Submitted:
23 April 2025
Posted:
23 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. History
2.1. Cajal’s Classification of Neurons and Astroglia
2.2. Hortega’s Classification of Microglia and Oligodendroglia
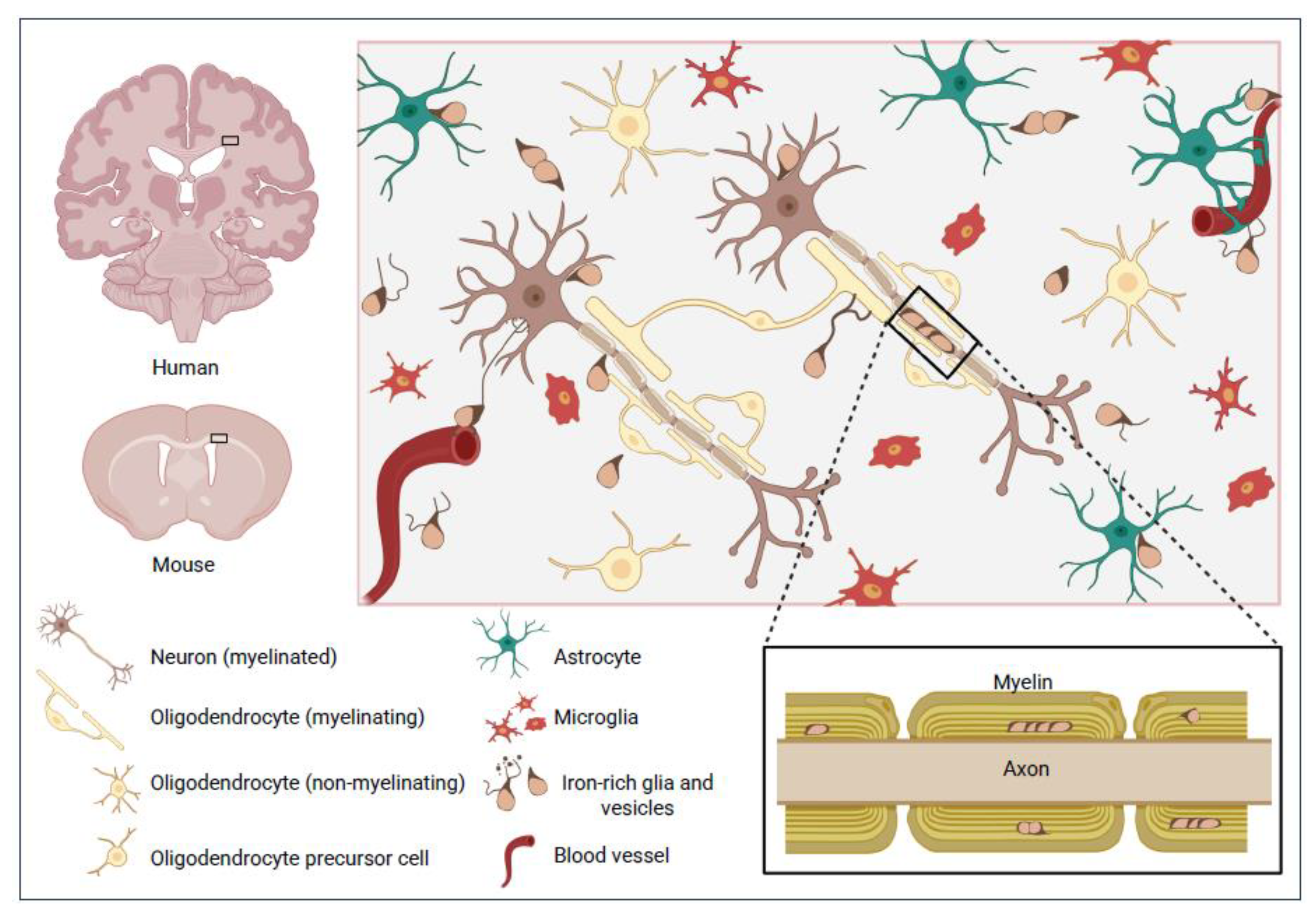
3. Paradoxical Properties of Cells Currently Classified as OLG
3.1. Invalid Presumptions Arising from Inferring Myelination or OLG Identity from Cell Size
3.1.1. Size
3.1.2. Other Morphological Features – Eccentric Nucleus, Shape and Fine, Sparse Processes
3.2. Invalid Presumptions Arising from Inferring Myelination or OLG Identity from Location
3.2.1. Nature and Roles of Myelin at Juxta-Neuronal or Other Non-Axonal Sites
3.2.2. Assumptions About the Nature and Roles of OLG-like Cells at Juxta-Myelin Sites
3.3. Non-Myelin Related Functions of Subsets of Cells Usually Assumed to Be OLG
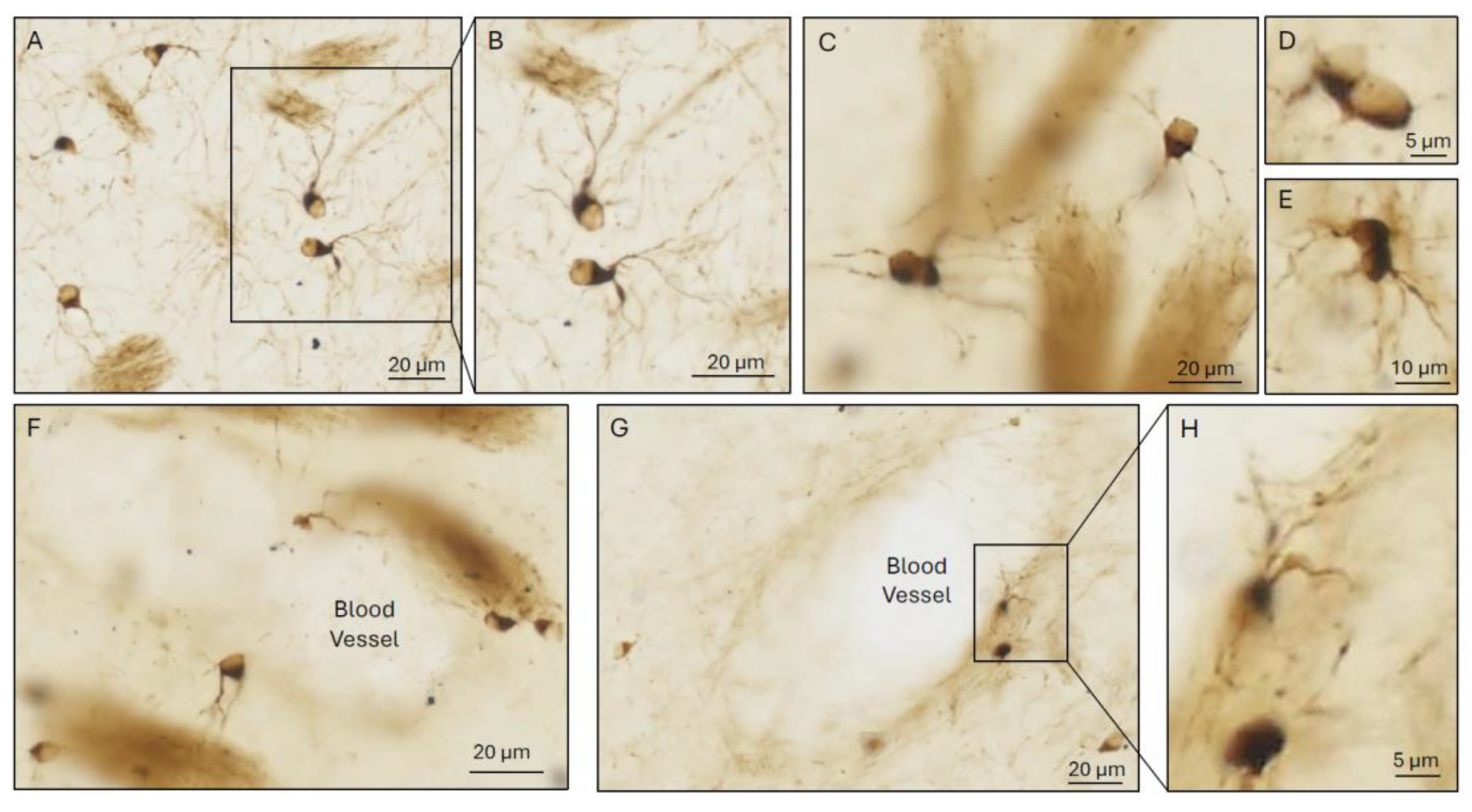
3.3.1. Juxta-Neuronal OLG
3.3.2. Juxta-Vascular OLG.
3.3.3. Interstitial OLG
3.3.4. Progenitor Cells of the OLG Lineage
3.3.5. Olfactory Ensheathing Glia
3.4. Limitations of Past Research
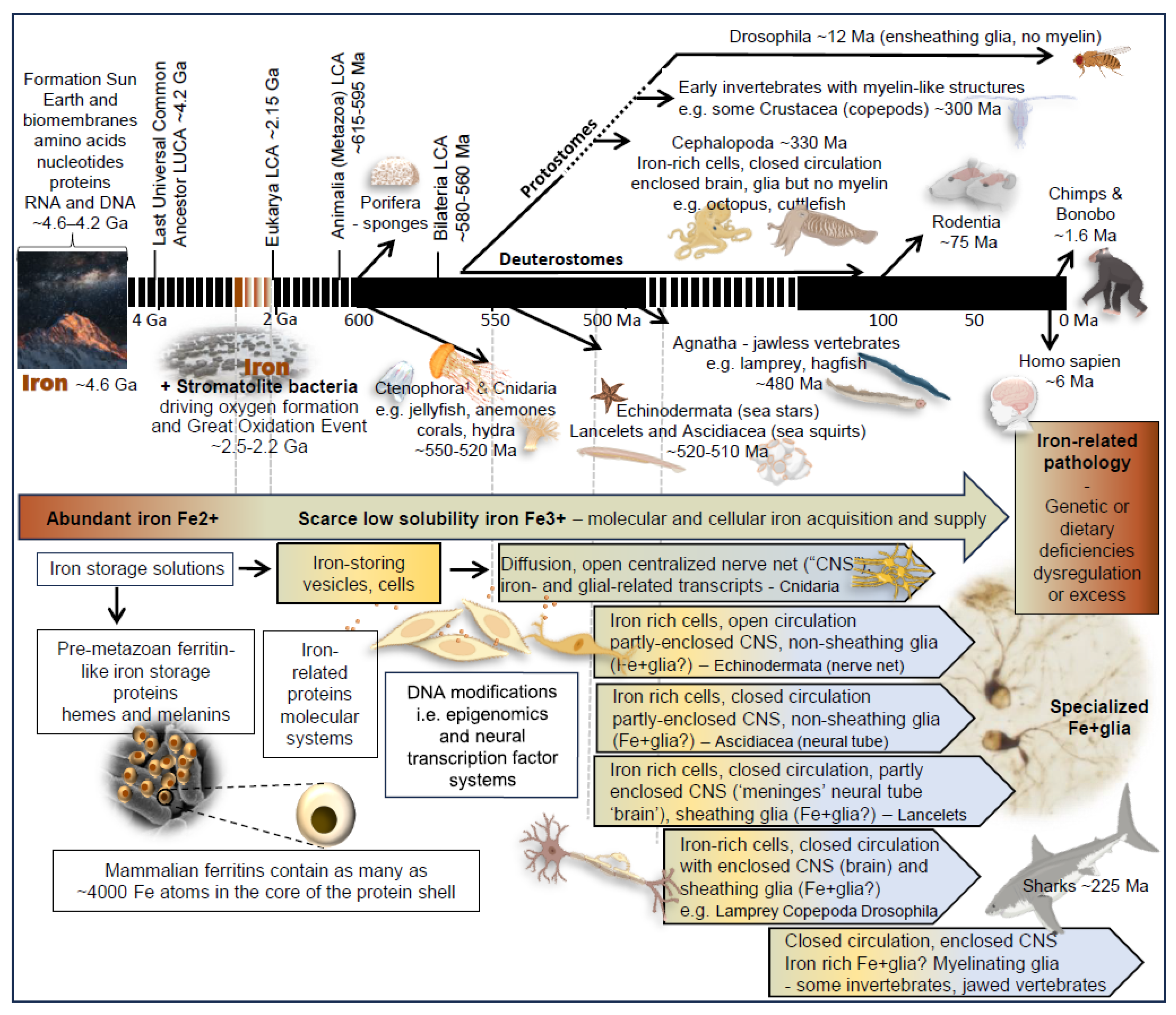
4. Rationales for Separation of Iron Regulation from, and Precedence over, Myelination
4.1. Evolutionary Advantages of Separating Iron Regulation and Myelination
4.2. Rethinking Relationships of Iron, Myelin and Iron-Rich, OLG-like Ferriglia
5. Evolutionary Emergence of Molecular and Cellular Mechanisms for Iron Regulation
5.1. Iron as a Powerful Driver of the Emergence and Evolution of Early Terrestrial Life
5.1.1. Iron and the Emergence of Early Macromolecules and Membranes
5.1.2. Iron in Non-Animal Organisms
5.2. Iron and the Evolution of the Nervous System in Animals (Animalia)
5.2.1. Iron in Sponges and Other Early Animals Without Neurons or Nervous Systems
5.2.2. Iron Regulation in Jellyfish and Other Early Animals with Early Nervous Systems
5.3. The Transition from Non-Myelinating to Myelinating Vertebrate Species
5.3.1. Iron Regulation in Ensheathing but Non-Myelinating Jawless Vertebrates (Agnatha)
5.3.2. Evidence for Myelin in Prehistoric Shark Fossils - the Oldest Known Jawed Vertebrate
5.4. Nervous System Enclosure and the Emergence of Ferriglia and Ensheathing Glia
5.4.1. Brain Barrier Systems
5.4.2. Brain Enclosure by Meninges and Related Structures
5.5. Evolution of Myelin-like Ensheathing Structures in Invertebrate Nervous Systems
5.5.1. Evolution of Invertebrate Myelin-like Structures
6. Technical Considerations
6.1. Challenges in Studying Iron and Other Metallic Elements in Brain Glial Cells
6.2. Small Molecule Fluorescence Probes and DNAzyme-Based Fluorescent Sensors of Labile Iron
