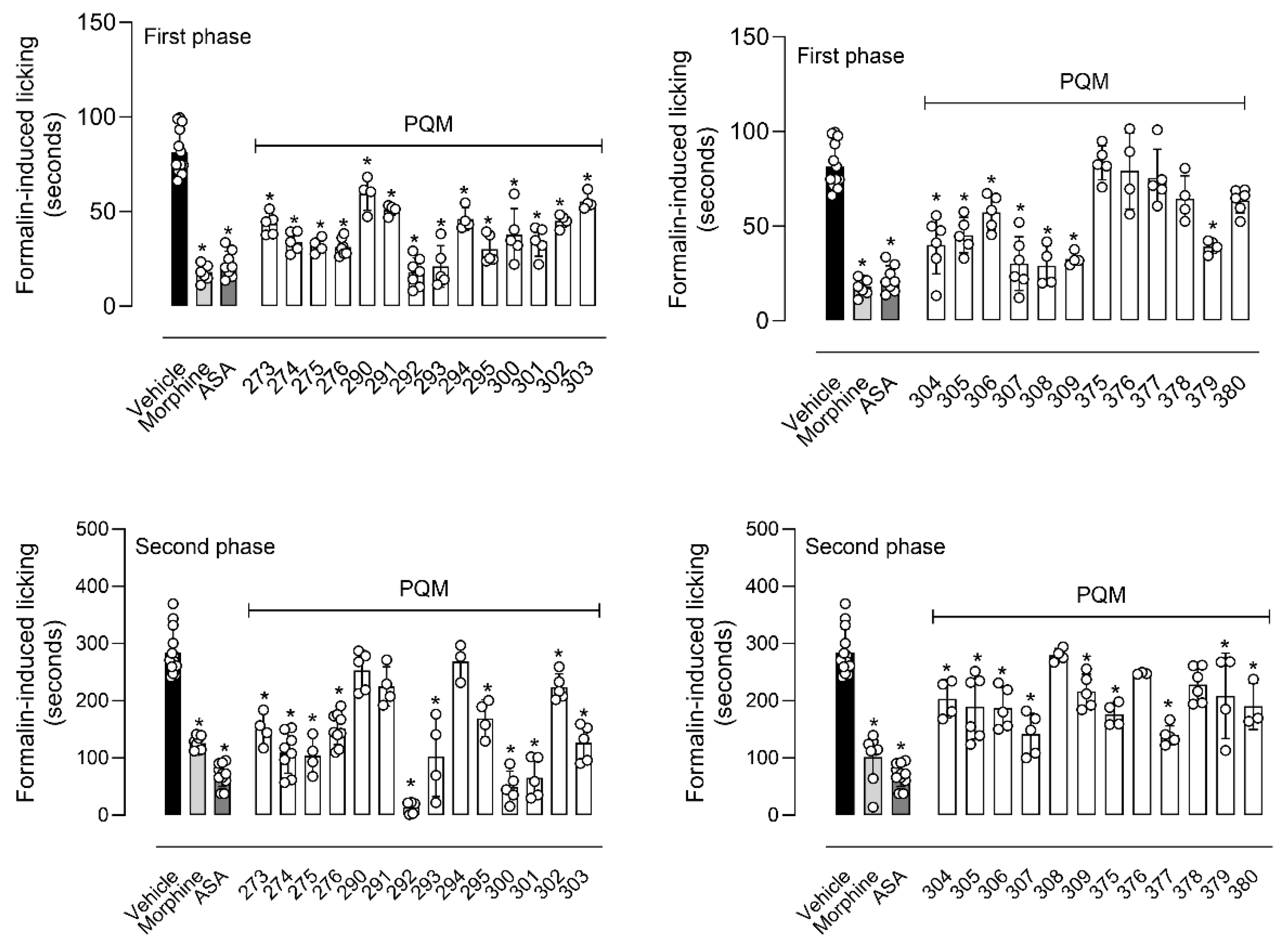
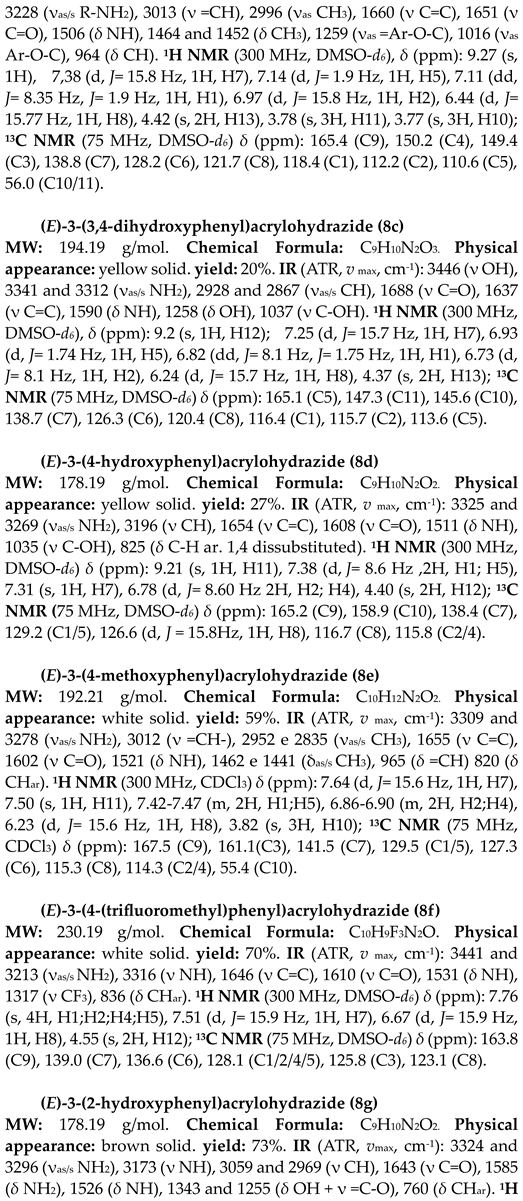
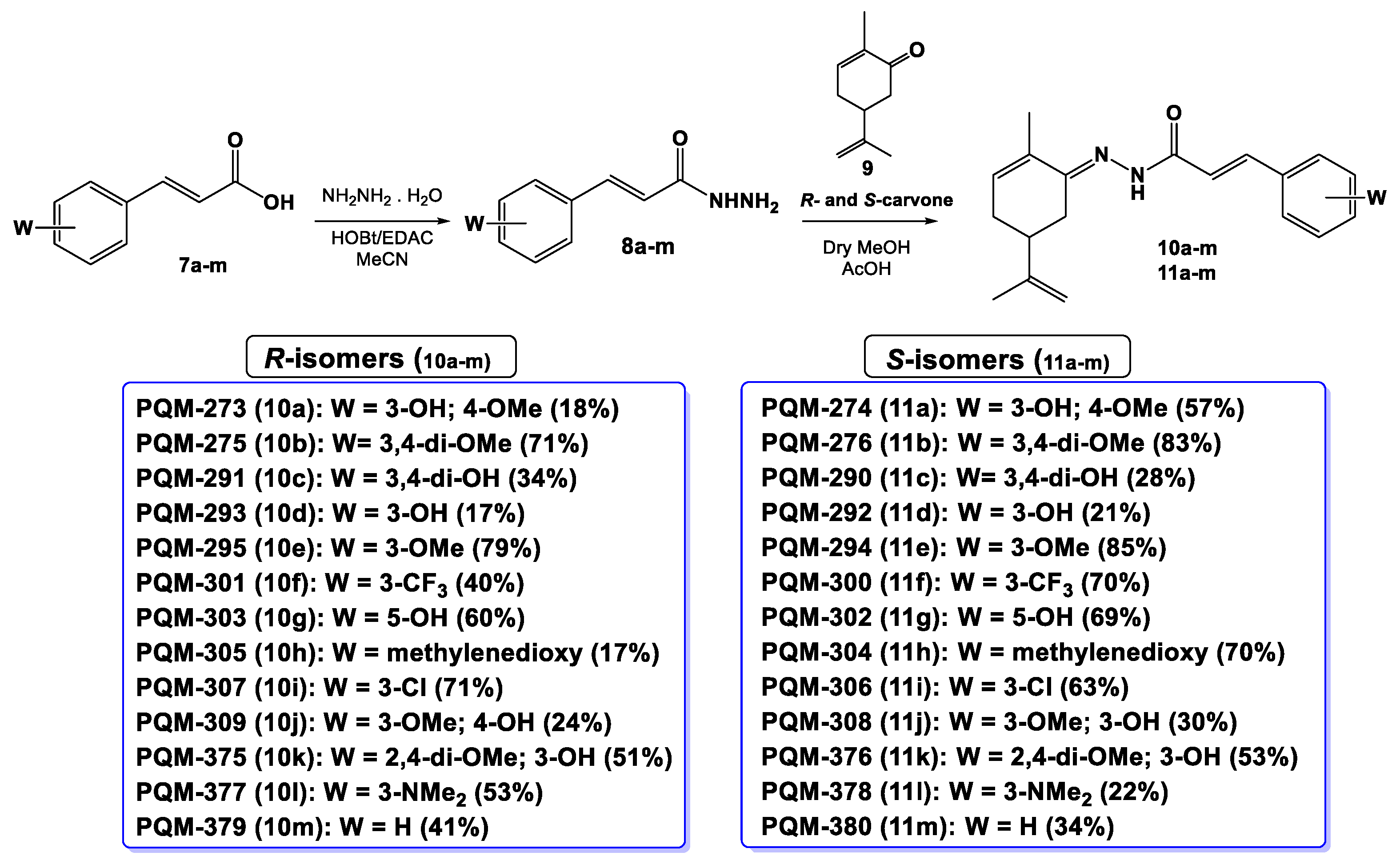
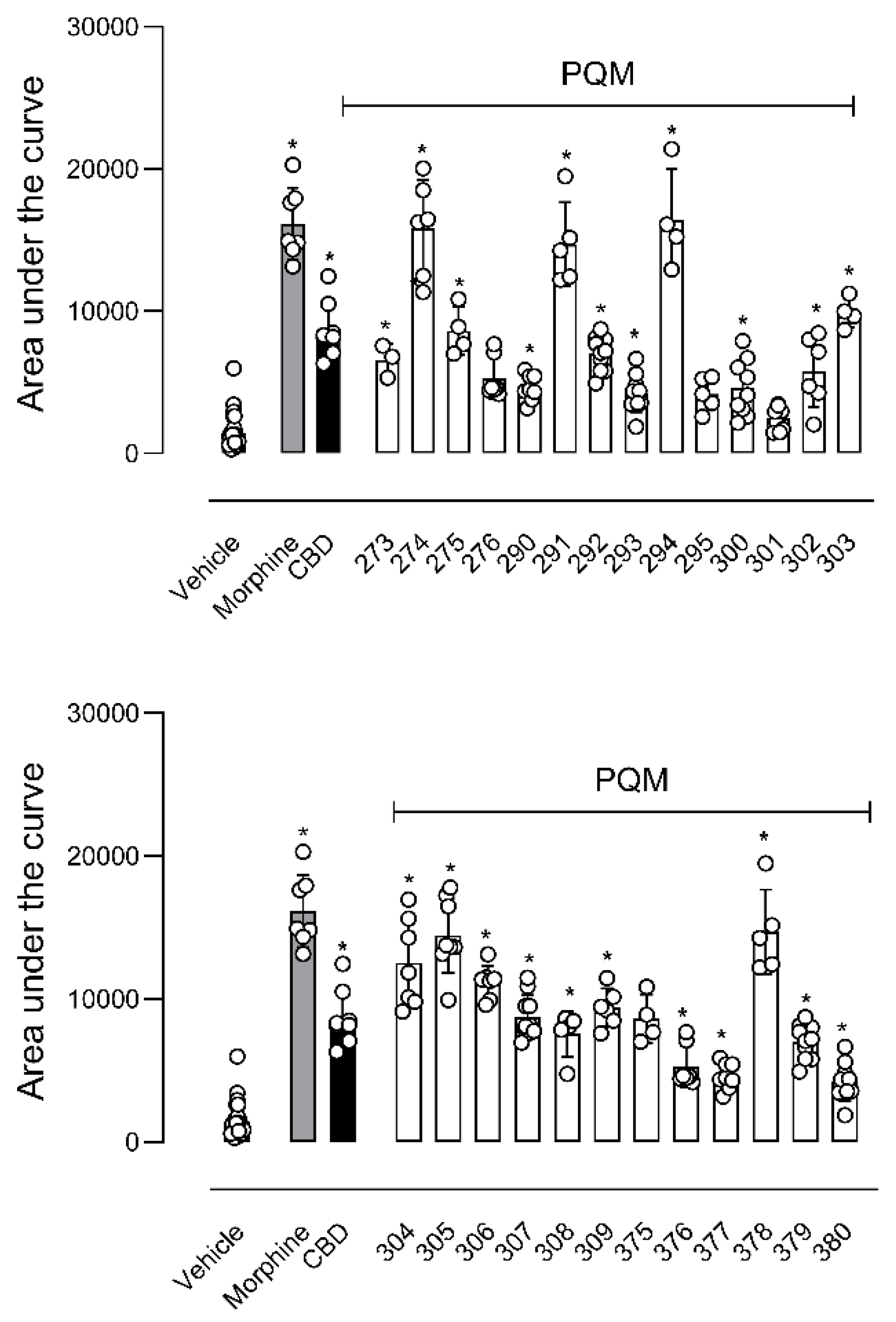
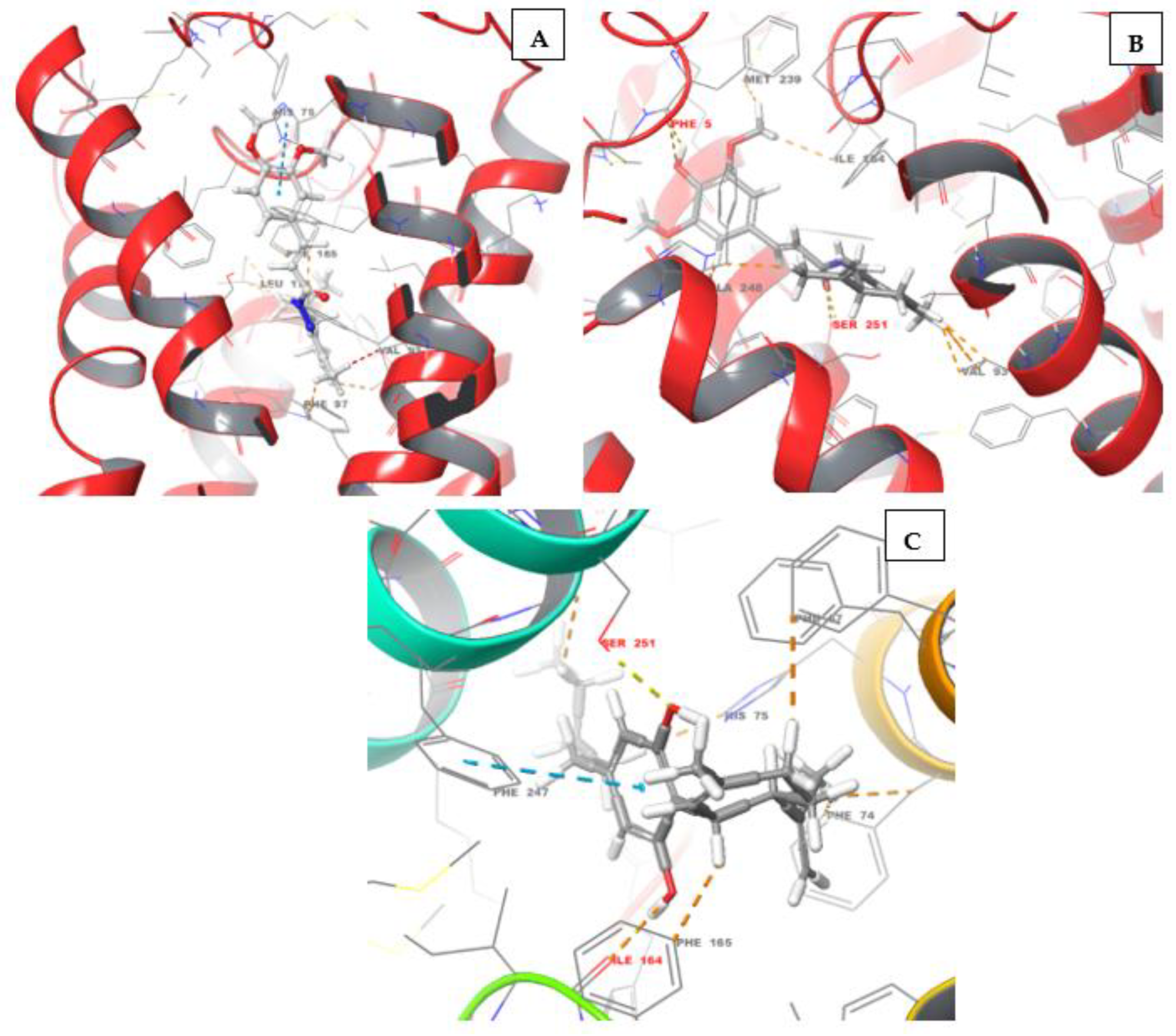
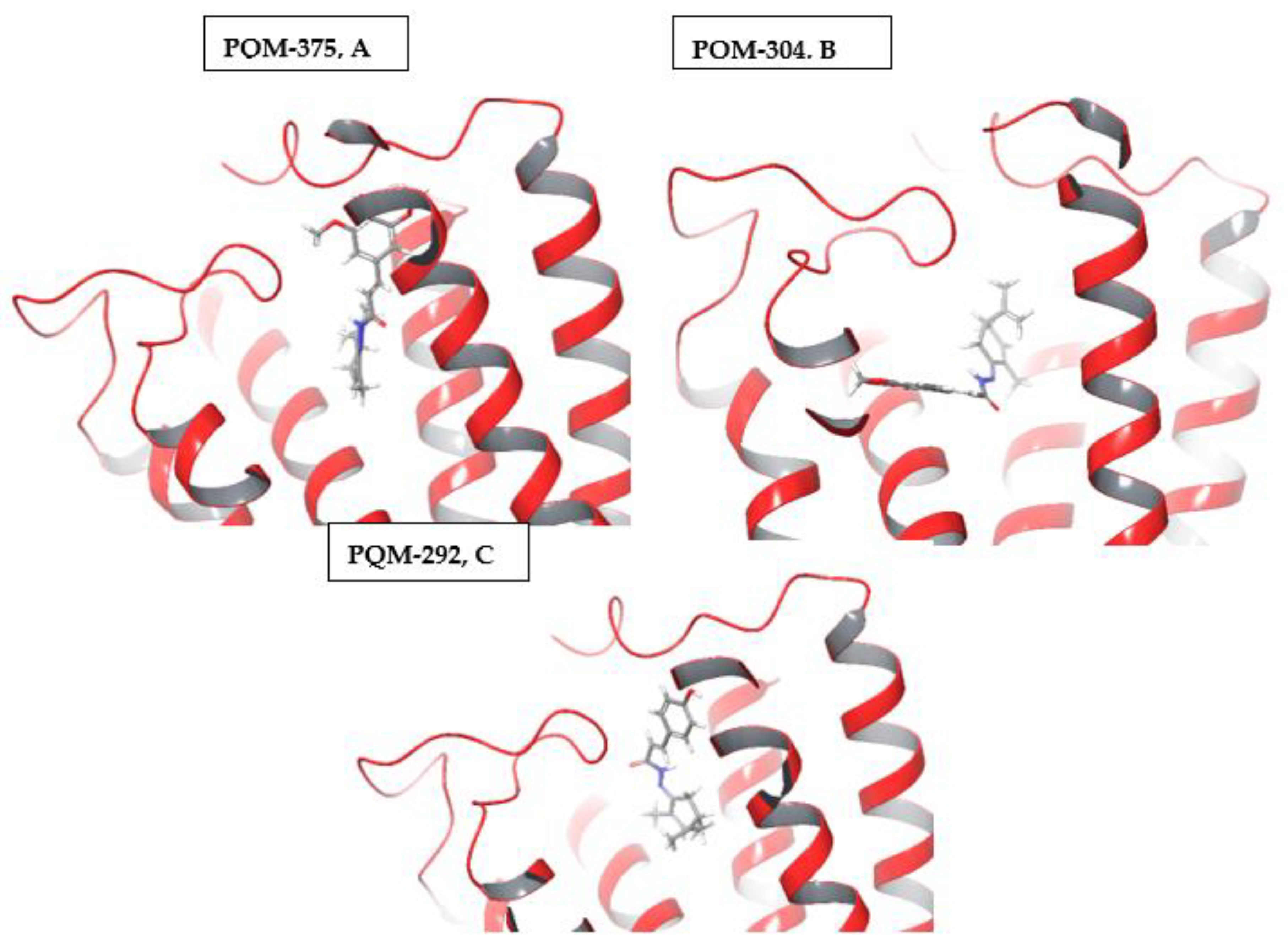
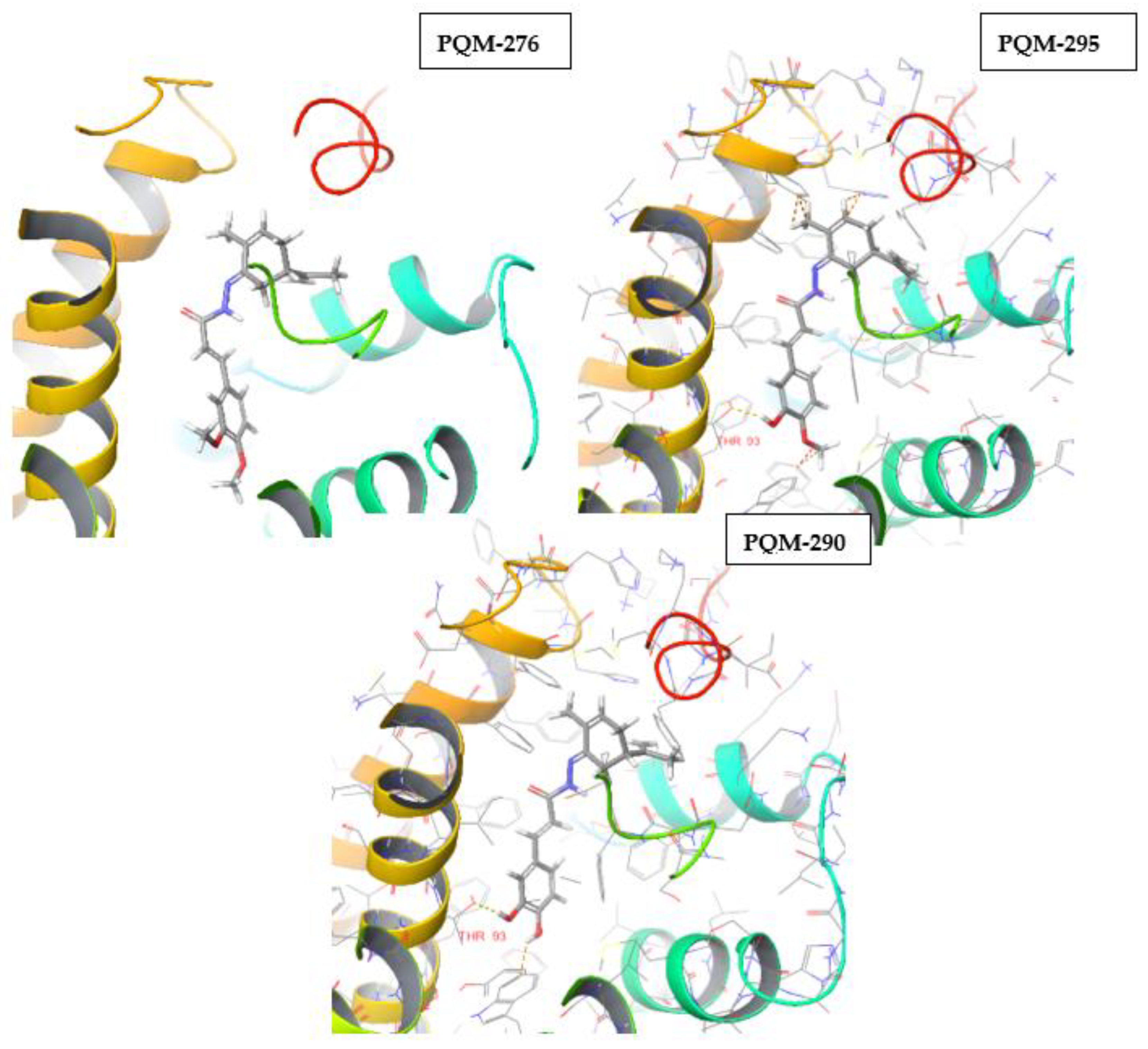
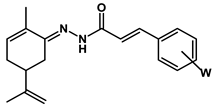
4.1.2. Synthesis of terpene-cinnamoyl-N-acyl-hydrazones (10a-m and 11a-m)
Freshly distilled R- or S-carvones (1 eq.) were diluted in anhydrous MeOH and stirred for 2 minutes, followed by addition of glacial AcOH (10-15 drops). Next, a solution of the correspondent hydrazide intermediates (8a-m,1 eq.) in dry MeOH was added to the reaction flask, and the reaction was maintained under stirring and room temperature, and the reaction was finished when TLC analysis showed the total consumption of the starting material. The reaction mixture was concentrated low pressure, and the solid material was filtered and/or precipitated with ice-cold MeOH, followed by washing with cold-water and cold-MeOH to remove the unreacted carvone amount. The purified products were obtained as solids in 17-79% yields.
(E)-3-(4-hydroxy-3-methoxyphenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-273, 10a)
