Submitted:
25 April 2025
Posted:
27 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
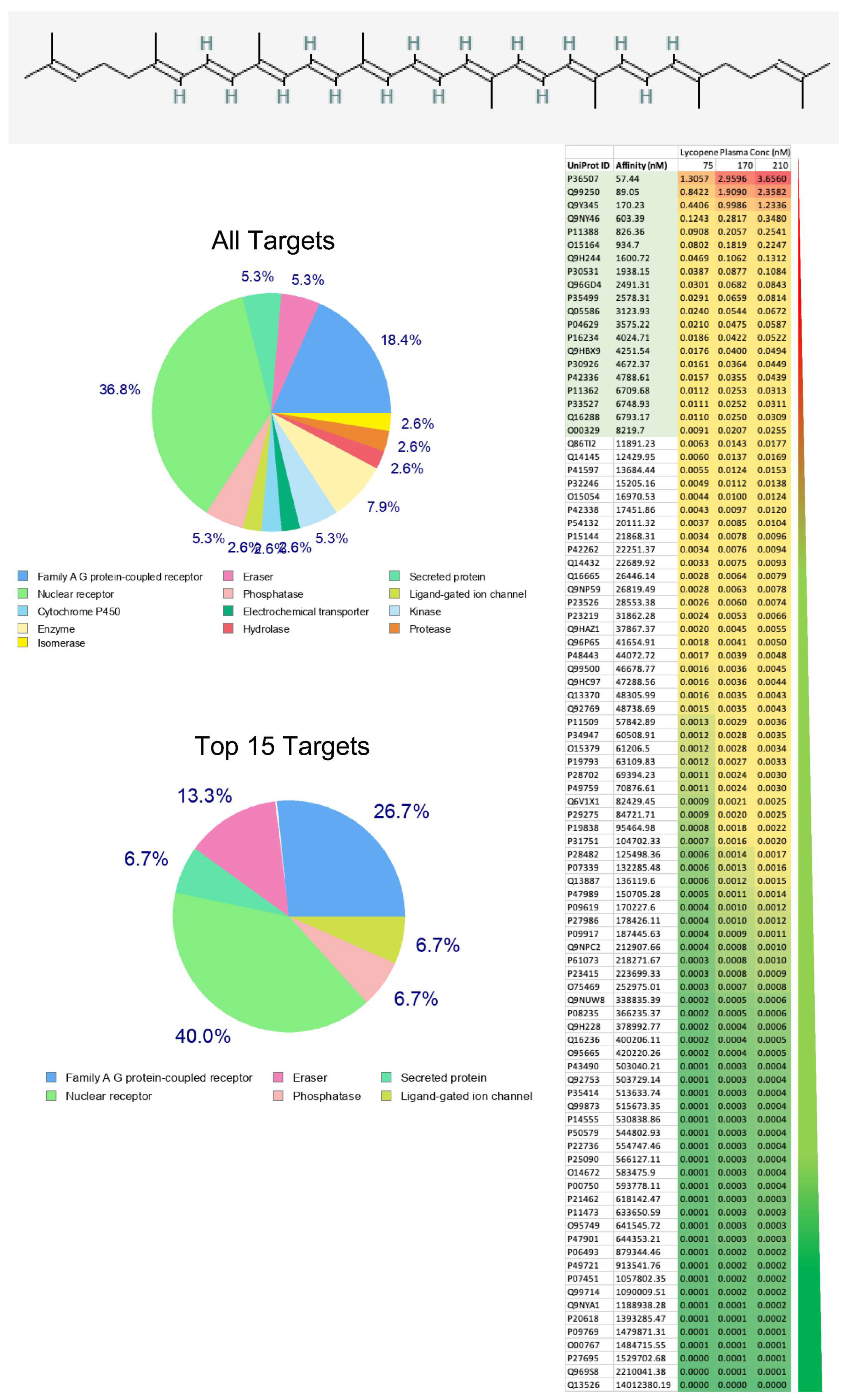
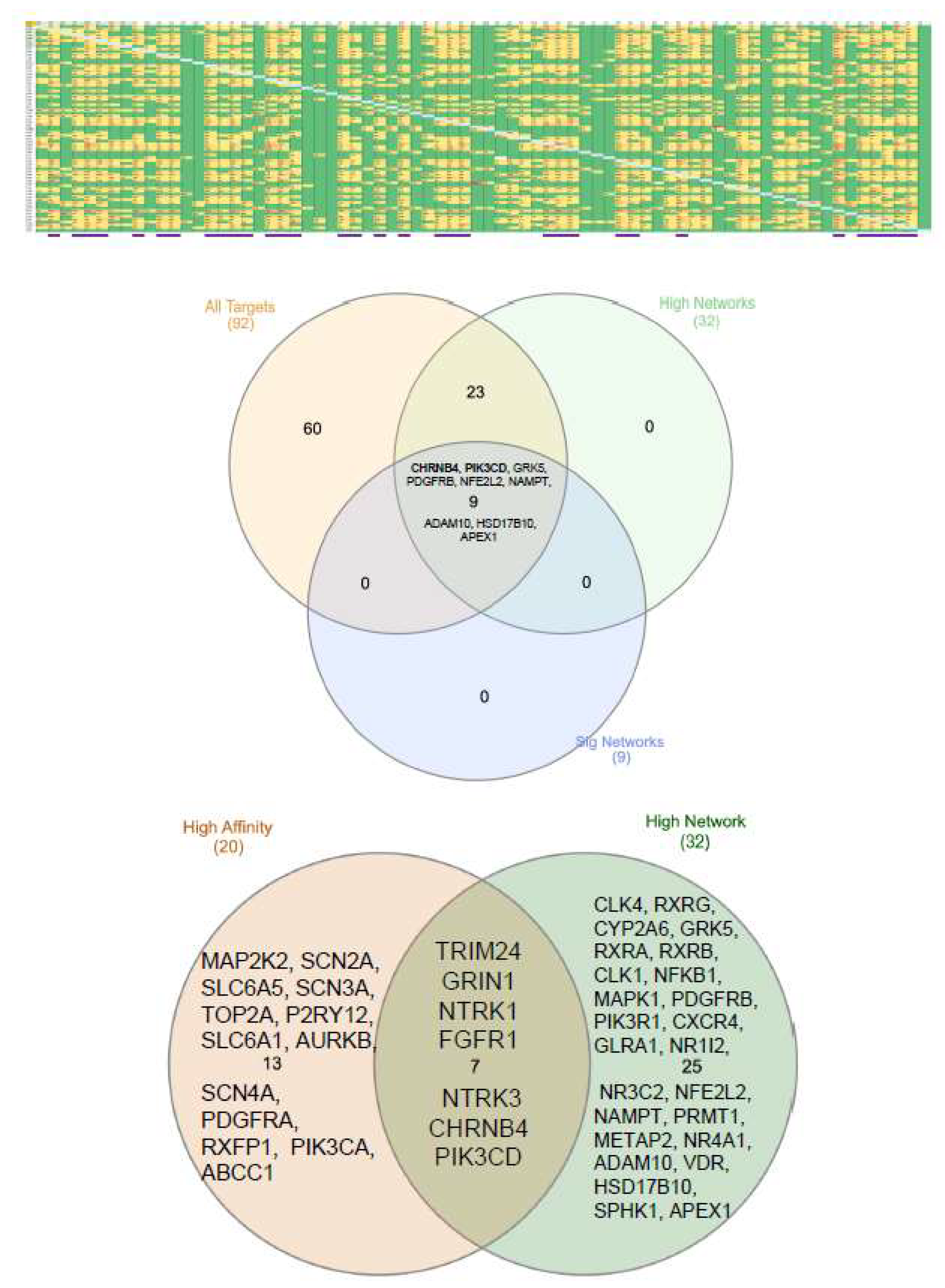
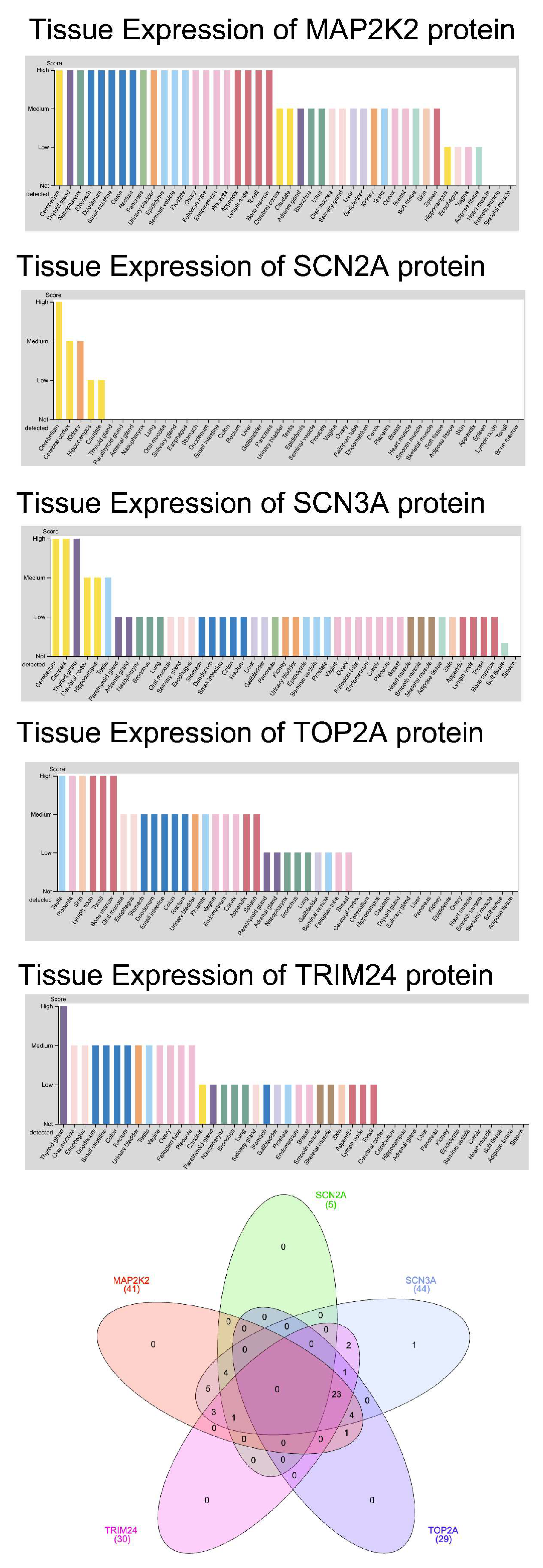
3. Results
4. Discussion
Acknowledgments
Declaration of Interest Statement
References
- Giovannucci, E.; Rimm, E.B.; Liu, Y.; Stampfer, M.J.; Willett, W.C. A Prospective Study of Tomato Products, Lycopene, and. Journal of the National Cancer Institute 2002, 94, 391. [Google Scholar] [CrossRef]
- Rao, A.; Waseem, Z.; Agarwal, S. Lycopene content of tomatoes and tomato products and their contribution to dietary lycopene. Food Research International 1998, 31, 737–741. [Google Scholar] [CrossRef]