6.3. Label-Free Imaging of Iron and Other Metallic Elements
7. Involvement of Iron-Rich Cells Resembling OLG in Neurological Conditions
7.1. Iron-Rich Glia in Brain Diseases Involving Mutations in Iron-Related Proteins
7.1.1. Hereditary Neuroferritinopathy
7.1.2. Friedreich’s Ataxia
7.2. Iron-Rich Glia in Brain Diseases Involving Amyloids
7.2.1. Alzheimer’s Disease
7.2.2. Superficial Siderosis with Amyloidosis
7.2.3. α-Synucleinopathies
7.3. Cerebrovascular Disease and Stroke
7.4. Brain Cancers
7.5. CNS Infections and Neuroimmune Disorders
8. Considerations for Iron Chelation and Other Therapies
9. Conclusions
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darwin, C.; Burkhardt, F.; Smith, S. The Correspondence of Charles Darwin: 1821-1836/[editors Frederick Burkhardt, Sydney Smith]; Cambridge University Press: 1985.
- Keynes, R.; Darwin, C. Charles Darwin’s zoology notes & specimen lists from HMS Beagle. 2001.
- Rozo, J.A.; Martínez-Gallego, I.; Rodríguez-Moreno, A. Cajal, the neuronal theory and the idea of brain plasticity. Front. Neuroanat. 2024, 18, 1331666. [CrossRef]
- García-Marín, V.; García-López, P.; Freire, M. Cajal's contributions to glia research. Trends Neurosci. 2007, 30, 479–487. [CrossRef]
- Tremblay, M.; Lecours, C.; Samson, L.; Sánchez-Zafra, V.; Sierra, A. From the Cajal alumni Achúcarro and Río-Hortega to the rediscovery of never-resting microglia. Front. Neuroanat. 2015, 9, 45–45. [CrossRef]
- y Cajal, R. Contribución al conocimiento de la neuroglia del cerebro humano. Trab Lab Invest Biol 1913, 11, 255.
- y Cajal, S.R. Sobre un nuevo proceder de impregnacion de la neuroglia y sus resultados en los centros nerviosos del hombre y animales; 1913.
- del Río-Hortega, P. La glía de escasas radiaciones (oligodendroglía) boletín de la real sociedad española de historia natural. Estud Neurol 1921, 21, 63-92.
- del Río-Hortega, P. El “tercer elemento” de los centros nerviosos [The “third element” of the nerve centers]. Bol Soc Esp Biol 1919, 9, 68-166.
- Kettenmann, H.; Verkhratsky, A. Neuroglia: the 150 years after. Trends Neurosci. 2008, 31, 653–659. [CrossRef]
- Verkhratsky, A.; Nedergaard, M. Physiology of Astroglia. Physiol. Rev. 2018, 98, 239–389. [CrossRef]
- Boullerne, A.I.; Feinstein, D.L. History of Neuroscience I. Pío del Río-Hortega (1882–1945): The Discoverer of Microglia and Oligodendroglia. ASN Neuro 2020, 12. [CrossRef]
- Delgado-García, J.M. Cajal and the Conceptual Weakness of Neural Sciences. Front. Neuroanat. 2015, 9. [CrossRef]
- James, O.G.; Mehta, A.R.; Behari, M.; Chandran, S. Centenary of the oligodendrocyte. The Lancet Neurology 2021, 20, 422.
