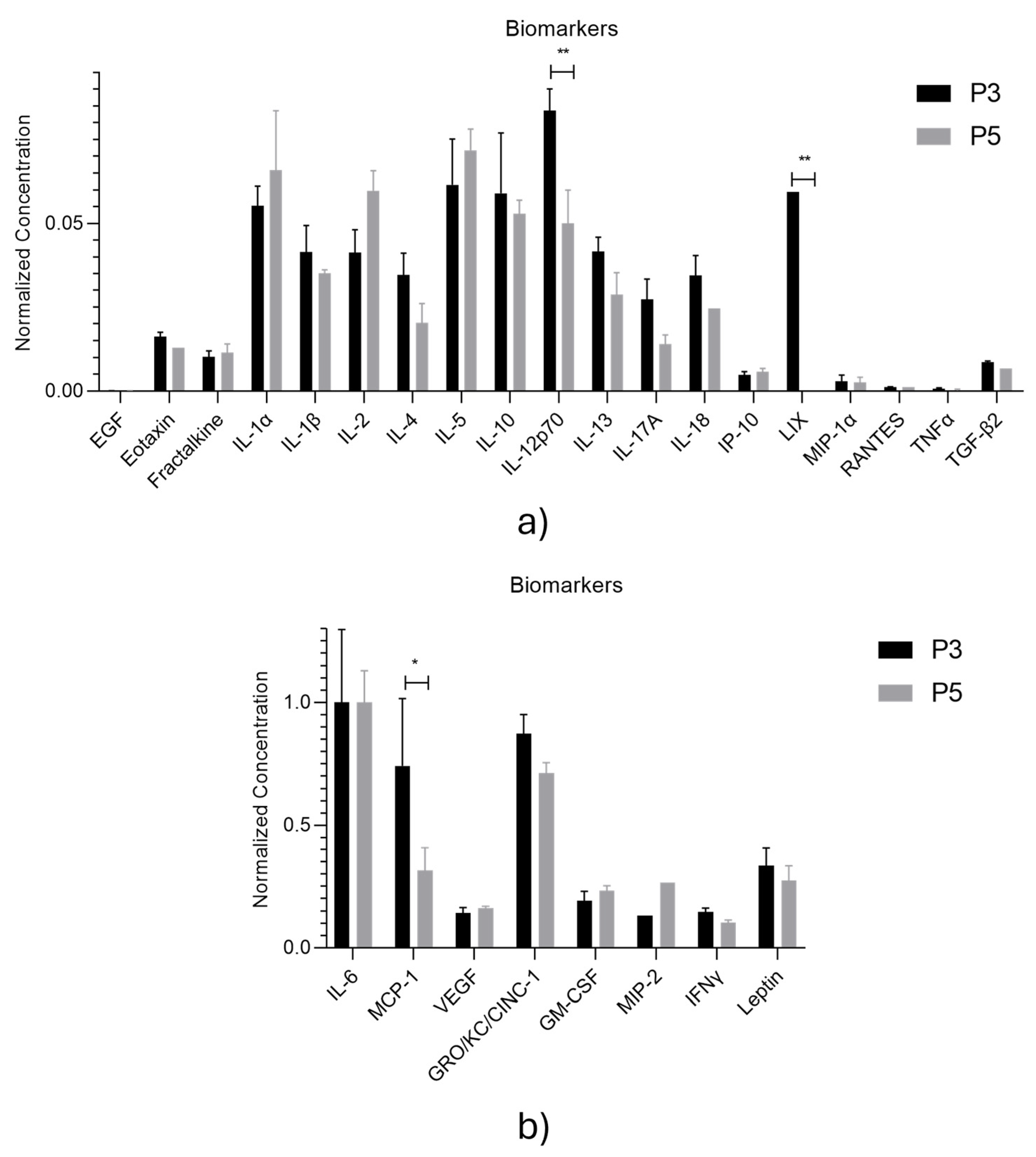
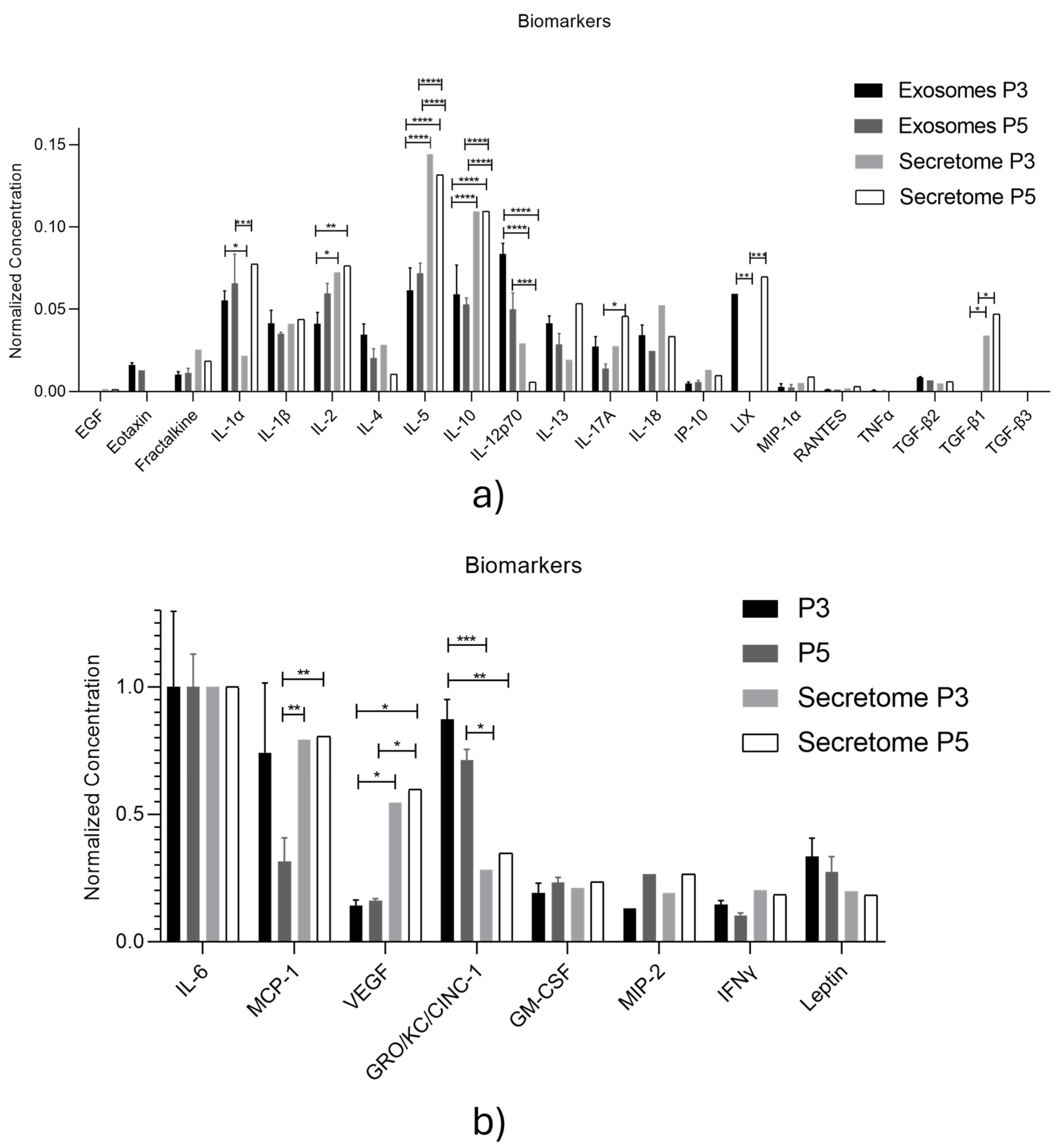
| Biomarker | Function in Wound Healing |
|---|---|
| EGF | Promotes keratinocyte and fibroblast proliferation, aiding re-epithelialization and collagen synthesis [34,35]. |
| G-CSF | Enhances neutrophil production, supporting debris clearance during the inflammatory phase [36,37]. |
| VEGF | Critical for angiogenesis, ensuring oxygen and nutrient delivery to healing tissues [38,39]. |
| IL-6, IL-1α and IL-1β | Key pro-inflammatory cytokines that regulate inflammation, recruit immune cells, and stimulate fibroblasts and keratinocytes [40]. |
| IL-2 and IL-12p70 | Primarily modulate immune responses, indirectly affecting wound healing [41,42]. |
| IL-4 and IL-13 | Promote fibroblast differentiation into myofibroblasts, impacting wound contraction and fibrosis [43,44]. |
| IL-5 and Eotaxin | Mainly recruit eosinophils, with limited direct impact on typical wound healing [45]. |
| IL-10 | Anti-inflammatory cytokine, crucial for resolving inflammation and minimizing scarring [46,47]. |
| IL-17A and IL-18 | Contribute to inflammation and influence keratinocyte activity and angiogenesis [48,49]. |
| RANTES (CCL5), MCP-1 (CCL2), MIP-1α (CCL3) and MIP-2 (CXCL2) | Chemokines that recruit immune cells to the wound site, supporting inflammation and repair [50,51]. |