MW: 340.42 g/mol. Chemical Formula: C20H24N2O3. Physical appearance: pale yellow solid. Melting range: 215-216°C. Purity: 100% (HPLC). Yield: 18%. IR (ATR, νmax, cm-1): 3280 (ν NH), 3055 (νas CH2), 2969 (νs CH2 ou νas CH3), 2919 (νs CH2), 2834 (νs CH3), 1661 (ν C=O), 1635 (C=N), 1515 (δ NH), 1251 (νas Ar-O-C), 1028 (νs Ar-O-C), 978 (δ C-H). 1H NMR (300 MHz, CDCl3), δ (ppm): 8.73 (s, 1H, H12), 7.75 (d, J = 15.9 Hz, 1H, H7), 7.40 (d, J = 15.9 Hz, 1H, H8), 7.17 (d, J = 6.92 Hz, 1H, H16), 7.08 (s, 1H, H5), 6.92 (d, J = 8.14 Hz, 1H, H2), 6.16 (d, J = 8.14 Hz, 1H, H15), 5.92 (s, 1H, H11), 4.82 (d, J = 13.0 Hz, 2H, H20), 3.93 (s, 3H, H10), 2.74 (dd, J= 4.0 Hz, J= 15.3 Hz 1H, H17), 2.29-2.45 (m, 2H, H17; H16), 2.07-2.18 (m, 2H, H18; H16), 1.97 (s, 3H, H21), 1.78 (s, 3H, H22); 13C NMR (75 MHz, CDCl3), δ (ppm): 167.8 (C9), 149.2 (C13), 147.7 (C3), 147.3 (C4), 146.6 (C19), 143.5 (C7), 133.0 (C15), 132.7 (C14), 127.9 (C6), 122.6 (C1), 114.7 (C2), 114.2 (C8), 110.4 (C20), 110.1 (C5), 55.9 (C10), 40.6 (C17), 30.0 (C18), 28.5 (C16), 20.8 (C22), 17.8 (C21). m/z (Rel. Int.): 341.18571.