- Hasel, P.; Rose, I.V.L.; Sadick, J.S.; Kim, R.D.; Liddelow, S.A. Neuroinflammatory astrocyte subtypes in the mouse brain. Nat. Neurosci. 2021, 24, 1475–1487. [CrossRef]
- Batiuk, M.Y.; Martirosyan, A.; Wahis, J.; de Vin, F.; Marneffe, C.; Kusserow, C.; Koeppen, J.; Viana, J.F.; Oliveira, J.F.; Voet, T.; et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 2020, 11, 1–15. [CrossRef]
- Tabata, H. Diverse subtypes of astrocytes and their development during corticogenesis. Front. Neurosci. 2015, 9, 114–114. [CrossRef]
- Stratoulias, V.; Ruiz, R.; Kanatani, S.; Osman, A.M.; Keane, L.; Armengol, J.A.; Rodríguez-Moreno, A.; Murgoci, A.-N.; García-Domínguez, I.; Alonso-Bellido, I.; et al. ARG1-expressing microglia show a distinct molecular signature and modulate postnatal development and function of the mouse brain. Nat. Neurosci. 2023, 26, 1008–1020. [CrossRef]
- Stratoulias, V.; Venero, J.L.; Tremblay, M.-È.; Joseph, B. Microglial subtypes: diversity within the microglial community. EMBO J. 2019, 38, e101997. [CrossRef]
- Tremblay, M. Microglial functional alteration and increased diversity in the challenged brain: Insights into novel targets for intervention. Brain, Behav. Immun. - Heal. 2021, 16, 100301. [CrossRef]
- Jäkel, S.; Agirre, E.; Falcão, A.M.; van Bruggen, D.; Lee, K.W.; Knuesel, I.; Malhotra, D.; Ffrench-Constant, C.; Williams, A.; Castelo-Branco, G. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 2019, 566, 543–547. [CrossRef]
- Marques, S.; Zeisel, A.; Codeluppi, S.; van Bruggen, D.; Falcão, A.M.; Xiao, L.; Li, H.; Häring, M.; Hochgerner, H.; Romanov, R.A.; et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 2016, 352, 1326–1329. [CrossRef]
- Prinz, M.; Jung, S.; Priller, J. Microglia Biology: One Century of Evolving Concepts. Cell 2019, 179, 292–311. [CrossRef]
- Giordano, K.R.; Denman, C.R.; Dubisch, P.S.; Akhter, M.; Lifshitz, J. An update on the rod microglia variant in experimental and clinical brain injury and disease. Brain Commun. 2021, 3, fcaa227. [CrossRef]
- Paolicelli, R.C.; Sierra, A.; Stevens, B.; Tremblay, M.-E.; Aguzzi, A.; Ajami, B.; Amit, I.; Audinat, E.; Bechmann, I.; Bennett, M.; et al. Microglia states and nomenclature: A field at its crossroads. Neuron 2022, 110, 3458–3483. [CrossRef]
- Kurtz, A.; Seltmann, S.; Bairoch, A.; Bittner, M.-S.; Bruce, K.; Capes-Davis, A.; Clarke, L.; Crook, J.M.; Daheron, L.; Dewender, J.; et al. A Standard Nomenclature for Referencing and Authentication of Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 1–6. [CrossRef]
- Konishi, H.; Koizumi, S.; Kiyama, H. Phagocytic astrocytes: Emerging from the shadows of microglia. Glia 2022, 70, 1009–1026. [CrossRef]
- Geyer, S.; Jacobs, M.; Hsu, N.-J. Immunity Against Bacterial Infection of the Central Nervous System: An Astrocyte Perspective. Front. Mol. Neurosci. 2019, 12, 57. [CrossRef]
- Akay, L.A.; Effenberger, A.H.; Tsai, L.-H. Cell of all trades: oligodendrocyte precursor cells in synaptic, vascular, and immune function. Genes Dev. 2021, 35, 180–198. [CrossRef]
- Asadollahi, E.; Trevisiol, A.; Saab, A.S.; Looser, Z.J.; Dibaj, P.; Ebrahimi, R.; Kusch, K.; Ruhwedel, T.; Möbius, W.; Jahn, O.; et al. Oligodendroglial fatty acid metabolism as a central nervous system energy reserve. Nat. Neurosci. 2024, 27, 1934–1944. [CrossRef]
- Omari, K.M.; John, G.; Lango, R.; Raine, C.S. Role for CXCR2 and CXCL1 on glia in multiple sclerosis. Glia 2005, 53, 24–31. [CrossRef]
- Omari, K.M.; John, G.R.; Sealfon, S.C.; Raine, C.S. CXC chemokine receptors on human oligodendrocytes: implications for multiple sclerosis. Brain 2005, 128, 1003–1015. [CrossRef]
- Pérez-Cerdá, F.; Sánchez-Gómez, M.V.; Matute, C. Pío del Río Hortega and the discovery of the oligodendrocytes. Front Neuroanat 2015, 9, 92.
- Ghasemi-Kasman, M.; Zare, L.; Baharvand, H.; Javan, M. In vivo conversion of astrocytes to myelinating cells by miR-302/367 and valproate to enhance myelin repair. J Tissue Eng Regen Med 2018, 12, e462-e472.
- Khanghahi, A.M.; Satarian, L.; Deng, W.; Baharvand, H.; Javan, M. In vivo conversion of astrocytes into oligodendrocyte lineage cells with transcription factor Sox10; Promise for myelin repair in multiple sclerosis. PLOS ONE 2018, 13, e0203785. [CrossRef]
- Meguro, R.; Asano, Y.; Odagiri, S.; Li, C.; Iwatsuki, H.; Shoumura, K. Nonheme-iron histochemistry for light and electron microscopy: a historical, theoretical and technical review. Arch. Histol. Cytol. 2007, 70, 1–19. [CrossRef]
- Bentivoglio, M.; Cotrufo, T.; Ferrari, S.; Tesoriero, C.; Mariotto, S.; Bertini, G.; Berzero, A.; Mazzarello, P. The Original Histological Slides of Camillo Golgi and His Discoveries on Neuronal Structure. Front. Neuroanat. 2019, 13, 3. [CrossRef]
- Chvátal, A.; Verkhratsky, A. An Early History of Neuroglial Research: Personalities. Neuroglia 2018, 1, 245–257. [CrossRef]
- y Cajal, S.R. Estructura de los centros nerviosos de las aves; 1888.
- Saceleanu, V.M.; Covache-Busuioc, R.-A.; Costin, H.-P.; Glavan, L.-A.; Ciurea, A.V. An Important Step in Neuroscience: Camillo Golgi and His Discoveries. Cells 2022, 11, 4112. [CrossRef]
- Parpura, V.; Verkhratsky, A. Astrocytes revisited: concise historic outlook on glutamate homeostasis and signaling. Croat. Med J. 2012, 53, 518–528. [CrossRef]
- Ramos, A.J. Astroglial heterogeneity: merely a neurobiological question? Or an opportunity for neuroprotection and regeneration after brain injury? Neural Regen Res 2016, 11, 1739-1741.
- del Rio-Hortega, P. Noticia de un nuevo y fácil método para la coloración de la neuroglia y del tejido conjuntivo. Trab Lab Invest Biol 1918, 15, 367-378.
- del Río-Hortega, P. Tercera aportación al conocimiento morfológico e interpretación funcional de la oligodendroglía; 1928.
- del Rio-Hortega, P. Cytology & cellular pathology of the nervous system; Hoeber: 1932.
- del Rio-Hortega, P. The microglia. 1939.
- del Rio-Hortega, P.; De Estudios, P.A. La microglia y su transformacion en células en bastoncito y cuerpos granulo-adiposos. Arch Neurobiol (Madr) 1920, 1, 171.
- Penfield, W. Oligodendroglia and its relation to classical neuroglia. Brain 1924, 47, 430–452. [CrossRef]
- Del Bigio, M.R. History of research concerning the ependyma: a view from inside the human brain. Front. Cell. Neurosci. 2024, 17, 1320369. [CrossRef]
- Butt, A.; Verkhratsky, A. Neuroglia: Realising their true potential. Brain Neurosci. Adv. 2018, 2. [CrossRef]
- for the UK Brain Expression Consortium (UKBEC); Heidari, M.; Johnstone, D.M.; Bassett, B.; Graham, R.M.; Chua, A.C.G.; House, M.J.; Collingwood, J.F.; Bettencourt, C.; Houlden, H.; et al. Brain iron accumulation affects myelin-related molecular systems implicated in a rare neurogenetic disease family with neuropsychiatric features. Mol. Psychiatry 2016, 21, 1599–1607. [CrossRef]
- Schnatz, A.; Müller, C.; Brahmer, A.; Krämer-Albers, E. Extracellular Vesicles in neural cell interaction and CNS homeostasis. FASEB BioAdvances 2021, 3, 577–592. [CrossRef]
- Pistono, C.; Bister, N.; Stanová, I.; Malm, T. Glia-Derived Extracellular Vesicles: Role in Central Nervous System Communication in Health and Disease. Front. Cell Dev. Biol. 2021, 8. [CrossRef]
- Granseth, B.; Odermatt, B.; Royle, S.J.; Lagnado, L. Clathrin-Mediated Endocytosis Is the Dominant Mechanism of Vesicle Retrieval at Hippocampal Synapses. Neuron 2006, 51, 773–786. [CrossRef]
- LoGiudice, L.; Matthews, G. The synaptic vesicle cycle: is kissing overrated? Neuron 2006, 51, 676-677.
- Ishikawa, K. On the Genesis of Hortega Cells.. Folia Anat. Jpn. 1932, 10, 229–248_1. [CrossRef]
- Iglesias-Rozas, J.R.; Garrosa, M. Río-Hortega’s third contribution to the morphological knowledge and functional interpretation of the oligodendroglia; Newnes: 2013.
- Connor, J.R.; Menzies, S.L. Relationship of iron to oligodendrocytes and myelination. Glia 1996, 17, 83-93.
- Connor, J.R.; Benkovic, S.A. Iron regulation in the brain: histochemical, biochemical, and molecular considerations. Ann Neurol 1992, 32, S51-S61.
- Connor, J.R.; Menzies, S.L.; Martin, S.M.S.; Mufson, E.J. Cellular distribution of transferrin, ferritin, and iron in normal and aged human brains. J. Neurosci. Res. 1990, 27, 595–611. [CrossRef]
- Battefeld, A.; Klooster, J.; Kole, M.H.P. Myelinating satellite oligodendrocytes are integrated in a glial syncytium constraining neuronal high-frequency activity. Nat. Commun. 2016, 7, 11298–11298. [CrossRef]
- Wood, P.; Bunge, R.P. The biology of the oligodendrocyte. In Oligodendroglia; Springer: 1984; pp. 1-46.
- Ludwin, S. The perineuronal satellite oligodendrocyte: A role in remyelination. Acta Neuropathol 1979, 47, 49-53.
- Belachew, S.; Yuan, X.; Gallo, V. Unraveling Oligodendrocyte Origin and Function by Cell-Specific Transgenesis. Dev. Neurosci. 2001, 23, 287–298. [CrossRef]
- Connor, J.R.; Menzies, S.L. Cellular management of iron in the brain. J. Neurol. Sci. 1995, 134, 33–44. [CrossRef]
- Takasaki, C.; Yamasaki, M.; Uchigashima, M.; Konno, K.; Yanagawa, Y.; Watanabe, M. Cytochemical and cytological properties of perineuronal oligodendrocytes in the mouse cortex. Eur. J. Neurosci. 2010, 32, 1326–1336. [CrossRef]
- Bernstein, H.-G.; Keilhoff, G.; Dobrowolny, H.; Guest, P.C.; Steiner, J. Perineuronal oligodendrocytes in health and disease: the journey so far. Prog. Neurobiol. 2019, 31, 89–99. [CrossRef]
- Radtke, C.; Sasaki, M.; Lankford, K.L.; Gallo, V.; Kocsis, J.D. CNPase Expression in Olfactory Ensheathing Cells. BioMed Res. Int. 2011, 2011, 608496. [CrossRef]
- Zhou, M.; Kiyoshi, C.M. Astrocyte syncytium: a functional reticular system in the brain. Neural Regen. Res. 2019, 14, 595–596. [CrossRef]
- Pacholko, A.G.; Wotton, C.A.; Bekar, L.K. Astrocytes - the ultimate effectors of long-range neuromodulatory networks? Front Cell Neurosci 2020, 14, 581075.