- Gerster, H. The potential role of lycopene for human health. Journal of the American College of Nutrition 1997, 16, 109–126. [Google Scholar] [CrossRef]
- Gupta, E.; Mishra, P.; Singh, P.; Mishra, N. Evidence and prospects of lycopene as powerful red superfood: modern approach to food science. Current Traditional Medicine 2024, 10, 50–60. [Google Scholar] [CrossRef]
- Yaqoob, S.; Riaz, M.; Shabbir, A.; Zia-Ul-Haq, M.; Alwakeel, S.S.; Bin-Jumah, M. Commercialization and marketing potential of carotenoids. Carotenoids: structure and function in the human body, 2021; 799–826. [Google Scholar]
- Kulawik, A.; Cielecka-Piontek, J.; Czerny, B.; Kamiński, A.; Zalewski, P. The Relationship Between Lycopene and Metabolic Diseases. Nutrients 2024, 16, 3708. [Google Scholar] [CrossRef]
- Przybylska, S.; Tokarczyk, G. Lycopene in the prevention of cardiovascular diseases. International journal of molecular sciences 2022, 23, 1957. [Google Scholar] [CrossRef]
- Hartsen, F. Beitrage zur Organischen Chemie. Chem. Zentra 1873, 204–207. [Google Scholar]
- Camara Hurtado, R.M.; Cáceres, J.O.; Cámara Hurtado, M.; Manzoor, S.; Fernández Ruiz, V.; Sánchez Mata, M.C. New developments in lycopene analysis by spectroscopic and chromatographic techniques, accompanied by mathematical modelling. 2014.