(E)-3-(4-hydroxy-3-methoxyphenyl)-N'-((S,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-274, 11a)
MW: 340.42 g/mol. Chemical Formula: C20H24N2O3. Physical appearance: yellow solid. Melting range: 209-201°C. Purity (HPLC): 100%. Yield: 57%. IR (ATR, vmax, cm-1): 3274 (ν NH), 2967 (νs =CH2 ou νas CH3), 2918 (νs =CH2), 1654 (ν C=O), 1616 (C=N), 1588 (δ NH), 1508 (ν C=Car)1270 (νas A-O-C), 1030 (νs Ar-O-C). 1H NMR (300 MHz, CDCl3), δ (ppm): 9.24 (s, 1H, H12), 7.74 (d, J= 15.9 Hz, 1H, H7), 7.42 (d, J= 15.9 Hz, 1H, H8), 7.16 (dd, J= 8.2 Hz, J= 1.4 Hz, 1H, H16), 7.08 (d, J= 1.4 Hz, 1H, H5), 6.94 (d, J= 8.2 Hz, 1H, H2), 6.16 (d, J= 5.2 Hz, 1H, H15), 6.06, (s, 1H, H11), 4.83 (d, J= 4.3 Hz, 2H, H20), 3.93 (s, 3H, H10), 2.83 (dd, J= 3.4 Hz, J= 16.1 Hz, 1H, H18), 2.45 (dt, J= 4.0 Hz, J= 11.9 Hz 1H, H17), 2.33 (dt, J= 5.0 Hz, J= 17.0 Hz 1H, H16), 2.05-2.19 (m, 2H, H18; H16), 1.97 (s, 3H, H21), 1.79 (s, 3H, H22); 13C NMR (75 MHz, CDCl3), δ (ppm): 167.8 (C9), 149.5 (C13), 147.7 (C3), 147.4 (C4), 146.6 (C19), 143.4 (C7), 133.0 (C15), 132.7 (C14), 127.9 (C6), 122.6 (C1), 114.8 (C2), 114.3 (C8), 110.4 (C20), 110.1 (C5), 55.9 (C10), 40.6 (C17), 30.1 (C18), 28.7 (C16), 20.8 (C22), 17.8 (C21). m/z (Rel. Int.): 341.18582.