- Szuchet, S.; Nielsen, J.A.; Lovas, G.; Domowicz, M.S.; de Velasco, J.M.; Maric, D.; Hudson, L.D. The genetic signature of perineuronal oligodendrocytes reveals their unique phenotype. Eur. J. Neurosci. 2011, 34, 1906–1922. [CrossRef]
- van Landeghem, F.K.; Weiss, T.; von Deimling, A. Expression of PACAP and glutamate transporter proteins in satellite oligodendrocytes of the human CNS. Regul Pept 2007, 142, 52-59.
- Xin, W.; Mironova, Y.A.; Shen, H.; Marino, R.A.; Waisman, A.; Lamers, W.H.; Bergles, D.E.; Bonci, A. Oligodendrocytes Support Neuronal Glutamatergic Transmission via Expression of Glutamine Synthetase. Cell Rep. 2019, 27, 2262–2271.e5. [CrossRef]
- Wellman, S.M.; Cambi, F.; Kozai, T.D. The role of oligodendrocytes and their progenitors on neural interface technology: A novel perspective on tissue regeneration and repair. Biomaterials 2018, 183, 200–217. [CrossRef]
- Palhol, J.S.C.; Balia, M.; Terán, F.S.-R.; Labarchède, M.; Gontier, E.; Battefeld, A. Direct association with the vascular basement membrane is a frequent feature of myelinating oligodendrocytes in the neocortex. Fluids Barriers CNS 2023, 20, 1–17. [CrossRef]
- Moos, T.; Møllgård, K. A sensitive post-DAB enhancement technique for demonstration of iron in the central nervous system. Histochem. 1993, 99, 471–475. [CrossRef]
- Morris, C.M.; Candy, J.M.; Keith, A.B.; Oakley, A.E.; Taylor, G.A.; Edwardson, J.A.; Bloxham, C.A.; Pullen, R.G.; Gocht, A. Brain iron homeostasis. J. Inorg. Biochem. 1992, 47, 257–265. [CrossRef]
- Wawrzyniak, A.; Balawender, K.; Lalak, R.; Staszkiewicz, R.; Boroń, D.; Grabarek, B.O. Oligodendrocytes in the periaqueductal gray matter and the corpus callosum in adult male and female domestic sheep. Brain Res. 2022, 1792, 148036. [CrossRef]
- Kirby, L.; Jin, J.; Cardona, J.G.; Smith, M.D.; Martin, K.A.; Wang, J.; Strasburger, H.; Herbst, L.; Alexis, M.; Karnell, J.; et al. Oligodendrocyte precursor cells present antigen and are cytotoxic targets in inflammatory demyelination. Nat. Commun. 2019, 10, 1–20. [CrossRef]
- Falcão, A.M.; van Bruggen, D.; Marques, S.; Meijer, M.; Jäkel, S.; Agirre, E.; Samudyata; Floriddia, E.M.; Vanichkina, D.P.; Ffrench-Constant, C.; et al. Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat. Med. 2018, 24, 1837–1844. [CrossRef]
- Richardson, W.D.; Young, K.M.; Tripathi, R.B.; McKenzie, I. NG2-glia as Multipotent Neural Stem Cells: Fact or Fantasy?. Neuron 2011, 70, 661–673. [CrossRef]
- Belachew, S.; Chittajallu, R.; Aguirre, A.A.; Yuan, X.; Kirby, M.; Anderson, S.; Gallo, V. Postnatal NG2 proteoglycan–expressing progenitor cells are intrinsically multipotent and generate functional neurons. J. Cell Biol. 2003, 161, 169–186. [CrossRef]
- Belachew, S.; Gallo, V. Synaptic and extrasynaptic neurotransmitter receptors in glial precursors' quest for identity. Glia 2004, 48, 185–196. [CrossRef]
- Ramón-Cueto, A.; Avila, J. Olfactory ensheathing glia: properties and function. Brain Res. Bull. 1998, 46, 175–187. [CrossRef]
- Mietto, B.S.; Jhelum, P.; Schulz, K.; David, S. Schwann Cells Provide Iron to Axonal Mitochondria and Its Role in Nerve Regeneration. J. Neurosci. 2021, 41, 7300–7313. [CrossRef]
- Valverde, F.; Lopez-Mascaraque, L. Neuroglial arrangements in the olfactory glomeruli of the hedgehog. J. Comp. Neurol. 1991, 307, 658–674. [CrossRef]
- Liu, L.; Tao-Cheng, J.-H.; Rallapalli, H.; Dodd, S.; Bouraoud, N.; Koretsky, A.P. High iron in mouse olfactory ensheathing cells at the glia limitans in the olfactory bulb underlies MRI T2* hypointensity. Imaging Neurosci. 2024, 2, 1–15. [CrossRef]
- Zhao, D.; Hu, M.; Liu, S. Glial cells in the mammalian olfactory bulb. Front. Cell. Neurosci. 2024, 18, 1426094. [CrossRef]
- Spassky, N.; Heydon, K.; Mangatal, A.; Jankovski, A.; Olivier, C.; Queraud-Lesaux, F.; Goujet-Zalc, C.; Thomas, J.L.; Zalc, B. Sonic hedgehog-dependent emergence of oligodendrocytes in the telencephalon: evidence for a source of oligodendrocytes in the olfactory bulb that is independent of PDGFRα signaling. Development 2001, 128, 4993–5004. [CrossRef]
- Wong, F.K.; Favuzzi, E. The brain's polymath: Emerging roles of microglia throughout brain development. Curr. Opin. Neurobiol. 2023, 79, 102700. [CrossRef]
- Connor, J.; Menzies, S.; St. Martin, S.; Mufson, E. A histochemical study of iron, transferrin, and ferritin in Alzheimer’s diseased brains. J Neurosci Res 1992, 31, 75-83.
- Morris, C.; Candy, J.; Oakley, A.; Bloxham, C.; Edwardson, J. Histochemical Distribution of Non-Haem Iron in the Human Brain. Acta Anat (Basel) 1992, 144, 235–257. [CrossRef]
- Hines, J.H. Evolutionary Origins of the Oligodendrocyte Cell Type and Adaptive Myelination. Front. Neurosci. 2021, 15. [CrossRef]
- Zhang, P.; Land, W.; Lee, S.; Juliani, J.; Lefman, J.; Smith, S.R.; Germain, D.; Kessel, M.; Leapman, R.; Rouault, T.A.; et al. Electron tomography of degenerating neurons in mice with abnormal regulation of iron metabolism. J. Struct. Biol. 2005, 150, 144–153. [CrossRef]
- Cheli, V.T.; Correale, J.; Paez, P.M.; Pasquini, J.M. Iron Metabolism in Oligodendrocytes and Astrocytes, Implications for Myelination and Remyelination. ASN Neuro 2020, 12. [CrossRef]
- Gerber, M.R.; Connor, J.R. Do oligodendrocytes mediate iron regulation in the human brain? Ann Neurol 1989, 26, 95-98.
- Werner, H.; Buscham, T.; Eichel, M.; Siems, S. Turning to myelin turnover. Neural Regen. Res. 2019, 14, 2063–2066. [CrossRef]
- Meyer, N.; Rinholm, J.E. Mitochondria in myelinating oligodendrocytes: slow and out of breath? Metabolites 2021, 11, 359.
- Reinert, A.; Morawski, M.; Seeger, J.; Arendt, T.; Reinert, T. Iron concentrations in neurons and glial cells with estimates on ferritin concentrations. BMC Neurosci. 2019, 20, 1–14. [CrossRef]
- Fünfschilling, U.; Supplie, L.M.; Mahad, D.; Boretius, S.; Saab, A.S.; Edgar, J.; Brinkmann, B.G.; Kassmann, C.M.; Tzvetanova, I.D.; Möbius, W.; et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 2012, 485, 517–521. [CrossRef]
- Carlisle, E.; Yin, Z.; Pisani, D.; Donoghue, P.C.J. Ediacaran origin and Ediacaran-Cambrian diversification of Metazoa. Sci. Adv. 2024, 10, eadp7161. [CrossRef]
- Wade, J.; Byrne, D.J.; Ballentine, C.J.; Drakesmith, H. Temporal variation of planetary iron as a driver of evolution. Proc. Natl. Acad. Sci. USA 2021, 118. [CrossRef]
- Nairz, M.; Schroll, A.; Sonnweber, T.; Weiss, G. The struggle for iron - a metal at the host-pathogen interface. Cell. Microbiol. 2010, 12, 1691–1702. [CrossRef]
- Deamer, D. The Role of Lipid Membranes in Life’s Origin. Life 2017, 7, 5. [CrossRef]
- Burcar, B.T.; Barge, L.M.; Trail, D.; Watson, E.B.; Russell, M.J.; McGown, L.B. RNA Oligomerization in Laboratory Analogues of Alkaline Hydrothermal Vent Systems. Astrobiology 2015, 15, 509–522. [CrossRef]
- Camprubi, E.; Jordan, S.F.; Vasiliadou, R.; Lane, N. Iron catalysis at the origin of life. IUBMB Life 2017, 69, 373–381. [CrossRef]
- Heard, A.W.; Bekker, A.; Kovalick, A.; Tsikos, H.; Ireland, T.; Dauphas, N. Oxygen production and rapid iron oxidation in stromatolites immediately predating the Great Oxidation Event. Earth Planet. Sci. Lett. 2022, 582. [CrossRef]
- Johnson, J.E.; Present, T.M.; Valentine, J.S. Iron: Life’s primeval transition metal. Proc Natl Acad Sci U S A 2024, 121, e2318692121.