- Bergougnoux, V. The history of tomato: from domestication to biopharming. Biotechnology advances 2014, 32, 170–189. [Google Scholar] [CrossRef]
- Shi, J.; Maguer, M.L. Lycopene in tomatoes: chemical and physical properties affected by food processing. Critical reviews in food science and nutrition 2000, 40, 1–42. [Google Scholar] [CrossRef]
- Clinton, S.K. Lycopene: chemistry, biology, and implications for human health and disease. Nutrition reviews 1998, 56, 35–51. [Google Scholar] [CrossRef]
- Srivastava, S.; Srivastava, A.K. Lycopene; chemistry, biosynthesis, metabolism and degradation under various abiotic parameters. Journal of Food Science and Technology 2015, 52, 41–53. [Google Scholar] [CrossRef]
- Valacchi, G.; Pecorelli, A. Role of Scavenger Receptor B1 (SR-B1) in Improving Food Benefits for Human Health. Annual Review of Food Science and Technology 2025, 16. [Google Scholar] [CrossRef]
- Moussa, M.; Landrier, J.-F.; Reboul, E.; Ghiringhelli, O.; Coméra, C.; Collet, X.; Fröhlich, K.; Böhm, V.; Borel, P. Lycopene absorption in human intestinal cells and in mice involves scavenger receptor class B type I but not Niemann-Pick C1-like 1. The Journal of nutrition 2008, 138, 1432–1436. [Google Scholar] [CrossRef]
- Arballo, J.; Amengual, J.; Erdman Jr, J.W. Lycopene: A critical review of digestion, absorption, metabolism, and excretion. Antioxidants 2021, 10, 342. [Google Scholar] [CrossRef]
- Botham, K.M.; Bravo, E. Lycopene and chylomicrons. In Lycopene; CRC Press: 2019; pp. 159-182.
- Gustin, D.M.; Rodvold, K.A.; Sosman, J.A.; Diwadkar-Navsariwala, V.; Stacewicz-Sapuntzakis, M.; Viana, M.; Crowell, J.A.; Murray, J.; Tiller, P.; Bowen, P.E. Single-dose pharmacokinetic study of lycopene delivered in a well-defined food-based lycopene delivery system (tomato paste-oil mixture) in healthy adult male subjects. Cancer Epidemiology Biomarkers & Prevention 2004, 13, 850–860. [Google Scholar]
- Khurana, S.; Sharma, S.; Goyal, P.K. Updates on the Anticancer Profile of Lycopene and its Probable Mechanism Against Breast and Gynecological Cancer. The Natural Products Journal 2025. [Google Scholar] [CrossRef]
- Ozkan, G.; Günal-Köroğlu, D.; Karadag, A.; Capanoglu, E.; Cardoso, S.M.; Al-Omari, B.; Calina, D.; Sharifi-Rad, J.; Cho, W.C. A mechanistic updated overview on lycopene as potential anticancer agent. Biomedicine & Pharmacotherapy 2023, 161, 114428. [Google Scholar]
- Khan, U.M.; Sevindik, M.; Zarrabi, A.; Nami, M.; Ozdemir, B.; Kaplan, D.N.; Selamoglu, Z.; Hasan, M.; Kumar, M.; Alshehri, M.M. Lycopene: food sources, biological activities, and human health benefits. Oxidative medicine and cellular longevity 2021, 2021, 2713511. [Google Scholar] [CrossRef]
- Heber, D.; Lu, Q.-Y. Overview of mechanisms of action of lycopene. Experimental biology and medicine 2002, 227, 920–923. [Google Scholar] [CrossRef]
- Mozos, I.; Stoian, D.; Caraba, A.; Malainer, C.; Horbańczuk, J.O.; Atanasov, A.G. Lycopene and vascular health. Frontiers in pharmacology 2018, 9, 521. [Google Scholar] [CrossRef]
- Lee, W.; Ku, S.-K.; Bae, J.W.; Bae, J.-S. Inhibitory effects of lycopene on HMGB1-mediated pro-inflammatory responses in both cellular and animal models. Food and Chemical Toxicology 2012, 50, 1826–1833. [Google Scholar] [CrossRef]
- Prema, A.; Janakiraman, U.; Manivasagam, T.; Thenmozhi, A.J. Neuroprotective effect of lycopene against MPTP induced experimental Parkinson’s disease in mice. Neuroscience letters 2015, 599, 12–19. [Google Scholar] [CrossRef]