(E)-3-(3,4-dimethoxyphenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-275, 10b)
MW: 354.45 g/mol. Chemical Formula: C21H26N2O3. Physical appearance: pale yellow solid. Melting range: 246-248°C. Purity (HPLC): 100%. Yield: 71%. IR (ATR, vmax, cm-1): 3158 (ν NH), 3063 (νas CH2), 2988 (νs CH2 ou νas CH3), 2907 (νs CH2), 2834 (νs CH3), 1659 (ν C=O), 1594 (δ NH), 1518 (ν C=Car), 1463 e 1443 (δas/s CH3), 1253 (νas Ar-O-C), 1023 (νs Ar-O-C), 980 (δ C-H). 1H NMR (300 MHz, CDCl3), δ (ppm): 9.36 (s, 1H, H12), 7.76 (d, J= 15.9 Hz, 1H, H7), 7.46 (d, J= 15.9 Hz, 1H, H8), 7.18 (d, J= 8.3 Hz, 1H, H1), 7.13 (s, 1H, H5), 6.89 (d, J= 8.3 Hz, 1H, H2), 6.16 (d, J= 5.3 Hz, 1H, H15), 4.84 (s, 2H, H20), 3.93 (s, 6H, H11;H10), 2.87 (d, J= 14.1 Hz, 1H, H18), 2.41 (dt, J= 9.2 Hz, J= 7.7 Hz 1H, H17), 2.29-2.32 (m, 1H, H16), 2.11-2.18 (m, 2H, H18; H16), 1.97 (s, 3H, H21), 1.80 (s, 3H, H20); 13C NMR (75 MHz, CDCl3), δ (ppm): 168.0 (C9), 150.8 (C3), 149.5 (C13), 149.1 (C4), 143.1 (C7), 133.0 (C14), 132.8 (C15), 128.4 (C6), 122.3 (C1), 114.7 (C2), 111.0 (C8), 110.4 (C5), 110.0 (C20), 56.0 (C11), 55.8 (C10), 40.6 (C17), 30.1 (C18), 28.8 (C16), 20.9 (C20), 17.8 (C21). m/z (Rel. Int.): 355.20161.
(E)-3-(3,4-dimethoxyphenyl)-N'-((S,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-276, 11b)
MW: 354.45 g/mol. Chemical Formula: C21H26N2O3. Physical appearance: pale yellow solid. Melting range: 239-240°C. Purity (HPLC): 100%. Yield: 83%. IR (ATR, v max, cm-1): 3158 (ν NH), 3063 (νas CH2), 2988 (νs CH2 ou νas CH3), 2907 (νs CH2), 2834 (νs CH3), 1660 (ν C=O), 1615 (ν C=N), 1595 (δ NH), 1518 (ν C=Car), 1463 and 1443 (δas/s CH3), 1253 (νas Ar-O-C), 1023 (νs Ar-O-C), 981 (δ C-H). 1H NMR (300 MHz, CDCl3), δ (ppm): 9.48 (s, 1H, H12), 7.75 (d, J= 15.9 Hz, 1H, H7), 7.46 (d, J= 15.9 Hz, 1H, H8), 7.18 (d, J= 8.2 Hz, 1H, H1), 7.13 (s, 1H, H5), 6.89 (d, J= 8.2 Hz, 1H, H2), 6.16 (d, J= 4.1 Hz, 1H, H15), 4.85 (s, 2H, H20), 3.93 (s, 6H, H10; H11), 2.88 (dd, J= 15.9 Hz, J= 3.1 Hz, 1H, H18), 2.45 (t, J= 11.9 Hz, 1H, H17), 2.34 (dd, J= 18.0 Hz, J= 5.0 Hz , 1H, H16), 2.12 (dd, J= 15.0 Hz, J= 13.4 Hz 2H, H18; H16), 1.97 (s, 3H, H21), 1.81 (s, 3H, H20); 13C NMR (75 MHz, CDCl3), δ (ppm): 168.0 (C9), 150.8 (C3), 149.6 (C13), 149.0 (C4), 143.1 (C7), 133.0 (C14), 132.8 (C15), 128.4 (C6), 122.3 (C1), 114.7 (C2), 111.0 (C8), 110.4 (C5), 110.0 (C20), 56.0 (C11), 55.8 (C10), 40.6 (C17), 30.1 (C18), 28.8 (C16), 20.9 (C20), 17.8 (C21). m/z (Rel. Int.): 355.20167.
(E)-3-(3,4-dihydroxyphenyl)-N'-((S,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-290, 11c)
MW: 326.40 g/mol. Chemical Formula: C19H22N2O3. Physical appearance: pale yellow solid. Melting range: 195-197ºC. Purity (HPLC): 100%. Yield: 28%. IR (ATR, vmax, cm-1): 3300 (ν OH), 3244 (ν NH), 1648 (ν C=O), 1616 (ν C=N), 1603 (δ NH), 1514 (ν C=Car), 1371 e 1120 (δ OH and ν =C-O), 979 (δ C=CH2), 817 (δ RC=CH). 1H NMR (300 MHz, DMSO-d6), δ (ppm): 10.45-10.41 (s, 1H, H12), 7.37 (dt, J= 16.0 Hz J= 30.08 Hz, 1H, H7), 7.02 (d, J= 13.2 Hz, 1H, H5), 6.91 (t, J= 6.8 Hz 1H, H1), 6.73 (dd, J= 11.9 Hz and J= 16.0 Hz, 2H, H2 e H8), 6.15 (s, 1H, H15), 4.80 (t, J= 7.7 Hz, 2H, H20), 2.92 (t, J= 12.7 Hz, 1H, H18), 2.21-2.36 (m, 2H, H17; H16), 2.01-2.14 (m, 2H, H18; H16), 1.91-1.83 (s, 3H, H22), 1.76 (s, 3H, H21); 13C (75 MHz, DMSO-d6), δ (ppm): 164.7/ 167.6 (C9,C9’), 149.7/153.5 (C13,C13’), 148.2 (C3), 146.1 (C4 e C19), 141.1/142.8 (C7, C7’), 133.8 (C6), 133.1 (C15), 132.7 (C14), 121.2 (C1), 117.7 (C8), 116.3 (C2), 114.5 (C5), 110.8 (C20), 18.3 (C22), 40.5 (C17), 30.1 (C18), 29.6 (C16), 20.8 (C21). m/z (Rel. Int.): 327.17020.
(E)-3-(3,4-dihydroxyphenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-291, 10c)
MW: 326.40 g/mol. Chemical Formula: C19H22N2O3. Physical appearance: yellow solid. Melting range: 204-205ºC. Purity (HPLC): 100%. Yield: 34%. IR (ATR, vmax, cm-1): 3299 (ν OH), 3243 (ν NH), 2972 (νs CH2 or νas CH3), 1648 (ν C=O), 1616 (ν C=N), 1603 (δ NH), 1514 (ν C=Car), 1370 and 1120 (δ OH and =C-O), 979 (δ C=CH2), 816 (δ RC=CH). 1H NMR (300 MHz, DMSO-d6), δ (ppm): 10.46-10.41 (s, 1H, H12), 9.46 (s, 1H, H11), 9.23 (s, 1H, H10), 7.37 (dt, J= 16.2 Hz J= 29.7 Hz, 1H, H7), 7.02 (d, J= 13.2 Hz, 1H, H5), 6.89-6.93 (m, 1H, H16), 6.74 (dd, J= 12.1 Hz and J= 16.41 Hz, 2H, H2; H8), 6.15 (s, 1H, H15), 4.80 (t, J= 7.2 Hz, 2H, H20), 2.93 (t, J= 13.2 Hz, 1H, H18), 2.21-2.36 (m, 2H, H17; H16), 2.03-2.14 (m, 2H, H18; H16), 1.91-1.83 (s, 3H, H22), 1.76 (s, 3H, H21); 13C NMR (75 MHz, DMSO-d6), δ (ppm): 162.7/ 167.6 (C9,C9’), 149.7/153.5 (C13,C13’), 148.1 (C3), 146.1 (C4), 146.0 (C19), 141.0/142.8 (C7, C7’), 133.8 (C6), 133.0 (C15), 132.7 (C14), 121.2 (C1), 117.7 (C8), 116.3 (C2), 114.1 (C5), 110.8 (C20), 40.5 (C17), 30.1 (C18), 29.6 (C16), 21.0 (C21), 18.5 (C22). m/z (Rel. Int.): 327.17022.
(E)-3-(4-hydroxyphenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-292, 11d)
MW: 310.40 g/mol. Chemical Formula: C19H22N2O2. Physical appearance: pale yellow solid. Melting range: 189-190ºC. Purity: 100%. Yield: 21%. IR (ATR, vmax, cm-1): 3299 (ν OH), 3066 (νas CH2), 3014 (νs CH2), 2953 and 2922 (νas/s CH3), 1654 (ν C=O), 1622 (ν C=N), 1601 (δ NH), 1516 (ν C=Car), 1442 (δs CH2), 1375 (δs CH3), 1274 (ν C-N), 1200 (ν C-O), 1166 (δ OH + =C-O), 974 (δ HC=CH), 887 (δ =CH), 827 (δ CHar). 1H NMR (300 MHz, DMSO-d6), δ (ppm): 10.4 (s, 1H, H10), 9.98 (s, 1H, H11), 7.31-7.59 (m, 3H, H1, H5; H7), 6.80 (t, J = 10.4 Hz, 3H, H2/H4; H8), 6.15 (s, 1H, H14), 4.80 (t, J= 7.65 Hz, 2H, H19), 2.93 (t, J= 13.4 Hz, 1H, H15), 2.32 (dd, J= 9.7 Hz, J= 15.9 Hz, 1H, H16), 2.02-2.22 (m, 3H, H17; H15), 1.85 (s,1H, H21), 1.75 (s, 1H, H20); 13C NMR (75 MHz, DMSO-d6), δ (ppm): 167.6 (C9), 159.6 (C3), 153.5 (C12), 133.8 (C14), 133.0 (C13), 129.9 (C1, C5), 126.4 (C6), 117.8 (C8), 116.3 (C2, C4), 110.8 (C19), 40.6 (C16), 30.1 (C17), 29.6 (C15), 20.9 (C20), 18.5 (C21). m/z (Rel. Int.): 311.17534.