| TNFα | It stimulates the production of other cytokines and chemokines, activates immune cells, and can influence fibroblast and keratinocyte behavior. Drives early inflammation but may impair healing if chronically elevated [52,53]. |
| Fractalkine (CX3CL1) | Aids immune cell recruitment and endothelial interaction [54,55]. |
| Leptin | Supports keratinocyte proliferation, angiogenesis, and collagen production [56,57]. |
| IFNγ and IP-10 (CXCL10) | Influence inflammation and ECM remodeling, with prolonged expression potentially impairing healing [58,59]. |
| GRO/KC/CINC-1 (CXCL1) and LIX (CXCL5) | Attract neutrophils during early wound responses [60]. |
| GM-CSF | Promotes differentiation of immune cells, supporting both inflammation and repair [61]. |
| TGFβ1 and TGFβ2 | Stimulate fibroblast proliferation, myofibroblast differentiation, and ECM production [62]. |
| TGFβ3 | Encourages regenerative healing with reduced scarring [63]. |
| Biomolecule | Exosomes Mean ± SEM (P3) |
Exosomes Mean ± SEM (P5) |
Secretome Mean ± SEM (P3) |
Secretome Mean ± SEM (P5) |
|---|---|---|---|---|
| EGF | 0.10 ± 0.01 | 0.1 ± 0.02 | 0.16 ± 0.03 | 0.18 ± 0.04 |
| Eotaxin | 1.79 ± 0.14 | 1.65 ± 0.00 | 0.16 ± 0.00 | 0.00 ± 0.00 |
| Fractalkine | 1.17 ± 0.19 | 1.47 ± 0.54 | 4.38 ± 0.29 | 3.06 ± 0.15 |
| GM-CSF | 20.10 ± 3.95 | 28.06 ± 4.02 | 35.39 ± 10.69 | 39.75 ± 11.55 |
| GRO/KC/CINC-1 | 90.97 ± 8.13 | 85.26 ± 8.73 | 47.32 ± 8.64 | 57.64 ± 7.24 |
| IFNγ | 15.31 ±1.68 | 12.31 ± 2.20 | 34.01 ± 2.26 | 30.67 ± 0.73 |