- Kaur, H.; Chauhan, S.; Sandhir, R. Protective effect of lycopene on oxidative stress and cognitive decline in rotenone induced model of Parkinson’s disease. Neurochemical research 2011, 36, 1435–1443. [Google Scholar] [CrossRef]
- Palozza, P.; Simone, R.; Catalano, A.; Parrone, N.; Monego, G.; Ranelletti, F.O. Lycopene regulation of cholesterol synthesis and efflux in human macrophages. The Journal of nutritional biochemistry 2011, 22, 971–978. [Google Scholar] [CrossRef]
- Özmen, Ö.; Şahi̇n, M.; Topsakal, Ş.; Taşan, Ş.; Şahi̇n, U. Lycopene ameliorates diabetes-induced pancreatic, hepatic, and renal damage by modulating the JAK/STAT/SOCS signaling pathway in rats. Iranian Journal of Basic Medical Sciences 2025, 28, 461. [Google Scholar]
- Liu, Y.; Tian, Y.; Dai, X.; Liu, T.; Zhang, Y.; Wang, S.; Shi, H.; Yin, J.; Xu, T.; Zhu, R. Lycopene ameliorates islet function and down-regulates the TLR4/MyD88/NF-κB pathway in diabetic mice and Min6 cells. Food & Function 2023, 14, 5090–5104. [Google Scholar]
- Tripathi, A.K.; Das, R.; Ray, A.K.; Mishra, S.K.; Anand, S. Recent insights on pharmacological potential of lycopene and its nanoformulations: an emerging paradigm towards improvement of human health. Phytochemistry Reviews 2024, 1–28. [Google Scholar] [CrossRef]
- Fang, Y.; Ou, S.; Wu, T.; Zhou, L.; Tang, H.; Jiang, M.; Xu, J.; Guo, K. Lycopene alleviates oxidative stress via the PI3K/Akt/Nrf2pathway in a cell model of Alzheimer’s disease. PeerJ 2020, 8, e9308. [Google Scholar] [CrossRef]
- Assar, E.A.; Vidalle, M.C.; Chopra, M.; Hafizi, S. Lycopene acts through inhibition of IκB kinase to suppress NF-κB signaling in human prostate and breast cancer cells. Tumor Biology 2016, 37, 9375–9385. [Google Scholar] [CrossRef]
- Simone, R.E.; Russo, M.; Catalano, A.; Monego, G.; Froehlich, K.; Boehm, V.; Palozza, P. Lycopene inhibits NF-kB-mediated IL-8 expression and changes redox and PPARγ signalling in cigarette smoke–stimulated macrophages. PloS one 2011, 6, e19652. [Google Scholar] [CrossRef]
- Marzocco, S.; Singla, R.K.; Capasso, A. Multifaceted effects of lycopene: a boulevard to the multitarget-based treatment for cancer. Molecules 2021, 26, 5333. [Google Scholar] [CrossRef]
- Han, H.; Lim, J.W.; Kim, H. Lycopene inhibits activation of epidermal growth factor receptor and expression of cyclooxygenase-2 in gastric cancer cells. Nutrients 2019, 11, 2113. [Google Scholar] [CrossRef]
- Kumar, A.H. Network Proteins of Human Sortilin1, Its Expression and Targetability Using Lycopene. Life 2024, 14, 137. [Google Scholar] [CrossRef]
- Khosravi, Z.; Kumar, A.H. Analysing the role of SERPINE1 network in the pathogenesis of human glioblastoma. Journal of Cancer Research 2023, 1, 1–6. [Google Scholar]
- Friend, J.; Kumar, A.H.S. A Network Pharmacology Approach to Assess the Comparative Pharmacodynamics of Pharmaceutical Excipient Trehalose in Human, Mouse, and Rat. Nature Cell Sci 2023, 1, 33–43. [Google Scholar] [CrossRef]
- Kumar, A.H. Pharmacological targets of Asundexian relevant to its therapeutic efficacy in treating cardiovascular diseases. Biology, Engineering, Medicine and Science Reports 2022, 8, 24–27. [Google Scholar] [CrossRef]
- Singh NK; Dhanasekaran M; AHS. K. Pharmacology of Berberine and Its Metabolites, Is It the Natures Ozempic or Imatinib? Arch Pharmacol Ther 2023, 5, 67–81. [Google Scholar] [CrossRef]