(E)-3-(4-hydroxyphenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-293, 10d)
MW: 310,40 g/mol. Chemical Formula: C19H22N2O2. Physical appearance: pale yellow solid. Melting range: 227-230ºC. Purity (HPLC): 100%. Yield: 17%. IR ( ATR, v max, cm-1): 3301 (ν OH), 3067 (νas CH2), 3014 e 2973 (νs/as C2), 2953 and 2921 (νas/s CH3), 1653 (ν C=O), 1622 (ν C=N), 1601 (δ NH), 1521 (ν C=Car), 1442 (δs CH2), 1375 (δs CH3), 1274 (ν C-N), 1201 (ν C-O), 1167 (δ OH + =C-O), 974 (δ HC=CH), 888 (δ =CH), 827 (δ CHar). 1H NMR (300 MHz, DMSO-d6), δ (ppm): 10.4 (s, 1H, H10), 9.96 (s, 1H, H11), 7.31-7.58 (m, 3H, H1, H5; H7), 6.79 (t, J= 9.95 Hz, 3H, H2/H4; H8), 6.15 (s, 1H, H14), 4.80 (t, J= 8.05 Hz, 2H, H19), 2,93 (t, J= 14.3 Hz, 1H, H15), 2.28-2.36 (m, 1H, H16), 1.98-2.22 (m, 3H, H17; H15), 1.85 (s,1H, H21), 1.75 (s, 1H, H20); 13C NMR (75 MHz, DMSO-d6), δ (ppm): 167.6 (C9), 159.6 (C3), 153.5 (C12), 133.8 (C14), 133.0 (C13), 130.0 (C1, C5), 126.4 (C6), 117.8 (C8), 116.3 (C2, C4), 110.8 (C19), 40.6 (C16), 30.1 (C17), 29.6 (C15), 20.9 (C20), 18.4 (C21). m/z (Rel. Int.): 311.17540.
(E)-3-(4-methoxyphenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-294, 11e)
MW: 324,42 g/mol. Chemical Formula: C20H24N2O2. Physical appearance: pale yellow solid. Melting range: 230-232ºC. Purity (HPLC): 100%. Yield: 85%. IR ( ATR, v max, cm-1): 3161 (ν NH), 3073 (νas CH2), 3031 (νs CH2), 2968 and 2918 (νas/s CH3), 2835 (νs OCH3), 1657 (ν C=O), 1617 (ν C=N), 1595 (δ NH), 1509 (ν C=Car), 1463 (δs CH2), 1374 (δs CH3), 1252 (νas C-O), 1166 (ν C-N), 984 (δ HC=CH), 892 (δ =CH), 817 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 1.80 (s, 3H, H20), 1.97 (S, 3H, H21), 2.07-2.17 (m, 2H, H17; H15), 2.28-2.48 (m, 2H, H16; H15), 2.89 (dd, J= 16.0.Hz, J= 3.5 Hz, 1H, H17), 3.84 (s, 3H, H10), 4.84 (s, 2H), 6.15 (d, J= 5.6 Hz, 1H, H14), 6.92 (d, J= 8.6 Hz, 2H, H2, H4), 7.48 (d, J= 16.0 Hz, 1H, H8), 7.54 (d, J= 8.6 Hz, 2H, H1; H5), 7.77 (d, J= 16.0 Hz, 1H, H7), 9.49 (s, 1H, H11). 13CNMR (75 MHz, CDCl3), δ (ppm): 168.2 (C9), 161.1 (C3), 149.6 (C18), 147.5 (C7), 142.9 (C13), 132.8 (C14), 129.8 (C1, C5), 128.1 (C6), 114.4 (C8), 114.2 (C2, C4), 110.3 (C19), 55.6 (C10), 40.6 (C16), 30.1 (C17), 28.8 (C15), 20.8 (C20), 17.9 (C21). m/z (Rel. Int.): 325.19118.
(E)-3-(4-methoxyphenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-295, 10e)
MW: 324,42 g/mol. Chemical Formula: C20H24N2O2. Physical appearance: pale yellow solid. Melting range: 215-216ºC. Purity (HPLC): 100%. Yield: 79%. IR (ATR, v max, cm-1): 3162 (ν NH), 3073 (νas CH2), 3032 (νs CH2), 2968 and 2918 (νas/s CH3), 2835 (νs OCH3), 1658 (ν C=O), 1617 (ν C=N), 1595 (δ NH), 1510 (ν C=Car), 1463 (δs CH2), 1375 (δs CH3), 1252 (νas C-O), 1166 (ν C-N), 984 (δ HC=CH), 893 (δ =CH), 817 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 9.50 (s, 1H, H11), 7.77 (d, J= 16.0 Hz, 1H, H7), 7.54 (d, J= 8.6 Hz, 2H, H1; H5), 7.46 (d, J= 16.0 Hz, 1H, H8), 6.92 (d, J= 8.6 Hz, 2H, H2, H4), 6.15 (d, J= 4.3 Hz, 1H, H14), 4.84 (s, 2H, H19), 3.84 (s, 3H, H10), 2.89 (dd, J= 16.1 Hz, J= 2.9 Hz, 1H, H17), 2.30-2.48 (m, 2H, H16; H15), 2.12 (dd, J= 14.4 Hz, J= 13.1 Hz, 2H, H17; H15), 1.98 (s, 3H, H21), 1.80 (s, 3H, H20); 13C NMR (75 MHz, CDCl3), δ (ppm): 168.2 (C9), 161.1 (C3), 149.6 (C18), 147.5 (C7), 142.9 (C13), 132.8 (C14), 129.8 (C1, C5), 128.1 (C6), 114.4 (C8), 114.2 (C2, C4), 110.3 (C19), 55.4 (C10), 40.6 (C16), 30.1 (C17), 28.9 (C15), 20.8 (C20), 17.9 (C21). m/z (Rel. Int.): 325.19113.
(E)-N'-((S,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)-3-(4-(trifluoromethyl)phenyl)acrylohydrazide (PQM-300, 11f)
MW: 362,40 g/mol. Chemical Formula: C20H21F3N2O. Physical appearance: white solid. Melting range: 220-221ºC. Purity (HPLC): 100%. Yield: 70%. IR (ATR, vmax, cm-1): 3168 (ν NH), 1688 (ν C=O), 1624 (δ NH), 1575 (ν C=N), 1318 and 1124 (ν CF3), 979 (δ HC=CH), 895 (δ =CH), 831 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 9.26 (s, 1H, H11), 7.81 (d, J = 16.1 Hz, 1H, H7), 7.62-7.70 (m, 5H, H1; H2; H4; H5; H8), 6.19 (d, J= 7.1 Hz, 1H, H14), 4,84 (d, J= 5.6 Hz 2H, H19), 2.83 (dd, J= 15.7 Hz, J= 4.0 Hz, 1H, H17), 2.30-2.48 (m, 2H, H16; H15), 2.07-2.18 (m, H17, H15), 1.97 (s, 3H, H21), 1.80 (s, 3H, H20); 13C NMR (75 MHz, CDCl3), δ (ppm): 167.1 (C9), 150.2 (C3), 147.3 (C18), 141.4 (C7), 138.7 (C6), 133.5 (C14), 132.7 (C13), 128.3 (C1; C2; C4; C5), 119.4 (C8), 110.5 (C19), 40.6 (C16), 30.1 (C17), 28.8 (C15), 20.8 (C20), 17.9 (C21). m/z (Rel. Int.): 363.16785.
(E)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)-3-(4-(trifluoromethyl)phenyl)acrylohydrazide (PQM-301, 10f)
MW: 362,40 g/mol. Chemical Formula: C20H21F3N2O. Physical appearance: white solid. Melting range: 102-103ºC. Purity (HPLC): 100%. Yield: 40%. IR (ATR, v max, cm-1): 3167 (ν NH), 2923 (νs CH2), 1666 (ν C=O), 1623 (δ NH), 1575 (ν C=N), 1318 (ν C-F3), 979 (δ HC=CH), 895 (δ =CH), 830 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 9.36 (s, 1H, H11), 7.80 (d, J= 16.1 Hz, 1H, H7), 7.61-7.69 (m, 5H, H1; H2; H4; H5; H8), 6.18 (d, J= 7.1 Hz, 1H, H14), 4.83 (d, J= 4.8 Hz 2H, H19), 2.83 (dd, J= 15.9 Hz, J= 3,8 Hz, 1H, H17), 2.31-2.49 (m, 2H, H16; H15), 2.03-2.21 (m, H17, H15), 1.96 (s, 3H, H21), 1.79 (s, 3H, H20); 13C NMR (75 MHz, CDCl3), δ (ppm): 167.2 (C9), 150.3 (C3), 147.3 (C18), 141.4 (C7), 138.7 (C6), 133.6 (C14), 132.7 (C13), 128.3 (C1; C2; C4; C5), 119.4 (C8), 110.4 (C19), 40.6 (C16), 30.1 (C17), 28.8 (C15), 20.8 (C20), 17.9 (C21). m/z (Rel. Int.): 363.16781.