- Wächtershäuser, G. From volcanic origins of chemoautotrophic life to Bacteria, Archaea and Eukarya. Philos. Trans. R. Soc. B: Biol. Sci. 2006, 361, 1787–1806; discussion 1806-1788. [CrossRef]
- Muchowska, K.B.; Varma, S.J.; Moran, J. Synthesis and breakdown of universal metabolic precursors promoted by iron. Nature 2019, 569, 104–107. [CrossRef]
- Barge, L.M.; Flores, E.; Baum, M.M.; VanderVelde, D.G.; Russell, M.J. Redox and pH gradients drive amino acid synthesis in iron oxyhydroxide mineral systems. Proc. Natl. Acad. Sci. 2019, 116, 4828–4833. [CrossRef]
- Sydow, C.; Sauer, F.; Siegle, A.F.; Trapp, O. Iron-Mediated Peptide Formation in Water and Liquid Sulfur Dioxide under Prebiotically Plausible Conditions**. ChemSystemsChem 2022, 5. [CrossRef]
- Shanker, U.; Bhushan, B.; Bhattacharjee, G.; Kamaluddin Formation of Nucleobases from Formamide in the Presence of Iron Oxides: Implication in Chemical Evolution and Origin of Life. Astrobiology 2011, 11, 225–233. [CrossRef]
- Georgieva, M.N.; Little, C.T.; Maslennikov, V.V.; Glover, A.G.; Ayupova, N.R.; Herrington, R.J. The history of life at hydrothermal vents. Earth-Science Rev. 2021, 217. [CrossRef]
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., Walter, P. The lipid bilayer. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, 2002.
- Suwalsky, M.; Martínez, F.; Cárdenas, H.; Grzyb, J.; Strzałka, K. Iron affects the structure of cell membrane molecular models. Chem. Phys. Lipids 2005, 134, 69–77. [CrossRef]
- Płaczkiewicz, J.; Gieczewska, K.; Musiałowski, M.; Adamczyk-Popławska, M.; Bącal, P.; Kwiatek, A. Availability of iron ions impacts physicochemical properties and proteome of outer membrane vesicles released by Neisseria gonorrhoeae. Sci. Rep. 2023, 13, 18733. [CrossRef]
- Gould, S.B.; Garg, S.G.; Martin, W.F. Bacterial Vesicle Secretion and the Evolutionary Origin of the Eukaryotic Endomembrane System. Trends Microbiol. 2016, 24, 525–534. [CrossRef]
- Cohen, Z.R.; Todd, Z.R.; Wogan, N.; Black, R.A.; Keller, S.L.; Catling, D.C. Plausible Sources of Membrane-Forming Fatty Acids on the Early Earth: A Review of the Literature and an Estimation of Amounts. ACS Earth Space Chem. 2022, 7, 11–27. [CrossRef]
- Weiss, M.C.; Sousa, F.L.; Mrnjavac, N.; Neukirchen, S.; Roettger, M.; Nelson-Sathi, S.; Martin, W.F. The physiology and habitat of the last universal common ancestor. Nat. Microbiol. 2016, 1, 16116. [CrossRef]
- Moody, E.R.R.; Álvarez-Carretero, S.; Mahendrarajah, T.A.; Clark, J.W.; Betts, H.C.; Dombrowski, N.; Szánthó, L.L.; Boyle, R.A.; Daines, S.; Chen, X.; et al. The nature of the last universal common ancestor and its impact on the early Earth system. Nat. Ecol. Evol. 2024, 8, 1654–1666. [CrossRef]
- Bradley, J.M.; Svistunenko, D.A.; Wilson, M.T.; Hemmings, A.M.; Moore, G.R.; Le Brun, N.E. Bacterial iron detoxification at the molecular level. J. Biol. Chem. 2020, 295, 17602–17623. [CrossRef]
- Philpott, C.C. Iron uptake in fungi: A system for every source. Biochim. et Biophys. Acta (BBA) - Mol. Cell Res. 2006, 1763, 636–645. [CrossRef]
- Bairwa, G.; Jung, W.H.; Kronstad, J.W. Iron acquisition in fungal pathogens of humans. Metallomics 2017, 9, 215–227. [CrossRef]
- Liu, Y.; Xiong, Z.; Wu, W.; Ling, H.-Q.; Kong, D. Iron in the Symbiosis of Plants and Microorganisms. Plants 2023, 12, 1958. [CrossRef]
- Sun, J.; Xiao, S.; Xue, C. The tug-of-war on iron between plant and pathogen. Phytopathol. Res. 2023, 5, 1–14. [CrossRef]
- Arosio, P.; Cairo, G.; Bou-Abdallah, F. A Brief History of Ferritin, an Ancient and Versatile Protein. Int. J. Mol. Sci. 2024, 26, 206. [CrossRef]
- Grant, R.; Filman, D.; Finkel, S.; Kolter, R.; Hogle, J. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Mol. Biol. 1998, 5, 294–303. [CrossRef]
- Peracino, B.; Buracco, S.; Bozzaro, S. The Nramp (Slc11) proteins regulate development, resistance to pathogenic bacteria and iron homeostasis in Dictyostelium discoideum. J. Cell Sci. 2013, 126, 301–311. [CrossRef]
- Peracino, B.; Monica, V.; Primo, L.; Bracco, E.; Bozzaro, S. Iron metabolism in the social amoeba Dictyostelium discoideum: A role for ferric chelate reductases. Eur. J. Cell Biol. 2022, 101, 151230. [CrossRef]
- Schoffman, H.; Lis, H.; Shaked, Y.; Keren, N. Iron–Nutrient Interactions within Phytoplankton. Front. Plant Sci. 2016, 7, 1223–1223. [CrossRef]
- Ros-Rocher, N.; Pérez-Posada, A.; Leger, M.M.; Ruiz-Trillo, I. The origin of animals: an ancestral reconstruction of the unicellular-to-multicellular transition. Open Biol. 2021, 11, 200359. [CrossRef]
- Müller, W.E. Review: How was metazoan threshold crossed? The hypothetical Urmetazoa. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 2001, 129, 433–460. [CrossRef]
- Delsuc, F.; Philippe, H.; Tsagkogeorga, G.; Simion, P.; Tilak, M.-K.; Turon, X.; López-Legentil, S.; Piette, J.; Lemaire, P.; Douzery, E.J.P. A phylogenomic framework and timescale for comparative studies of tunicates. BMC Biol. 2018, 16, 39. [CrossRef]
- Moroz, L.L.; Kohn, A.B. Independent origins of neurons and synapses: insights from ctenophores. Philos. Trans. R. Soc. B: Biol. Sci. 2016, 371, 20150041. [CrossRef]
- Kapli, P.; Telford, M.J. Topology-dependent asymmetry in systematic errors affects phylogenetic placement of Ctenophora and Xenacoelomorpha. Sci. Adv. 2020, 6, eabc5162. [CrossRef]
- Holland, L.Z. Evolution of basal deuterostome nervous systems. J. Exp. Biol. 2015, 218, 637–645. [CrossRef]
- Satoh, N.; Rokhsar, D.; Nishikawa, T. Chordate evolution and the three-phylum system. Proc. R. Soc. B: Biol. Sci. 2014, 281, 20141729. [CrossRef]
- Miguel-Tomé, S.; Llinás, R.R. Broadening the definition of a nervous system to better understand the evolution of plants and animals. Plant Signal. Behav. 2021, 16, 1927562. [CrossRef]
- Mackie, G.O.; Meech, R.W. Central Circuitry in the Jellyfish Aglantha Digitale: I. The Relay System. J. Exp. Biol. 1995, 198, 2261–2270. [CrossRef]
- Watanabe, H.; Fujisawa, T.; Holstein, T.W. Cnidarians and the evolutionary origin of the nervous system. Dev. Growth Differ. 2009, 51, 167–183. [CrossRef]
- Evans, S.D.; Droser, M.L.; Erwin, D.H. Developmental processes in Ediacara macrofossils. Proc. R. Soc. B: Biol. Sci. 2021, 288, 20203055. [CrossRef]
- Gavilán, B.; Perea-Atienza, E.; Martínez, P. Xenacoelomorpha: a case of independent nervous system centralization?. Philos. Trans. R. Soc. B: Biol. Sci. 2016, 371, 20150039. [CrossRef]
- Arendt, D.; Bertucci, P.Y.; Achim, K.; Musser, J.M. Evolution of neuronal types and families. Curr. Opin. Neurobiol. 2019, 56, 144–152. [CrossRef]
- Liebeskind, B.J.; Hillis, D.M.; Zakon, H.H.; Hofmann, H.A. Complex Homology and the Evolution of Nervous Systems. Trends Ecol. Evol. 2016, 31, 127–135. [CrossRef]
- Hejnol, A.; Rentzsch, F. Neural nets. Curr. Biol. 2015, 25, R782–R786. [CrossRef]
- Moroz, L.L.; Romanova, D.Y. Alternative neural systems: What is a neuron? (Ctenophores, sponges and placozoans). Front. Cell Dev. Biol. 2022, 10, 1071961. [CrossRef]
- Natalio, F.; Wiese, S.; Friedrich, N.; Werner, P.; Tahir, M.N. Localization and Characterization of Ferritin in Demospongiae: A Possible Role on Spiculogenesis. Mar. Drugs 2014, 12, 4659–4676. [CrossRef]
- Finoshin, A.D.; Adameyko, K.I.; Mikhailov, K.V.; Kravchuk, O.I.; Georgiev, A.A.; Gornostaev, N.G.; Kosevich, I.A.; Mikhailov, V.S.; Gazizova, G.R.; Shagimardanova, E.I.; et al. Iron metabolic pathways in the processes of sponge plasticity. PLOS ONE 2020, 15, e0228722. [CrossRef]