- Landrier, J.-F.; Breniere, T.; Sani, L.; Desmarchelier, C.; Mounien, L.; Borel, P. Effect of tomato, tomato-derived products and lycopene on metabolic inflammation: from epidemiological data to molecular mechanisms. Nutrition Research Reviews 2023, 1–17. [Google Scholar]
- Puah, B.-P.; Jalil, J.; Attiq, A.; Kamisah, Y. New insights into molecular mechanism behind anti-cancer activities of lycopene. Molecules 2021, 26, 3888. [Google Scholar] [CrossRef] [PubMed]
- Tufail, T.; Bader Ul Ain, H.; Noreen, S.; Ikram, A.; Arshad, M.T.; Abdullahi, M.A. Nutritional benefits of lycopene and beta-carotene: A comprehensive overview. Food Science & Nutrition 2024, 12, 8715–8741. [Google Scholar]
- Shafe, M.O.; Gumede, N.M.; Nyakudya, T.T.; Chivandi, E. Lycopene: A Potent Antioxidant with Multiple Health Benefits. Journal of Nutrition and Metabolism 2024, 2024, 6252426. [Google Scholar]
- Wu, S.; Chen, R.; Chen, J.; Yang, N.; Li, K.; Zhang, Z.; Zhang, R. Study of the anti-inflammatory mechanism of β-carotene based on network pharmacology. Molecules 2023, 28, 7540. [Google Scholar] [CrossRef]
- Mukherjee, S.; Yun, J.W. β-Carotene stimulates browning of 3T3-L1 white adipocytes by enhancing thermogenesis via the β3-AR/p38 MAPK/SIRT signaling pathway. Phytomedicine 2022, 96, 153857. [Google Scholar] [CrossRef]
- Moran, N.E.; Thomas-Ahner, J.M.; Smith, J.W.; Silva, C.; Hason, N.A.; Erdman Jr, J.W.; Clinton, S.K. β-Carotene oxygenase 2 genotype modulates the impact of dietary lycopene on gene expression during early TRAMP prostate carcinogenesis. The Journal of nutrition 2022, 152, 950–960. [Google Scholar] [CrossRef]
- Tan, H.; Wang, F.; Hu, J.; Duan, X.; Bai, W.; Wang, X.; Wang, B.; Su, Y.; Hu, J. Inhibitory interaction of flavonoids with organic cation transporter 2 and their structure–activity relationships for predicting nephroprotective effects. Journal of Applied Toxicology 2023, 43, 1421–1435. [Google Scholar] [CrossRef]
- Mittal, S.; Sawarkar, S.; Doshi, G.; Pimple, P.; Shah, J.; Bana, T. Pharmacokinetics and bioavailability of nutraceuticals. Industrial Application of Functional Foods, Ingredients and Nutraceuticals, 2023; 725–783. [Google Scholar]
- Neves-Silva, S.; Xavier-de-Britto, I.; Gomes-da-Silva, N.C.; França, Á.R.S.; Pedrochi, F.; Queiroz, M.N.; Moura-Silva, J.; Majerowicz, D.; Ricci-Junior, E.; Ferreira, T.P.T. Enhancing therapeutic efficacy: In vivo mechanisms and biochemical effects of lycopene encapsulated in nanomicelles for acute inflammation and lipid metabolism. European Journal of Pharmaceutics and Biopharmaceutics 2025, 207, 114585. [Google Scholar]
- Báo, S.N.; Machado, M.; Da Silva, A.L.; Melo, A.; Cunha, S.; Sousa, S.S.; Malheiro, A.R.; Fernandes, R.; Leite, J.R.S.; Vasconcelos, A.G. Potential biological properties of lycopene in a self-emulsifying drug delivery system. Molecules 2023, 28, 1219. [Google Scholar] [CrossRef]
- Binsuwaidan, R.; Sultan, A.A.; Negm, W.A.; Attallah, N.G.; Alqahtani, M.J.; Hussein, I.A.; Shaldam, M.A.; El-Sherbeni, S.A.; Elekhnawy, E. Bilosomes as nanoplatform for oral delivery and modulated in vivo antimicrobial activity of lycopene. Pharmaceuticals 2022, 15, 1043. [Google Scholar] [CrossRef]
- Guo, Z.; Liu, Y.; Luo, Y. Mechanisms of carotenoid intestinal absorption and the regulation of dietary lipids: lipid transporter-mediated transintestinal epithelial pathways. Critical reviews in food science and nutrition 2024, 64, 1791–1816. [Google Scholar] [PubMed]
- Li, Y.; Zhan, M.; Li, J.; Zhang, W.; Shang, X. Lycopene alleviates lipopolysaccharide-induced testicular injury in rats by activating the PPAR signaling pathway to integrate lipid metabolism and the inflammatory response. Translational Andrology and Urology 2023, 12, 271. [Google Scholar] [PubMed]