| IL-1α | 5.85 ±0.61 | 7.97 ± 3.67 | 3.79 ± 2.27 | 12.83 ± 5.50 |
| IL-1β | 4.42 ± 0.83 | 4.29 ± 0.23 | 6.99 ± 0.86 | 7.27 ± 0.63 |
| IL-2 | 4.40 ± 0.71 | 7.24 ± 1.23 | 12.21 ± 1.23 | 12.65 ± 0.58 |
| IL-4 | 3.71 ± 0.68 | 2.54 ± 1.17 | 4.88 ± 1.30 | 2.63 ± 0.00 |
| IL-5 | 6.50 ± 1.42 | 8.68 ± 1.32 | 24.13 ± 2.69 | 21.13 ± 2.46 |
| IL-6 | 104.20 ± 30.96 | 119.66 ± 21.91 | 166.36 ± 50.22 | 165.62 ± 0.00 |
| IL-10 | 6.24 ± 1.87 | 6.42 ± 0.83 | 18.37 ± 1.87 | 18.17 ± 0.88 |
| IL-12p70 | 8.80 ± 0.68 | 6.08 ± 2.04 | 4.34 ± 1.80 | 1.42 ± 0.00 |
| IL-13 | 4.43 ± 0.45 | 3.54 ± 1.34 | 3.39 ± 0.43 | 8.84 ± 0.51 |
| IL-17A | 2.94 ± 0.63 | 1.77 ± 0.58 | 4.77 ± 0.30 | 7.57 ± 1.11 |
| IL-18 | 3.68 ± 0.63 | 3.05 ± 0.00 | 8.89 ± 1.31 | 5.55 ± 1.24 |
| IP-10 | 0.61 ± 0.1 | 0.79 ± 0.21 | 2.33 ± 0.31 | 1.96 ± 0.38 |
| Leptin | 35.04 ± 7.41 | 32.97 ± 12.35 | 32.31 ± 8.97 | 30.49 ± 4.08 |
| LIX | 6.29 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| MCP-1 | 77.34 ± 28.53 | 37.68 ± 19.27 | 131.64 ± 38.26 | 137.21 ± 19.77 |
| MIP-1α | 0.40 ± 0.20 | 0.40 ± 0.34 | 1.70 ± 0.57 | 2.19 ± 0.39 |
| MIP-2 | 13.81 ± 0.00 | 31.83 ± 0.00 | 32.78 ± 5.77 | 44.02 ± 7.97 |
| RANTES | 0.23 ± 0.01 | 0.24 ± 0.00 | 0.51 ± 0.03 | 0.50 ± 0.01 |
| TNFα | 0.17 ± 0.03 | 0.13 ± 0.07 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| VEGF | 14.77 ± 2.43 | 19.47 ± 1.60 | 91.75 ± 3.22 | 98.88 ± 1.06 |