- Müller, W.E.; Korzhev, M.; Le Pennec, G.; Müller, I.M.; Schröder, H.C. Origin of metazoan stem cell system in sponges: first approach to establish the model (Suberites domuncula). Biomol. Eng. 2003, 20, 369–379. [CrossRef]
- Krasko, A.; Schröder, H.C.; Batel, R.; Grebenjuk, V.A.; Steffen, R.; Müller, I.M.; Müller, W.E. Iron Induces Proliferation and Morphogenesis in Primmorphs from the Marine SpongeSuberites domuncula. DNA Cell Biol. 2002, 21, 67–80. [CrossRef]
- Müller, W.E. The stem cell concept in sponges (Porifera): Metazoan traits. Semin. Cell Dev. Biol. 2006, 17, 481–491. [CrossRef]
- Lechauve, C.; Jager, M.; Laguerre, L.; Kiger, L.; Correc, G.; Leroux, C.; Vinogradov, S.; Czjzek, M.; Marden, M.C.; Bailly, X. Neuroglobins, Pivotal Proteins Associated with Emerging Neural Systems and Precursors of Metazoan Globin Diversity. J. Biol. Chem. 2013, 288, 6957–6967. [CrossRef]
- Ho, V.R.; Goss, G.G.; Leys, S.P. ATP and glutamate coordinate contractions in the freshwater sponge Ephydatia muelleri. J. Exp. Biol. 2025, 228. [CrossRef]
- Adameyko, K.I.; Burakov, A.V.; Finoshin, A.D.; Mikhailov, K.V.; Kravchuk, O.I.; Kozlova, O.S.; Gornostaev, N.G.; Cherkasov, A.V.; Erokhov, P.A.; Indeykina, M.I.; et al. Conservative and Atypical Ferritins of Sponges. Int. J. Mol. Sci. 2021, 22, 8635. [CrossRef]
- Riesgo, A.; Farrar, N.; Windsor, P.J.; Giribet, G.; Leys, S.P. The Analysis of Eight Transcriptomes from All Poriferan Classes Reveals Surprising Genetic Complexity in Sponges. Mol. Biol. Evol. 2014, 31, 1102–1120. [CrossRef]
- Srivastava, M.; Simakov, O.; Chapman, J.; Fahey, B.; Gauthier, M.E.A.; Mitros, T.; Richards, G.S.; Conaco, C.; Dacre, M.; Hellsten, U.; et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 2010, 466, 720–726. [CrossRef]
- Mayorova, T.D.; Koch, T.L.; Kachar, B.; Jung, J.H.; Reese, T.S.; Smith, C.L. Placozoan secretory cell types implicated in feeding, innate immunity and regulation of behavior. PLOS ONE 2025, 20, e0311271. [CrossRef]
- Romanova, D.Y.; Povernov, A.A.; Nikitin, M.A.; Borman, S.I.; Frank, Y.A.; Moroz, L.L. Long-term dynamics of placozoan culture: emerging models for population and space biology. Front. Cell Dev. Biol. 2025, 12, 1514553. [CrossRef]
- Najera, D.G.; Dittmer, N.T.; Weber, J.J.; Kanost, M.R.; Gorman, M.J. Phylogenetic and sequence analyses of insect transferrins suggest that only transferrin 1 has a role in iron homeostasis. Insect Sci. 2020, 28, 495–508. [CrossRef]
- Najle, S.R.; Grau-Bové, X.; Elek, A.; Navarrete, C.; Cianferoni, D.; Chiva, C.; Cañas-Armenteros, D.; Mallabiabarrena, A.; Kamm, K.; Sabidó, E.; et al. Stepwise emergence of the neuronal gene expression program in early animal evolution. Cell 2023, 186, 4676–4693.e29. [CrossRef]
- Jékely, G.; Keijzer, F.; Godfrey-Smith, P. An option space for early neural evolution. Philos. Trans. R. Soc. B: Biol. Sci. 2015, 370, 20150181. [CrossRef]
- Steinmetz, P.R.H. A non-bilaterian perspective on the development and evolution of animal digestive systems. Cell Tissue Res. 2019, 377, 321–339. [CrossRef]
- A Schwarz, J.; Brokstein, P.B.; Voolstra, C.; Terry, A.Y.; Manohar, C.F.; Miller, D.J.; Szmant, A.M.; Coffroth, M.A.; Medina, M. Coral life history and symbiosis: Functional genomic resources for two reef building Caribbean corals, Acropora palmata and Montastraea faveolata. BMC Genom. 2008, 9, 97–97. [CrossRef]
- Zhou, Y.; Liu, H.; Feng, C.; Lu, Z.; Liu, J.; Huang, Y.; Tang, H.; Xu, Z.; Pu, Y.; Zhang, H. Genetic adaptations of sea anemone to hydrothermal environment. Sci. Adv. 2023, 9, eadh0474. [CrossRef]
- Jain, A.; Connolly, E.L. Mitochondrial iron transport and homeostasis in plants. Front. Plant Sci. 2013, 4, 348. [CrossRef]
- Smith, H.L.; Pavasovic, A.; Surm, J.M.; Phillips, M.J.; Prentis, P.J. Evidence for a Large Expansion and Subfunctionalization of Globin Genes in Sea Anemones. Genome Biol. Evol. 2018, 10, 1892–1901. [CrossRef]
- Sprecher, S.G. Neural Cell Type Diversity in Cnidaria. Front. Neurosci. 2022, 16, 909400. [CrossRef]
- Koizumi, O.; Hamada, S.; Minobe, S.; Hamaguchi-Hamada, K.; Kurumata-Shigeto, M.; Nakamura, M.; Namikawa, H. The nerve ring in cnidarians: its presence and structure in hydrozoan medusae. Zoology 2014, 118, 79–88. [CrossRef]
- Moroz, L.L. Convergent evolution of neural systems in ctenophores. J. Exp. Biol. 2015, 218, 598–611. [CrossRef]
- Kass-Simon, G.; Pierobon, P. Cnidarian chemical neurotransmission, an updated overview. Comp Biochem Physiol A Mol Integr Physiol 2007, 146, 9-25.
- Carlberg, M.; Anctil, M. Biogenic amines in coelenterates. Comp Biochem Physiol C Comp Pharmacol Toxicol 1993, 106, 1-9.
- De Robertis, E.M.; Tejeda-Muñoz, N. Evo-Devo of Urbilateria and its larval forms. Dev. Biol. 2022, 487, 10–20. [CrossRef]
- Katsuki, T.; Greenspan, R.J. Jellyfish nervous systems. Curr. Biol. 2013, 23, R592–R594. [CrossRef]
- Satterlie, R.A. The search for ancestral nervous systems: an integrative and comparative approach. J. Exp. Biol. 2015, 218, 612–617. [CrossRef]
- Chari, T.; Weissbourd, B.; Gehring, J.; Ferraioli, A.; Leclère, L.; Herl, M.; Gao, F.; Chevalier, S.; Copley, R.R.; Houliston, E.; et al. Whole-animal multiplexed single-cell RNA-seq reveals transcriptional shifts acrossClytiamedusa cell types. Sci. Adv. 2021, 7, eabh1683. [CrossRef]
- Oren, M.; Brikner, I.; Appelbaum, L.; Levy, O. Fast Neurotransmission Related Genes Are Expressed in Non Nervous Endoderm in the Sea Anemone Nematostella vectensis. PLOS ONE 2014, 9, e93832. [CrossRef]
- Sheloukhova, L.; Watanabe, H. Evolution of glial cells: a non-bilaterian perspective. Neural Dev. 2024, 19, 10. [CrossRef]
- Garm, A.; Poussart, Y.; Parkefelt, L.; Ekström, P.; Nilsson, D.-E. The ring nerve of the box jellyfish Tripedalia cystophora. Cell Tissue Res. 2007, 329, 147–157. [CrossRef]
- Rey, S.; Zalc, B.; Klämbt, C. Evolution of glial wrapping: A new hypothesis. Dev. Neurobiol. 2020, 81, 453–463. [CrossRef]
- Gehring, W.J. New Perspectives on Eye Development and the Evolution of Eyes and Photoreceptors. J. Hered. 2005, 96, 171–184. [CrossRef]
- O'Connor, M.; Garm, A.; Marshall, J.N.; Hart, N.S.; Ekström, P.; Skogh, C.; Nilsson, D.-E.; Megan, O.; Anders, G.; N., M.J.; et al. Visual pigment in the lens eyes of the box jellyfish Chiropsella bronzie. Proc. R. Soc. B: Biol. Sci. 2010, 277, 1843–1848. [CrossRef]
- Gehring, W.; Seimiya, M. Eye evolution and the origin of Darwin's eye prototype. Ital. J. Zoöl. 2010, 77, 124–136. [CrossRef]
- Wolkow, N.; Li, Y.; Maminishkis, A.; Song, Y.; Alekseev, O.; Iacovelli, J.; Song, D.; Lee, J.C.; Dunaief, J.L. Iron upregulates melanogenesis in cultured retinal pigment epithelial cells. Exp. Eye Res. 2014, 128, 92–101. [CrossRef]
- Yuan, T.; York, J.R.; McCauley, D.W. Gliogenesis in lampreys shares gene regulatory interactions with oligodendrocyte development in jawed vertebrates. Dev. Biol. 2018, 441, 176–190. [CrossRef]
- Bullock, T.H.; Moore, J.K.; Fields, R. Evolution of myelin sheaths: Both lamprey and hagfish lack myelin. Neurosci. Lett. 1984, 48, 145–148. [CrossRef]
- Zalc, B. The acquisition of myelin: An evolutionary perspective. Brain Res. 2016, 1641, 4–10. [CrossRef]
- Clark, A.J.; Uyeno, T.A. Feeding in jawless fishes. In Feeding in Vertebrates: Evolution, Morphology, Behavior, Biomechanics, Bels, V., Whishaw, I.Q., Eds.; Springer International Publishing: Cham, 2019; pp. 189-230.