- Georgiadou, E.C.; Antoniou, C.; Majak, I.; Goulas, V.; Filippou, P.; Smolińska, B.; Leszczyńska, J.; Fotopoulos, V. Tissue-specific elucidation of lycopene metabolism in commercial tomato fruit cultivars during ripening. Scientia Horticulturae 2021, 284, 110144. [Google Scholar] [CrossRef]
- Borrageiro, G.; Haylett, W.; Seedat, S.; Kuivaniemi, H.; Bardien, S. A review of genome-wide transcriptomics studies in Parkinson's disease. European Journal of Neuroscience 2018, 47, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Shi, E.; Chmielecki, J.; Tang, C.-M.; Wang, K.; Heinrich, M.C.; Kang, G.; Corless, C.L.; Hong, D.; Fero, K.E.; Murphy, J.D. FGFR1 and NTRK3 actionable alterations in “Wild-Type” gastrointestinal stromal tumors. Journal of translational medicine 2016, 14, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Amatu, A.; Sartore-Bianchi, A.; Siena, S. NTRK gene fusions as novel targets of cancer therapy across multiple tumour types. ESMO open 2016, 1, e000023. [Google Scholar] [CrossRef]
- Appikonda, S.; Thakkar, K.N.; Barton, M.C. Regulation of gene expression in human cancers by TRIM24. Drug Discovery Today: Technologies 2016, 19, 57–63. [Google Scholar] [CrossRef]
- Chambon, M.; Orsetti, B.; Berthe, M.-L.; Bascoul-Mollevi, C.; Rodriguez, C.; Duong, V.; Gleizes, M.; Thénot, S.; Bibeau, F.; Theillet, C. Prognostic significance of TRIM24/TIF-1α gene expression in breast cancer. The American journal of pathology 2011, 178, 1461–1469. [Google Scholar] [CrossRef]
- Venkatesan, S.; Binko, M.A.; Mielnik, C.A.; Ramsey, A.J.; Lambe, E.K. Deficits in integrative NMDA receptors caused by Grin1 disruption can be rescued in adulthood. Neuropsychopharmacology 2023, 48, 1742–1751. [Google Scholar] [CrossRef]
- Xu, Y.; Song, R.; Chen, W.; Strong, K.; Shrey, D.; Gedela, S.; Traynelis, S.F.; Zhang, G.; Yuan, H. Recurrent seizure-related GRIN1 variant: molecular mechanism and targeted therapy. Annals of clinical and translational neurology 2021, 8, 1480–1494. [Google Scholar] [CrossRef]
- Mohammadi, R.; Ghiasi, M.; Mehdizadeh, S.; Mohammadi, J.; Mohammad Ganji, S. A comprehensive bioinformatic evaluation of the NTRK family’s potential as prognostic biomarkers in breast cancer. Bioinformatics Advances 2025, 5, vbaf030. [Google Scholar] [CrossRef]
- Stravodimou, A.; Voutsadakis, I.A. Neurotrophic receptor tyrosine kinase family members in secretory and non-secretory breast carcinomas. World journal of clinical oncology 2022, 13, 135. [Google Scholar] [CrossRef]
- Becchetti, A.; Grandi, L.C.; Cerina, M.; Amadeo, A. Nicotinic acetylcholine receptors and epilepsy. Pharmacological Research 2023, 189, 106698. [Google Scholar] [CrossRef]
- Kanda, M.; Shimizu, D.; Nakamura, S.; Sawaki, K.; Umeda, S.; Miwa, T.; Tanaka, H.; Inokawa, Y.; Hattori, N.; Hayashi, M. Blockade of CHRNB2 signaling with a therapeutic monoclonal antibody attenuates the aggressiveness of gastric cancer cells. Oncogene 2021, 40, 5495–5504. [Google Scholar] [CrossRef] [PubMed]