| G-CSF | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| TGF-β1 | 0.00 ± 0.00 | 0.00 ± 0.00 | 5.82 ± 1.07 | 7.14 ± 1.02 |
| TGF-β2 | 0.99 ± 0.07 | 0.90 ± 0.00 | 0.97 ± 0.04 | 0.98 ± 0.00 |
| TGF-β3 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.19 ± 0.00 | 0.00 ± 0.00 |
| Exosomes P3 vs Exosomes P5 |
Exosomes P3 vs Secretome P3 |
Exosomes P3 vs Secretome P5 |
Exosomes P5 vs Secretome P3 |
Exosomes P5 vs Secretome P5 |
Secretome P3 vs Secretome P5 |
|
|---|---|---|---|---|---|---|
| IL-1α | ns | * | ns | *** | ns | **** |
| IL-2 | ns | * | ** | ns | ns | ns |
| IL-5 | ns | **** | **** | **** | **** | ns |
| IL-10 | ns | **** | **** | **** | **** | ns |
| IL-12p70 | ** | **** | **** | ns | *** | ns |
| IL-13 | ns | ns | ns | ns | ns | ** |
| IL-17a | ns | ns | ns | ns | * | ns |
| LIX | ** | ** | ns | ns | *** | ** |
| TGF-β1 | ns | ns | * | ns | * | ns |
| MCP-1 | * | ns | ns | ** | ** | ns |
| VEGF | ns | * | * | ns | * | ns |
| GRO/KC/CINC-1 | ns | *** | ** | * | ns | ns |
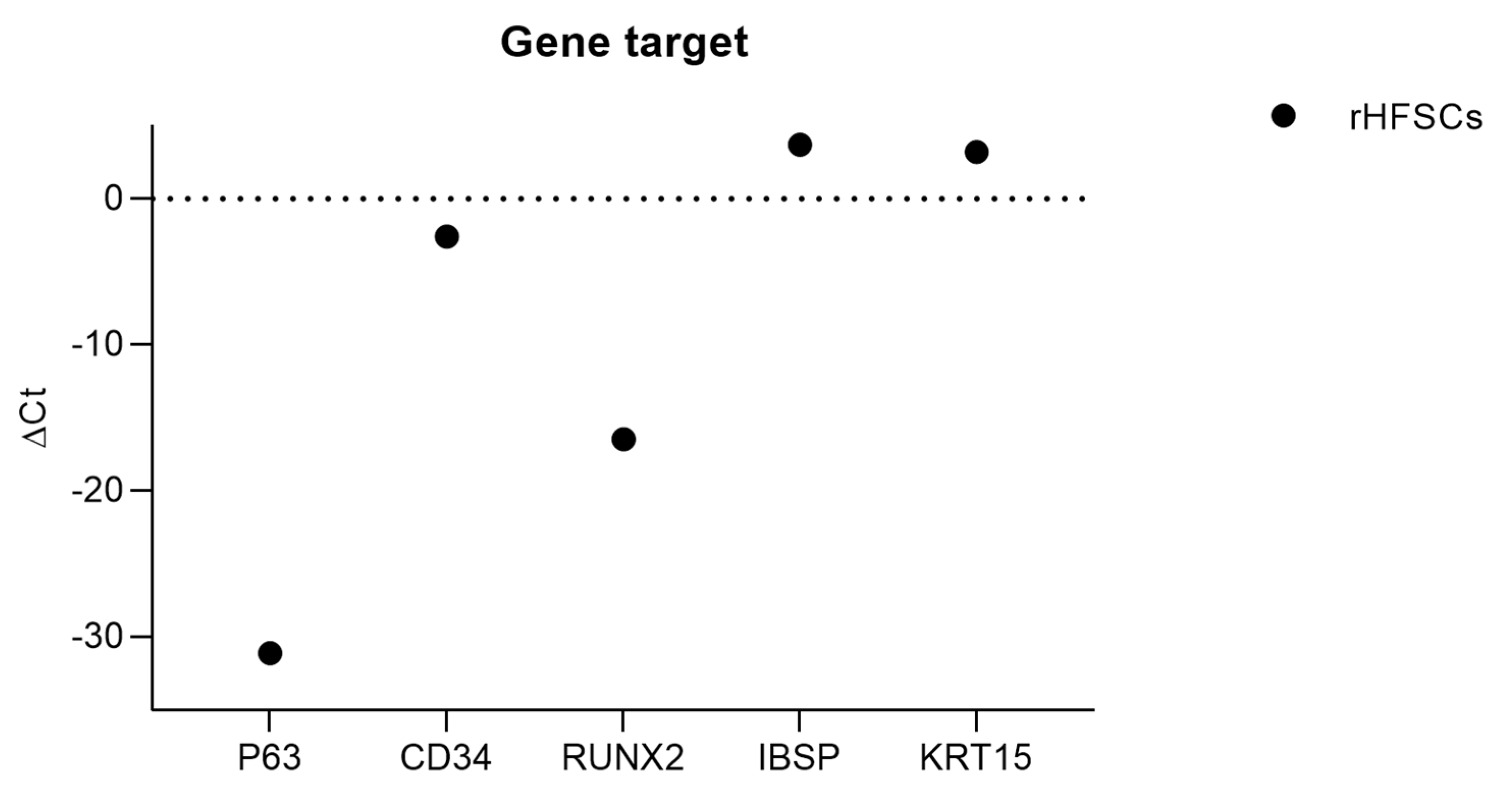
| Target Gene | Ct Average | ΔCt |
|---|---|---|
| KRT14 | nd | nd |
| p63 | 4.92±0.00 | -31.1 |
| CD34 | 33.40±1.41 | -2.6 |
| COL2A1 | nd | nd |
| ITGα6 | nd | nd |
| ACAN | nd | nd |
| ITGβ1 | nd | nd |
| RUNX2 | 19.45±0.00 | -16.5 |
| KRT10 | nd | nd |
| IBSP | 39.67±0.00 | 3.7 |
| KRT15 | 39.19±0.00 | 3.2 |
| ADIPOQ | nd | nd |
| AAK1 | nd | nd |
| KRT19 | nd | nd |