(E)-3-(2-hydroxyphenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-302, 11g)
MW: 310,39 g/mol. Chemical Formula: C19H22N2O2. Physical appearance: yellow solid. Melting range: 215-217ºC. Purity (HPLC): 100%. Yield: 69%. IR (ATR, v max, cm-1): 3208 (ν OH), 1644 (ν C=O), 1599 (δ NH), 1451 (δ CH3), 1200 (ν C-OH), 989 (δ HC=CH), 888 (δ =CH), 754 (δ CHar). 1H NMR (300 MHz, DMSO-d6), δ (ppm): 10.13-10.46 (s, 1H, H11; H11’), 7.82 (t, J= 16.9 Hz, 1H, H7), 7.47-7.66 (m, 1H, H8), 7.20 (t, J= 7.8 Hz, 1H, H3), 6.82-7.01 (m, 3H, H2; H4; H5), 6.16 (s, 1H, H14), 4.78-4.83 (m, 2H, H19), 2.94 (t, J= 12.5 Hz, 1H, H17), 2.21-2.36 (m, 2H, H16; H15), 2.03-2.11 (m, 2H, H17, H15), 1.89-1.84 (s, 3H, H21), 1.76 (s, 3H, H20). 13C NMR (75 MHz, DMSO-d6), δ (ppm): 167.9 (C9), 157.2 (C5), 153.8 (C12), 149.7 (C7), 148.3 (C18), 138.7 (C6), 138.0 (C13), 136.2 (C3), 133.1 (C14), 133.9 (C1), 122.2 (C8), 121.7 (C6), 120.6 (C2), 117.7 (C6), 116.7 (C4), 110.8 (C19), 40.6 (C16), 30.2 (C17), 29.6 (C15), 21.0 (C20), 18.5 (C21). m/z (Rel. Int.): 311.17541.
(E)-3-(2-hydroxyphenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-303, 10g)
MW: 310,39 g/mol. Chemical Formula: C19H22N2O2. Physical appearance: pale yellow solid. Melting range: 231-233ºC. Purity (HPLC): 100%. Yield: 60%. IR (ATR, v max, cm-1): 3208 (ν OH), 1644 (ν C=O), 1651 (ν C=N), 1599 (δ NH), 1451 (δ CH3), 1200 (ν C-OH), 989 (δ HC=CH), 889 (δ =CH), 754 (δ CHar). 1H NMR (300 MHz, DMSO-d6), δ (ppm): 10.41-10.52 (s, 1H, H10), 10.13 (s, 1H, H11), 7.81 (t, J= 17.1 Hz, 1H, H7), 7.56 (dd, J= 11.2 Hz and J= 44.2 Hz 1H, H8), 7,20 (t, J = 7.75.Hz, 1H, H3), 6.84-6.92 (m, 3H, H2; H4; H1), 6.16 (s, 1H, H14), 4.78-4.83 (m, 2H, H19), 2.94 (t, J= 13.6 Hz, 1H, H17), 2.29 (dd, J= 14.7 Hz e J= 26.2 Hz, 2H, H16; H15), 2.09 (dd, J= 15.5 Hz and J= 28.0 Hz, 2H, H17; H15), 1.88-1.84 (s, 3H, H21;H21’), 1.76 (s, 3H, H20); 13C NMR (75 MHz, DMSO-d6), δ (ppm): 167.4 (C9), 156.4 (C5), 153.3 (C12), 149.2 (C7), 147.8 (C18), 138.7 (C6), 137.5 (C13), 135.7 (C3), 133.4 (C14), 132.6 (C1), 130.8 (C8), 121.7 (C6), 119.4 (C2), 116.2 (C4), 110.3 (C19), 40.6 (C16), 29.7 (C15, C17), 20.5 (C20), 18.0 (C21). m/z (Rel. Int.): 311.17543.
(E)-3-(benzo[d][
1,
3]
dioxol-5-yl)-N'-((S,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-304, 11h)
MW: 338,41 g/mol. Chemical Formula: C20H22N2O3. Physical appearance: pale yellow solid. Melting range: 244-245ºC. Purity (HPLC): 100%. Yield: 70%. IR (ATR, v max, cm-1): 3155 (ν NH), 2916 (νs CH2), 1669 (ν C=O), 1629 (C=N), 1609 (δ NH), 1485 (δ CH2), 1361 (δ CH3), 1239 (νass C-O-C), 1035 (νs C-O), 973 (δ HC=CH). 1H NMR (300 MHz, CDCl3), δ (ppm): 9,31 (s, 1H, H11), 7,72 (d, J = 15,87Hz, 1H, H7), 7,40 (d, J = 15,90Hz, 1H, H8), 7,11 (s, 1H, H1), 7,06 (d, J = 8,80Hz, 1H, H5), 6,83 (d, J = 7,98Hz, 1H, H4), 6,15 (d, J = 5,98Hz, 1H, H14), 6,01 (s, 2H, H10), 4,83 (s, 2H, H19), 2,85 (dd, J = 3,74Hz e J = 15,89Hz 1H, H17), 2,37-2,48 (m, 1H, H16), 2,28-2,36 (m, 1H, H15), 2,10-2,18 (m, 2H, H17, H15), 1,97 (s, 3H, H21), 1,80 (s, 3H, H20); 13C NMR (75 MHz, CDCl3), δ (ppm): 167,9 (C9), 149,6 (C3), 149,3 (C12), 148,2 (C2), 147,4 (C18), 143,0 (C7), 133,0 (C13), 132,9 (C6), 129,8 (C5), 124,5 (C8), 114,8 (C1), 110,4 (C4), 108,5 (C19), 106,6 (C14), 101,5 (C10), 40,6 (C16), 30,1(C17), 28,8 (C15), 20,8 (C20), 17,9 (C21). m/z (Rel. Int.): 339.17039.
(E)-3-(benzo[d][
1,
3]
dioxol-5-yl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-305, 10h)
MW: 338,41 g/mol. Chemical Formula: C20H22N2O3. Physical appearance: pale yellow solid. Melting range: 237-239ºC. Purity: 100%. Yield: 17%. IR (ATR, v max, cm-1): 3355 (ν NH), 1653 (ν C=O), 1610 (C=N), 1595 (δ NH), 1489 (δ CH2), 1374 (δ CH3), 1254 (νass C-O-C), 1102 (νs C-O), 985 (δ HC=CH). 1H NMR (300 MHz, CDCl3), δ (ppm): 9,08 (s, 1H, H11), 7,73 (d, J = 15,9Hz, 1H, H7), 7,39 (d, J = 15,95Hz, 1H, H8), 7,12 (s, 1H, H1), 7,06 (dd, J = 8,11Hz, J = 1,03Hz, 1H, H5), 6,83 (d, J = 7,98Hz, 1H, H4), 6,16 (d, J = 5,68Hz, 1H, H14), 6,02 (s, 2H, H10), 4,84 (d, J = 5,62Hz, 2H, H19), 2,80 (dd, J = 3,84Hz e J = 15,28Hz 1H, H17), 2,29-2,46 (m, 2H, H16 e H15), 2,12 (dd, J=6,24Hz e J=8,34Hz, J=15,54Hz 2H, H17, H15), 1,97 (s, 3H, H21), 1,79 (s, 3H, H20), 13C NMR (75 MHz, CDCl3), δ (ppm): 167,8 (C9), 149,5 (C3), 149,3 (C12), 148,2 (C2), 147,4 (C18), 143,1 (C7), 133,0 (C13), 132,8 (C6), 129,8 (C5), 124,5 (C8), 114,7 (C1), 110,4 (C4), 108,6 (C19), 106,6 (C14), 101,5 (C10), 40,6 (C16), 30,1 (C17), 28,7 (C15), 20,8 (C20), 17,9 (C21). m/z (Rel. Int.): 339.17044.
(E)-3-(4-chlorophenyl)-N'-((S,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-306, 11i)
MW: 328,84 g/mol. Chemical Formula: C19H21ClN2O. Physical appearance: white solid. Melting range: 215-216ºC. Purity: 100%. Yield: 63%. IR (ATR, v max, cm-1): 3167 (ν NH), 2919 (νs CH2), 1663 (ν C=O), 1617 (C=N), 1489 (δ CH2), 1361 (δ CH3), 1088 (νs C-Cl), 976 (δ HC=CH), 815 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 8,97 (s, 1H, H10), 7,76 (d, J = 16,37Hz, 1H, H7), 7,50-7,56 (m, 3H, H2, H4, H8), 7,37 (d, J = 8,43, 2H, H1, H5), 6,18 (s, 1H, H13), 4,83 (d, J = 10,57Hz, 2H, H18), 2,78 (dd, J = 3,94Hz e J = 15,79Hz 1H, H16), 2,31-2,46 (m, 2H, H14, H15), 2,11 (dd, J = 13,93Hz e J = 25,80Hz, 2H, H16, H14), 1,97 (s, 3H, H20), 1,79 (s, 3H, H19); 13C NMR (75 MHz, CDCl3), δ (ppm): 167,3 (C9), 149,7 (C11), 147,3 (C17), 141,9 (C7), 135,8 (C12), 133,8 (C13), 133,3 (C6), 132,7 (C3), 129,3 (C2 e C4), 129,1 (C1 e C5), 117,3 (C8), 110,5 (C18), 40,6 (C15), 30,1 (C16), 28,6 (C14), 20,8 (C19), 18,0 (C20). m/z (Rel. Int.): 329.14164.