- Labate, A.; Mumoli, L.; Fratto, A.; Quattrone, A.; Gambardella, A. Hippocampal sclerosis worsens autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE) phenotype related to CHRNB2 mutation. European Journal of Neurology 2013, 20, 591–593. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhang, H.; Cheng, H.; Wen, J.; Li, D. PIK3CD correlates with prognosis, epithelial–mesenchymal transition and tumor immune infiltration in breast carcinoma. Discover Oncology 2023, 14, 187. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.-R.; Lin, Q.-Y.; Adu, M.; Huang, H.-L.; Yan, Z.-H.; Shao, F.; Zhong, G.-Y.; Zhang, Z.-L.; Sang, Z.-P.; Cao, L. The sources, properties, extraction, biosynthesis, pharmacology, and application of lycopene. Food & Function 2023, 14, 9974–9998. [Google Scholar]
- Yeung, A.W.K.; Choudhary, N.; Tewari, D.; El-Demerdash, A.; Tomczyk, M.; Das, N.; Pirgozliev, V.; Lucarini, M.; Durazzo, A.; Souto, E.B. Lycopene: total-scale literature landscape analysis of a valuable nutraceutical with numerous potential applications in the promotion of human and animal health. Animal Science Papers and Reports 2022, 40, 119–134. [Google Scholar]
- Tang, J.; Li, Y.; Liu, Z.; Wei, M.; Shi, Q.; Yang, F. Integrated transcriptomics and metabolomics analyses reveal the molecular mechanisms of red-light on carotenoids biosynthesis in tomato fruit. Food Quality and Safety 2022, 6, fyac009. [Google Scholar] [CrossRef]
- Sun, S.; Meng, Q.; Bai, Y.; Cao, C.; Li, J.; Cheng, B.; Shi, B.; Shan, A. Lycopene improves maternal reproductive performance by modulating milk composition and placental antioxidative and immune status. Food & Function 2021, 12, 12448–12467. [Google Scholar]
- Fernandes, M.; Barcelos, D.; Carapeto, F.C.L.; Cardili, L.; Comodo, A.N.; Mazloum, S.F.; Marins, M.M.; Mendes, A.R.; Pesquero, J.B.; Landman, G. Evaluation of Heterogeneity in the Coding Region of BRAF, MAP2K1, and MAP2K2 Genes in Primary and Metastatic Melanomas. Journal of Cutaneous Pathology 2024. [Google Scholar] [CrossRef]
- Mizuno, S.; Ikegami, M.; Koyama, T.; Sunami, K.; Ogata, D.; Kage, H.; Yanagaki, M.; Ikeuchi, H.; Ueno, T.; Tanikawa, M. High-throughput functional evaluation of MAP2K1 variants in cancer. Molecular Cancer Therapeutics 2023, 22, 227–239. [Google Scholar] [CrossRef]
- Tzialla, C.; Arossa, A.; Mannarino, S.; Orcesi, S.; Veggiotti, P.; Fiandrino, G.; Zuffardi, O.; Errichiello, E. SCN2A and arrhythmia: A potential correlation? A case report and literature review. European Journal of Medical Genetics 2022, 65, 104639. [Google Scholar] [CrossRef] [PubMed]
- Asadollahi, R.; Delvendahl, I.; Muff, R.; Tan, G.; Rodríguez, D.G.; Turan, S.; Russo, M.; Oneda, B.; Joset, P.; Boonsawat, P. Pathogenic SCN2A variants cause early-stage dysfunction in patient-derived neurons. Human Molecular Genetics 2023, 32, 2192–2204. [Google Scholar] [CrossRef] [PubMed]
- Brunklaus, A.; Brünger, T.; Feng, T.; Fons, C.; Lehikoinen, A.; Panagiotakaki, E.; Vintan, M.-A.; Symonds, J.; Andrew, J.; Arzimanoglou, A. The gain of function SCN1A disorder spectrum: novel epilepsy phenotypes and therapeutic implications. Brain 2022, 145, 3816–3831. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, A.; Pour Abbasi, M.; Khorrami, S.; Khodamoradi, S.; Mohammadi Goldar, Z.; Ebrahimzadeh, F. The TRIM proteins in cancer: from expression to emerging regulatory mechanisms. Clinical and Translational Oncology 2022, 1–11. [Google Scholar] [CrossRef]
- Boyson, S.P.; Gao, C.; Quinn, K.; Boyd, J.; Paculova, H.; Frietze, S.; Glass, K.C. Functional roles of bromodomain proteins in cancer. Cancers 2021, 13, 3606. [Google Scholar] [CrossRef]
- Kulawik, A.; Cielecka-Piontek, J.; Zalewski, P. The importance of antioxidant activity for the health-promoting effect of lycopene. Nutrients 2023, 15, 3821. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