| Secretome P3 | Secretome P5 | Exosomes P3 | Exosomes P5 | |
| Secretome P3 | ns | * | ns | |
| Secretome P5 | ns | * | ||
| Exosomes P3 | ns |
| 24 h | 72 h | 168 h | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exosomes | Secretome | DMEM 10% | DMSO 10% | Exosomes | Secretome | DMEM 10% | DMSO 10% | Exosomes | Secretome | DMEM 10% | DMSO 10% | |
| Exosomes | * | **** | **** | **** | **** | **** | **** | **** | **** | |||
| Secretome | **** | **** | ns | **** | **** | **** | ||||||
| DMEM 10% | **** | **** | **** | |||||||||
| 24h | 72h | 168h | ||||
|---|---|---|---|---|---|---|
| Secretome | DMSO 10% | Secretome | DMSO 10% | Secretome | DMSO 10% | |
| Exosomes | ns | **** | ** | **** | **** | ** |
| Secretome | *** | **** | *** | |||
| No MMC | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2h | 4h | 6h | 8h | 10h | 12h | 24h | 32h | 53h | |
| Control vs Exosomes |
ns | ns | ns | ns | ns | * | ns | ns | ns |
| Control vs Secretome |
ns | ** | * | * | ns | * | ns | ns | ns |
| Exosomes vs Secretome |
ns | ns | ns | ns | ns | ns | ns | ns | ns |
| With MMC | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2h | 4h | 6h | 8h | 10h | 12h | 24h | 32h | 53h | |
| Control vs Exosomes |
ns | ns | * | ns | ns | ns | ns | ns | ns |
| Control vs Secretome |
ns | ns | ns | ns | ns | ** | ns | ns | ns |
| Exosomes vs Secretome |
ns | ns | ns | ns | ns | ns | ns | ns | ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