- Glover, C.N.; Niyogi, S.; Blewett, T.A.; Wood, C.M. Iron transport across the skin and gut epithelia of Pacific hagfish: Kinetic characterisation and effect of hypoxia. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 2016, 199, 1–7. [CrossRef]
- Tsioros, K.; Youson, J. Intracellular distribution of iron (and associated elements) in various cell types of larvae and juveniles of the sea lamprey (Petromyzon marinus). Tissue Cell 1997, 29, 137–162. [CrossRef]
- Hall, S.J.; Youson, J.H. Distribution of ferric iron in larval lampreys, Petromyzon marinus L. Histol Histopathol. 1988, 3, 7–20.
- Macey, D.J.; Youson, J.H. Occurrence and Structure of Iron Inclusions in Adipocytes of Larval Lampreys. Acta Zoöl. 1990, 71, 69–76. [CrossRef]
- González-Llera, L.; Shifman, M.I.; Barreiro-Iglesias, A. Repulsive guidance molecule A (RGMA) is widely expressed in the CNS of the sea lamprey Petromyzon marinus during early development. MicroPubl Biol. 2024, 2024. [CrossRef]
- Youson, J.H.; Sargent, P.A.; Pearce, G.W. Iron and aluminum deposition in the meninges of the lamprey: Identification of an aluminum-ferritin inclusion body. Anat. Rec. 1989, 223, 13–20. [CrossRef]
- Barreiro-Iglesias, A.; Laramore, C.; Shifman, M.; Anadón, R.; Selzer, M.; Rodicio, M. The sea lamprey tyrosine hydroxylase: cDNA cloning and in situ hybridization study in the brain. Neuroscience 2010, 168, 659–669. [CrossRef]
- Andersen, Ø.; Pantopoulos, K.; Kao, H.; Muckenthaler, M.; Youson, J.H.; Pieribone, V. Regulation of iron metabolism in the sanguivore lamprey Lampetra fluviatilis --molecular cloning of two ferritin subunits and two iron-regulatory proteins (IRP) reveals evolutionary conservation of the iron-regulatory element (IRE)/IRP regulatory system. Eur. J. Biochem. 1998, 254, 223–229. [CrossRef]
- Macey, D.J.; Cake, M.H.; Potter, I.C. Exceptional iron concentrations in larval lampreys (Geotria australis) and the activities of superoxide radical detoxifying enzymes. Biochem. J. 1988, 252, 167–172. [CrossRef]
- Webster, R.; Pollara, B. Isolation and partial characterization of transferrin in the sea lamprey, Petromyzon marinus. Comp. Biochem. Physiol. 1969, 30, 509–527. [CrossRef]
- Smalley, S.R.; Macey, D.J.; Potter, I.C. Changes in the amount of nonhaem iron in the plasma, whole body, and selected organs during the postlarval life of the lampreyGeotria australis. J. Exp. Zoöl. 1986, 237, 149–157. [CrossRef]
- Yang, W.; Mu, B.; You, J.; Tian, C.; Bin, H.; Xu, Z.; Zhang, L.; Ma, R.; Wu, M.; Zhang, G.; et al. Non-classical ferroptosis inhibition by a small molecule targeting PHB2. Nat. Commun. 2022, 13, 7473. [CrossRef]
- Guo, J.; Lyu, S.; Qi, Y.; Chen, X.; Yang, L.; Zhao, C.; Wang, H. Molecular evolution and gene expression of ferritin family involved in immune defense of lampreys. Dev. Comp. Immunol. 2023, 146, 104729. [CrossRef]
- Birceanu, O.; Ferreira, P.; Neal, J.; Sunga, J.; Anthony, S.; Davidson, S.M.; Edwards, S.L.; Wilson, J.M.; Youson, J.H.; Vijayan, M.M.; et al. Divergent Pathways of Ammonia and Urea Production and Excretion during the Life Cycle of the Sea Lamprey. Physiol. Biochem. Zoöl. 2022, 95, 551–567. [CrossRef]
- McCauley, D.W.; Bronner-Fraser, M. Conservation and divergence of BMP2/4 genes in the lamprey: expression and phylogenetic analysis suggest a single ancestral vertebrate gene. Evol. Dev. 2004, 6, 411–422. [CrossRef]
- González-Llera, L.; Shifman, M.I.; Barreiro-Iglesias, A. Neogenin expression in ependymo-radial glia of the larval sea lamprey Petromyzon marinus spinal cord. MicroPubl Biol 2023, 2023. [CrossRef]
- Lamanna, F.; Hervas-Sotomayor, F.; Oel, A.P.; Jandzik, D.; Sobrido-Cameán, D.; Santos-Durán, G.N.; Martik, M.L.; Stundl, J.; Green, S.A.; Brüning, T.; et al. A lamprey neural cell type atlas illuminates the origins of the vertebrate brain. Nat. Ecol. Evol. 2023, 7, 1714–1728. [CrossRef]
- Robertson, B.; Kardamakis, A.; Capantini, L.; Pérez-Fernández, J.; Suryanarayana, S.M.; Wallén, P.; Stephenson-Jones, M.; Grillner, S. The lamprey blueprint of the mammalian nervous system. Prog Brain Res 2014, 212, 337-349. [CrossRef]
- Ryczko, D.; Cone, J.J.; Alpert, M.H.; Goetz, L.; Auclair, F.; Dubé, C.; Parent, M.; Roitman, M.F.; Alford, S.; Dubuc, R. A descending dopamine pathway conserved from basal vertebrates to mammals. Proc. Natl. Acad. Sci. 2016, 113, E2440–E2449. [CrossRef]
- Foo, L.C.; Dougherty, J.D. Aldh1L1 is expressed by postnatal neural stem cells in vivo. Glia 2013, 61, 1533–1541. [CrossRef]
- Weil, M.-T.; Heibeck, S.; Töpperwien, M.; Dieck, S.T.; Ruhwedel, T.; Salditt, T.; Rodicio, M.C.; Morgan, J.R.; Nave, K.-A.; Möbius, W.; et al. Axonal Ensheathment in the Nervous System of Lamprey: Implications for the Evolution of Myelinating Glia. J. Neurosci. 2018, 38, 6586–6596. [CrossRef]
- Möbius, W.; Patzig, J.; Nave, K.-A.; Werner, H.B. Phylogeny of proteolipid proteins: divergence, constraints, and the evolution of novel functions in myelination and neuroprotection. Neuron Glia Biol. 2008, 4, 111–127. [CrossRef]
- Kitagawa, K.; Sinoway, M.; Yang, C.; Gould, R.; Colman, D. A proteolipid protein gene family: Expression in sharks and rays and possible evolution from an ancestral gene encoding a pore-forming polypeptide. Neuron 1993, 11, 433–448. [CrossRef]
- Zalc, B.; Colman, D.R. Origins of Vertebrate Success. Science 2000, 288, 271–271. [CrossRef]
- Grassé, P. Le système nerveux des insectes. Traité de Zoologie. Anatomi, Systematique, Biologie. T. VIII, fasc. III 1975, 321-510.
- Hartline, D.K. The evolutionary origins of glia. Glia 2011, 59, 1215–1236. [CrossRef]
- Tomassy, G.S.; Dershowitz, L.B.; Arlotta, P. Diversity Matters: A Revised Guide to Myelination. Trends Cell Biol. 2016, 26, 135–147. [CrossRef]
- Knowles, L. The Evolution of Myelin: Theories and Application to Human Disease. J. Evol. Med. 2017, 5, 1–23. [CrossRef]
- Roots, B.I. The phylogeny of invertebrates and the evolution of myelin. Neuron Glia Biol. 2008, 4, 101–109. [CrossRef]
- Helm, C.; Karl, A.; Beckers, P.; Kaul-Strehlow, S.; Ulbricht, E.; Kourtesis, I.; Kuhrt, H.; Hausen, H.; Bartolomaeus, T.; Reichenbach, A.; et al. Early evolution of radial glial cells in Bilateria. Proc. R. Soc. B: Biol. Sci. 2017, 284, 20170743. [CrossRef]
- Abbott, N.J. Dynamics of CNS Barriers: Evolution, Differentiation, and Modulation. Cell. Mol. Neurobiol. 2005, 25, 5–23. [CrossRef]
- Bundgaard, M.; Abbott, N.J. All vertebrates started out with a glial blood-brain barrier 4–500 million years ago. Glia 2008, 56, 699–708. [CrossRef]
- Abbott, N.J.; Bundgaard, M. Electron-dense tracer evidence for a blood?brain barrier in the cuttlefishSepia officinalis. J. Neurocytol. 1992, 21, 276–294. [CrossRef]
- Bundgaard, M.; Abbott, N.J. Fine structure of the blood?brain interface in the cuttlefishSepia officinalis (Mollusca, Cephalopoda). J. Neurocytol. 1992, 21, 260–275. [CrossRef]
- Zueva, O.; Khoury, M.; Heinzeller, T.; Mashanova, D.; Mashanov, V. The complex simplicity of the brittle star nervous system. Front. Zoöl. 2018, 15, 1–26. [CrossRef]
- Märkel, K.; Röser, U. Ultrastructure and organization of the epineural canal and the nerve cord in sea urchins (Echinodermata, Echinoida). Zoomorphology 1991, 110, 267–279. [CrossRef]
- Mashanov, V.; Zueva, O. Radial glia in echinoderms. Developmental neurobiology 2019, 79, 396-405.