(E)-3-(4-chlorophenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-307, 10i)
MW: 328,84 g/mol. Chemical Formula: C19H21ClN2O. Physical appearance: pale yellow solid. Melting range: 234-236ºC. Purity: 100%. Yield: 71%. IR (ATR, v max, cm-1): 3166 (ν NH), 2916 (νs CH2), 1663 (ν C=O), 1617 (C=N), 1489 (δ CH2), 1360 (δ CH3), 1087 (νs C-Cl), 976 (δ HC=CH), 815 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 8,92 (s, 1H, H10), 7,76 (d, J = 16,05Hz, 1H, H7), 7,51-7,55 (m, 3H, H2, H4, H8), 7,37 (d, J = 8,49Hz, 2H, H1, H5), 6,18 (d, J = 4,84Hz, 1H, H13), 4,82 (d, J = 11,28Hz, 2H, H18), 2,76 (dd, J = 3,86Hz e J = 15,90Hz 1H, H16), 2,29-2,49 (m, 2H, H14, H15), 2,13 (m, J = 7,47Hz, J = 10,063Hz e J = 28,31Hz, 2H, H16, H14), 1,97 (s, 3H, H20), 1,79 (s, 3H, H19), 13C NMR (75 MHz, CDCl3), δ (ppm): 167,2 (C9), 149,7 (C11), 147,3 (C17), 141,9 (C7), 135,8 (C12), 133,8 (C13), 133,2 (C6), 132,7 (C3), 129,3 (C2 e C4), 129,1 (C1 e C5), 117,3 (C8), 110,5 (C28), 40,6 (C15), 30,0 (C16), 28,6 (C14), 20,8 (C19), 17,9 (C20). m/z (Rel. Int.): 341.18622.
(E)-3-(3-hydroxy-4-methoxyphenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-308, 11j)
MW: 340,42 g/mol. Chemical Formula: C20H24N2O3. Physical appearance: yellow solid. Melting range: 190-192ºC. Purity: 100%. Yield: 30%. IR (ATR, v max, cm-1): 3362 (ν OH), 2911 (νs CH2), 2835 (νs CH3), 1647 (C=O), 1598 (δ NH), 1506 (δ C=Car), 1274 (ν C-O-C), 974 (δ CH), 795 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 9,21 (s, 1H, H12), 7,72 (d, J = 16,0Hz, 1H, H7), 7,42 (d, J = 16,0Hz, 1H, H8), 7,23 (s, 1H, H5), 7,08 (d, J = 8,8Hz, 1H, H1), 6,86 (d, J = 8,2Hz, 1H, H2), 6,15 (d, J = 4,9Hz, 1H, H15), 5,85 (s, 1H, H10), 4,83 (d, J = 5,2Hz, 2H, H20), 3,93 (s, 3H, H11), 2,82 (dd, J = 2,5Hz, J=15,3Hz, 1H, H18), 2,44 (t, J = 11,7Hz 1H, H17), 2,32 (dd, J = 5,0Hz, J = 16,9Hz, 1H, H16), 2,11 (dd, J = 11,1Hz, J = 26,3Hz, 2H, H18, H16), 1,97 (s, 3H, H22), 1,79 (s, 3H, H21); 13C NMR (75 MHz, CDCl3), δ (ppm): 168,0 (C9), 149,5 (C3), 148,3 (C4), 147,4 (C19), 145,8 (C14), 143,2 (C7), 132,9 (C15), 129,0 (C6), 122,2 (C1), 114,8 (C8), 113,0 (C5), 110,4 (C2), 110,5 (C20), 56,0 (C11), 40,6 (C17), 30,1 (C18), 28,7 (C16), 20,8 (C21), 17,9 (C22). m/z (Rel. Int.): 341.18600.
(E)-3-(3-hydroxy-4-methoxyphenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-309, 10j)
MW: 340,42 g/mol. Chemical Formula: C20H24N2O3. Physical appearance: yellow solid. Melting range: 194-195ºC. Purity: 100%. Yield: 24%. IR (ATR, v max, cm-1): 3243 (ν OH), 2911 (νs CH2), 2836 (νs CH3), 1647 (C=O), 1598 (δ NH), 1557 (ν C=N), 1506 (δ C=Car), 1274 (ν C-O-C), 975 (δ CH), 795 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 9,03 (s, 1H, H12), 7,73 (d, J = 15,8Hz, 1H, H7), 7,41 (d, J = 16,0Hz, 1H, H8), 7,24 (d, J = 1,6Hz, 1H, H5), 7,08 (dd, J = 8,8Hz, J = 1,7Hz, 1H, H1), 6,86 (d, J = 8,3Hz, 1H, H2), 6,15 (d, J = 5,6Hz, 1H, H15), 5,78 (s, 1H, H10), 4,82 (d, J = 8,6Hz, 2H, H20), 3,93 (s, 3H, H11), 2,79 (dd, J = 3,6Hz, J = 16,1Hz, 1H, H18), 2,28-2,48 (m, 2H, H17 e H16), 2,12 (dd, J = 8,1Hz, J = 22,0Hz, 2H, H18, H16), 1,97 (s, 3H, H22), 1,79 (s, 3H, H21); 13C NMR (75 MHz, CDCl3), δ (ppm): 167,9 (C9), 149,4 (C3), 148,3 (C4), 147,4 (C19), 145,7 (C14), 143,3 (C7), 132,9 (C15), 129,0 (C6), 122,2 (C1), 114,8 (C8), 112,9 (C5), 110,4 (C2), 110,5 (C20), 56,0 (C11), 40,6 (C17), 30,1 (C18), 28,6 (C16), 20,8 (C21), 17,9 (C22). m/z (Rel. Int.): 341.18580.
(E)-3-(4-hydroxy-3,5-dimethoxyphenyl)-N'-((S,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-375, 10k)
MW: 370,45 g/mol. Chemical Formula: C21H26N2O4. Physical appearance: pale yellow solid. Melting range: 217-219ºC. Purity: 100%. Yield: 51%. IR (ATR, v max, cm-1): 1652 (C=O), 1609 (δ NH), 1510 (ν C=N), 1456 (δ C=Car), 1322 (ν C-O-C), 981 (δ CH), 605 (δ CHar). 1H NMR (300 MHz, DMSO-d6), δ (ppm): 10,34 (s, 1H, H13), 7,37-7,57 (m, 1H, H7), 6,69 (s, 2H, H1 e H5), 6,80 (d, J = 15,4Hz, 1H, H8), 6,15 (s, 1H, H16), 4,82 (d, J = 10,2 Hz, 2H, H21), 3,80 (s, 6H, H10 e H12), 2,91 (d, J = 14,5Hz, 1H, H19), 2,30 (dd, J = 14,9Hz, J = 32,1Hz, 2H, H18 e H17), 2,10 (dd, J = 13,2Hz, J = 26,3Hz, 2H, H19, H17), 1,84 (s, 3H, H22), 1,76 (s, 3H, H23); 13C NMR (75 MHz, DMSO-d6), δ (ppm): 167,1 e 162,1 (C9), 153,1 (C14), 148,1 (C2, C4), 147,8 (C20), 140,9 (C7), 137,6 (C3), 133,4 (C15), 132,6 (C16), 125,3 (C8), 117,8 (C6), 110,4 (C21), 105,5 (C1, C5), 56,0 (C10, C12), 40,4 (C18), 29,6 (C19), 29,1 (C17), 20,5 (C22), 18,0 (C23). m/z (Rel. Int.): 371.19604.
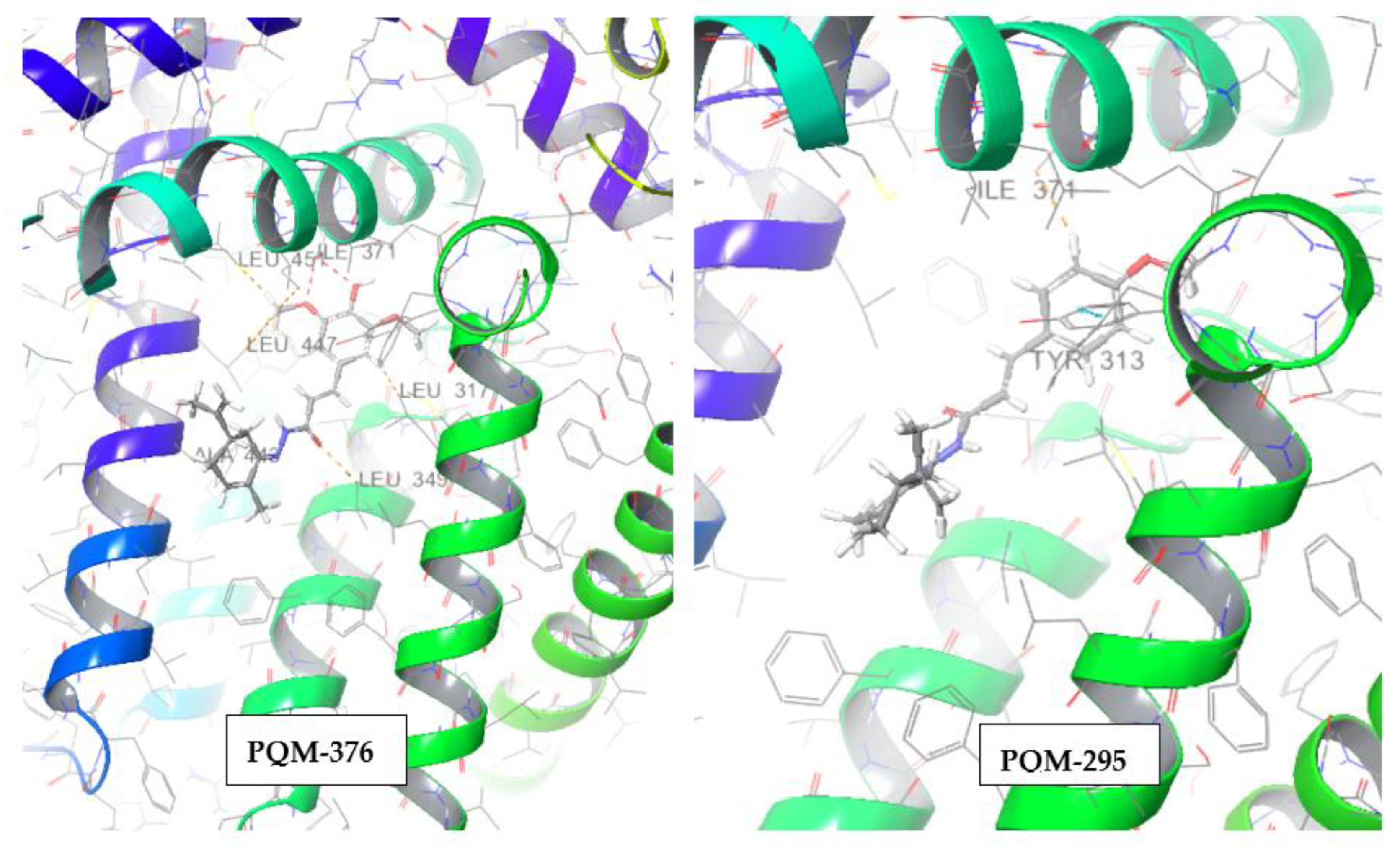
(E)-3-(4-hydroxy-3,5-dimethoxyphenyl)-N'-((S,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-376, 11k)
MW: 370,45 g/mol. Chemical Formula: C21H26N2O4. Physical appearance: pale yellow solid. Melting range: 196-197ºC. Purity: 95%. Yield: 53%. IR (ATR, v max, cm-1): 1652 (C=O), 1612 (δ NH), 1512 (ν C=N), 1456 (δ C=Car), 1323 (ν C-O-C), 980 (δ CH), 604 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 8,85 (s, 1H, H13), 7,73 (d, J = 15,8 Hz, 1H, H7), 7,41 (d, J = 15,7 Hz, 1H, H8), 6,84 (s, 2H, H1 e H5), 6,16 (d, J = 7,1Hz 1H, H16), 5,81 (s, 1H, H11), 4,82 (d, J = 10,9 Hz, 2H, H21), 3,93 (s, 6H, H10 e H12), 2,76 (dd, J = 4,1Hz, J = 15,4Hz 1H, H19), 2,44 (dd, J = 8,4Hz, J = 19,8Hz, 1H, H18), 2,28-2,38 (m,1H, H17), 2,04-2,18 (m, 2H, H19, H17), 1,96 (s, 3H, H22), 1,79 (s, 3H, H23); 13C NMR (75 MHz, CDCl3), δ (ppm): 167,7 (C9), 149,3 (C12), 147,3 (C20), 147,2 (C2, C4), 143,7 (C7), 136,9 (C3), 133,1 (C16), 132,7 (C15), 126,9 (C6), 114,7 (C8), 110,5 (C21), 105,2 (C1, C5), 56,3 (C10, C12), 40,6 (C18), 30,1 (C19), 28,6 (C17), 20,8 (C22), 17,7 (C23). m/z (Rel. Int.): 371.19626.
(E)-3-(4-(dimethylamino)phenyl)-N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-377, 10l)
MW: 337,46 g/mol. Chemical Formula: C21H27N3O. Physical appearance: pale yellow solid. Melting range: 236-237ºC. Purity: 72%. Yield: 53%. IR (ATR, v max, cm-1): 3155 e 2914 (δ NH), 1651 (C=O), 1591 (δ NH), 1553 (ν C=N), 1350 (δ C=Car), 1227 (ν C-O-C), 937 (δ CH), 645 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 9,01 (s, 1H, H12), 7,76 (d, J = 15,8 Hz, 1H, H7), 7,49 (d, J = 8,6Hz, 2H, H1 e H5), 7,35 (d, J = 15,9 Hz, 1H, H8), 6,69 (d, J = 8,6 Hz, 2H, H2 e H4), 6,13 (d, J = 5,7 Hz 1H, H15), 4,82 (d, J = 6,4 Hz, 2H, H20), 3,0 (s, 6H, H10 e H11), 2,81 (dd, J = 3,9 Hz, J = 15,6Hz 1H, H18), 2,43 (dd, J = 7,8Hz, J = 19,8Hz, 1H, H17), 2,31 (dt, J = 4,9 Hz, J = 10,3Hz,1H, H16), 2,03-2,17 (m, 2H, H18, H16), 1,97 (s, 3H, H21), 1,79 (s, 3H, H22), 13C NMR (75 MHz, CDCl3), δ (ppm): 168,5 (C9), 151,6 (C3), 148,8 (C13), 147,5 (C19), 143,9 (C7), 133,0 (C14), 132,5 (C15), 129,8 (C1 e C5), 123,3 (C6), 111,9 (C2 e C4), 111,3 (C8), 110,3 (C20), 40,7 (C17), 40,2 (C10 e C11), 30,1 (C18), 28,7 (C16), 20,8 (C22), 17,9 (C21). m/z (Rel. Int.): 338.22232.
(E)-3-(4-(dimethylamino)phenyl)-N'-((S,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)acrylohydrazide (PQM-378, 11l)
MW: 337,46 g/mol. Chemical Formula: C21H27N3O. Physical appearance: pale yellow solid. Melting range: 220-223ºC. Purity: 75%. Yield: 22%. IR (ATR, v max, cm-1): 3154 e 2914 (δ NH), 1651 (C=O), 1590 (δ NH), 1552 (ν C=N), 1360 (δ C=Car), 1227 (ν C-O-C), 987 (δ CH), 645 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 9,07 (s, 1H, H12), 7,76 (d, J = 15,9 Hz, 1H, H7), 7,49 (d, J = 8,6 Hz, 2H, H1 e H5), 7,35 (d, J = 15,9 Hz, 1H, H8), 6,69 (d, J = 8,6 Hz, 2H, H2 e H4), 6,13 (d, J = 6,1 Hz 1H, H15), 4,82 (d, J = 5,3 Hz, 2H, H20), 3,01 (s, 6H, H10 e H11), 2,82 (dd, J = 4,0 Hz, J = 15,7 Hz 1H, H18), 2,44 (dt, J = 4,4Hz, J = 12,5Hz, 1H, H17), 2,31 (dt, J = 5,50 Hz, J = 11,4Hz, 1H, H16), 2,04-2,17 (m, 2H, H18, H16), 1,97 (s, 3H, H21), 1,79 (s, 3H, H22); 13C NMR (75 MHz, CDCl3), δ (ppm): 168,5 (C9), 151,6 (C3), 148,8 (C13), 147,5 (C19), 143,9 (C7), 133,0 (C14), 132,5 (C15), 129,8 (C1 e C5), 123,3 (C6), 111,9 (C2 e C4), 111,3 (C8), 110,3 (C20), 40,7 (C17), 40,2 (C10 e C11), 30,1 (C18), 28,7 (C16), 20,8 (C22), 17,9 (C21). m/z (Rel. Int.): 338.22235.
N'-((R,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)cinnamohydrazide (PQM-379, 10m)
MW: 294,39 g/mol. Chemical Formula: C19H22N2O. Physical appearance: pale yellow solid. Melting range: 210-211ºC. Purity: 100%. Yield: 41%. IR (ATR, v max, cm-1): 3168, 3025 e 2916 (δ NH), 1660 (C=O), 1447 (ν C=N), 1358 (δ C=Car), 1218 (ν C-O-C), 886 (δ CH), 760 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 9,32 (s, 1H, H10), 7,81 (d, J = 16,0 Hz, 1H, H7), 7,55-7,60 (m, 3H, H1, H5, H3), 7,40 (d, J = 6,2 Hz, 3H, H2, H4, H8), 6,16 (d, J = 5,5 Hz 1H, H13), 4,83 (s, 2H, H19), 2,86 (dd, J = 4,0 Hz, J = 15,8 Hz 1H, H16), 2,40-2,50 (m, 1H, H15) , 2,28-2,37 (m, 1H, H14), 2,06-2,18 (m, 2H, H16, H14), 1,97 (s, 3H, H18), 1,80 (s, 3H, H20); 13C NMR (75 MHz, CDCl3), δ (ppm): 167,8 (C9), 149,7 (C11), 147,4 (C17), 143,3 (C7), 135,4 (C6), 133,1 (C13), 132,8 (C12), 129,9 (C3), 128,8 (C2, C4), 128,2 (C1 e C5), 116,9 (C8), 110,4 (C19), 40,6 (C15), 30,1 (C16), 28,8 (C14), 20,8 (C20), 17,9 (C18). m/z (Rel. Int.): 295.18033.
N'-((S,E)-2-methyl-5-(prop-1-en-2-yl)cyclohex-2-en-1-ylidene)cinnamohydrazide (PQM308, 11m)
MW: 294,39 g/mol. Chemical Formula: C19H22N2O. Physical appearance: pale yellow solid. Melting range: 185-186ºC. Purity: 100%. Yield: 34%. IR (ATR, v max, cm-1): 3171, 3057 e 2916 (δ NH), 1663 (C=O), 1447 (ν C=N), 1359 (δ C=Car), 1220 (ν C-O-C), 886 (δ CH), 760 (δ CHar). 1H NMR (300 MHz, CDCl3), δ (ppm): 9,28 (s, 1H, H10), 7,81 (d, J = 16,0 Hz, 1H, H7), 7,55-7,60 (m, 3H, H1, H5, H3), 7,40 (d, J = 6,1 Hz, 3H, H2, H4, H8), 6,16 (d, J = 5,8 Hz 1H, H13), 4,84 (d, J = 3,7 Hz, 2H, H19), 2,85 (dd, J = 3,9 Hz, J = 15,8 Hz 1H, H16), 2,40-2,51 (m, 1H, H14); 13C NMR (75 MHz, CDCl3), δ (ppm): 167,7 (C9), 149,7 (C11), 147,4 (C17), 143,3 (C7), 35,4 (C6), 133,1 (C13, C12), 129,9 (C3), 128,8 (C2, C4), 128,2 (C1 e C5), 116,9 (C8), 110,4 (C19), 40,6 (C15), 30,1 (C16), 28,8 (C14), 20,8 (C20), 17,9 (C18). m/z (Rel. Int.): 295.18027.