- Langlet, F. Tanycyte Gene Expression Dynamics in the Regulation of Energy Homeostasis. Front. Endocrinol. 2019, 10, 286. [CrossRef]
- Quiroga, S.Y.; Bonilla, E.C.; Bolaños, D.M.; Carbayo, F.; Litvaitis, M.K.; Brown, F.D. Evolution of flatworm central nervous systems: Insights from polyclads. Genet. Mol. Biol. 2015, 38, 233–248. [CrossRef]
- Bailly, X.; Reichert, H.; Hartenstein, V. The urbilaterian brain revisited: novel insights into old questions from new flatworm clades. Dev. Genes Evol. 2012, 223, 149–157. [CrossRef]
- Riva, A.; Orrù, B.; Riva, F.T. Giuseppe Sterzi (1876–1919) of the University of Cagliari: A brilliant neuroanatomist and medical historian. The Anatomical Record 2000, 261, 105-110. [CrossRef]
- Benito-Gutiérrez, È. A gene catalogue of the amphioxus nervous system. Int. J. Biol. Sci. 2006, 2, 149–160. [CrossRef]
- Holland, N.D.; Somorjai, I.M.L. The sensory peripheral nervous system in the tail of a cephalochordate studied by serial blockface scanning electron microscopy. J. Comp. Neurol. 2020, 528, 2569–2582. [CrossRef]
- Bozzo, M.; Lacalli, T.C.; Obino, V.; Caicci, F.; Marcenaro, E.; Bachetti, T.; Manni, L.; Pestarino, M.; Schubert, M.; Candiani, S. Amphioxus neuroglia: Molecular characterization and evidence for early compartmentalization of the developing nerve cord. Glia 2021, 69, 1654–1678. [CrossRef]
- Gattoni, G.; Andrews, T.G.R.; Benito-Gutiérrez, È. Restricted Proliferation During Neurogenesis Contributes to Regionalisation of the Amphioxus Nervous System. Front. Neurosci. 2022, 16, 812223. [CrossRef]
- He, C.; Han, T.; Liao, X.; Zhou, Y.; Wang, X.; Guan, R.; Tian, T.; Li, Y.; Bi, C.; Lu, N.; et al. Phagocytic intracellular digestion in amphioxus ( Branchiostoma ). Proc. R. Soc. B: Biol. Sci. 2018, 285, 20180438. [CrossRef]
- He, C.; Han, T.; Liao, X.; Zhou, Y.; Wang, X.; Guan, R.; Tian, T.; Li, Y.; Bi, C.; Lu, N.; et al. Correction to ‘Phagocytic intracellular digestion in amphioxus ( Branchiostoma )’. Proc. R. Soc. B: Biol. Sci. 2018, 285, 20181277. [CrossRef]
- Bosch-Queralt, M.; Fledrich, R.; Stassart, R.M. Schwann cell functions in peripheral nerve development and repair. Neurobiol. Dis. 2022, 176, 105952. [CrossRef]
- Clausen, J. The Effect of Vitamin A Deficiency on Myelination in the Central Nervous System of the Rat. Eur. J. Biochem. 1969, 7, 575–582. [CrossRef]
- Popescu, D.C.; Huang, H.; Singhal, N.K.; Shriver, L.; McDonough, J.; Clements, R.J.; Freeman, E.J. Vitamin K enhances the production of brain sulfatides during remyelination. PLOS ONE 2018, 13, e0203057. [CrossRef]
- Gomez-Pinedo, U.; Cuevas, J.A.; Benito-Martín, M.S.; Moreno-Jiménez, L.; Esteban-Garcia, N.; Torre-Fuentes, L.; Matías-Guiu, J.A.; Pytel, V.; Montero, P.; Matias-Guiu, J. Vitamin D increases remyelination by promoting oligodendrocyte lineage differentiation. Brain Behav. 2019, 10, e01498. [CrossRef]
- Goudarzvand, M.; Javan, M.; Mirnajafi-Zadeh, J.; Mozafari, S.; Tiraihi, T. Vitamins E and D3 Attenuate Demyelination and Potentiate Remyelination Processes of Hippocampal Formation of Rats Following Local Injection of Ethidium Bromide. Cell. Mol. Neurobiol. 2009, 30, 289–299. [CrossRef]
- Stadelmann, C.; Timmler, S.; Barrantes-Freer, A.; Simons, M. Myelin in the Central Nervous System: Structure, Function, and Pathology. Physiol. Rev. 2019, 99, 1381–1431. [CrossRef]
- Asadollahi, E.; Trevisiol, A.; Saab, A.S.; Looser, Z.J.; Dibaj, P.; Ebrahimi, R.; Kusch, K.; Ruhwedel, T.; Möbius, W.; Jahn, O.; et al. Oligodendroglial fatty acid metabolism as a central nervous system energy reserve. Nat. Neurosci. 2024, 27, 1934–1944. [CrossRef]
- Jeffery, W.R. Closing the wounds: One hundred and twenty five years of regenerative biology in the ascidian Ciona intestinalis. Genesis 2014, 53, 48–65. [CrossRef]
- Osugi, T.; Sasakura, Y.; Satake, H. The nervous system of the adult ascidian Ciona intestinalis Type A (Ciona robusta): Insights from transgenic animal models. PLOS ONE 2017, 12, e0180227–e0180227. [CrossRef]
- de Abreu, I.S.; Wajsenzon, I.J.R.; Dias, J.C.; Allodi, S.; Monteiro-De-Barros, C. Central nervous system regeneration in ascidians: cell migration and differentiation. Cell Tissue Res. 2022, 390, 335–354. [CrossRef]
- Stacey, J.; Driedzic, W. Temporal variability in, and impact of food availability on vanadium and iron concentrations in Ciona intestinalis tissues (Tunicata, Ascidiacea). J. Exp. Mar. Biol. Ecol. 2010, 386, 11–18. [CrossRef]
- Waggett, R.J.; Buskey, E.J. Escape reaction performance of myelinated and non-myelinated calanoid copepods. J. Exp. Mar. Biol. Ecol. 2008, 361, 111–118. [CrossRef]
- Hama, K. Some observations on the fine structure of the giant synapse in the stellate ganglk of the squid, Doryteuphis bleekeri. Z Zellforsch Mikrosk Anat 1962, 56, 437–444. [CrossRef]
- Bear, R.S.; Schmitt, F.O.; Young, J.Z. The sheath components of the giant nerve fibres of the squid. Proc. R. Soc. London. Ser. B. Biol. Sci. 1937, 123, 496–504. [CrossRef]
- Pumphrey, R.J.; Young, J.Z. The Rates of Conduction of Nerve Fibres of Various Diameters in Cephalopods. J. Exp. Biol. 1938, 15, 453–466. [CrossRef]
- Peracchia, C. Direct communication between axons and sheath glial cells in crayfish. Nature 1981, 290, 597–598. [CrossRef]
- Hsu), K.X.(.; Terakawa, S. Fenestration nodes and the wide submyelinic space form the basis for the unusually fast impulse conduction of shrimp myelinated axons. J. Exp. Biol. 1999, 202, 1979–1989. [CrossRef]
- Wang, S.S.-H.; Shultz, J.R.; Burish, M.J.; Harrison, K.H.; Hof, P.R.; Towns, L.C.; Wagers, M.W.; Wyatt, K.D. Functional Trade-Offs in White Matter Axonal Scaling. J. Neurosci. 2008, 28, 4047–4056. [CrossRef]
- Wilson, C.H.; Hartline, D.K. Novel organization and development of copepod myelin. ii. nonglial origin. J. Comp. Neurol. 2011, 519, 3281–3305. [CrossRef]
- Wilson, C.H.; Hartline, D.K. Novel organization and development of copepod myelin. i. ontogeny. J. Comp. Neurol. 2011, 519, 3259–3280. [CrossRef]
- Sidon, E.W.; Youson, J.H. Morphological changes in the liver of the sea lamprey, Petromyzon marinus L., during metamorphosis: I. Atresia of the bile ducts. J. Morphol. 1983, 177, 109–124. [CrossRef]
- Herdson, P.B.; Kaltenbach, J.P. Electron microscope studies on enzyme activity and the isolation of thiohydantoin-induced myelin figures in rat liver. J. Cell Biol. 1965, 25, 485–493. [CrossRef]
- Choung, H.Y.G.; Jean-Gilles, J.; Goldman, B. Myeloid bodies is not an uncommon ultrastructural finding. Ultrastruct. Pathol. 2022, 46, 130–138. [CrossRef]
- Jauregui, H.; Bradford, W.D.; Arstila, A.U.; Kinney, T.D.; Trump, B.F. Iron metabolism and cell membranes. III. Iron-induced alterations in HeLa cells. Am J Pathol. 1975, 80, 33–52.
- Ostrowski, S.; A Dierick, H.; Bejsovec, A. Genetic Control of Cuticle Formation During Embryonic Development ofDrosophila melanogaster. Genetics 2002, 161, 171–182. [CrossRef]
- Wang, Y.; Farine, J.-P.; Yang, Y.; Yang, J.; Tang, W.; Gehring, N.; Ferveur, J.-F.; Moussian, B. Transcriptional Control of Quality Differences in the Lipid-Based Cuticle Barrier in Drosophila suzukii and Drosophila melanogaster. Front. Genet. 2020, 11, 887. [CrossRef]
- Limmer, S.; Weiler, A.; Volkenhoff, A.; Babatz, F.; Klã¤Mbt, C. The Drosophila blood-brain barrier: development and function of a glial endothelium. Front. Neurosci. 2014, 8, 365–365. [CrossRef]
- Yildirim, K.; Petri, J.; Kottmeier, R.; Klämbt, C. Drosophila glia: Few cell types and many conserved functions. Glia 2018, 67, 5–26. [CrossRef]
- Banerjee, S.; Bhat, M.A. Glial ensheathment of peripheral axons in Drosophila. J. Neurosci. Res. 2007, 86, 1189–1198. [CrossRef]
- Beckers, P.; Helm, C.; Purschke, G.; Worsaae, K.; Hutchings, P.; Bartolomaeus, T. The central nervous system of Oweniidae (Annelida) and its implications for the structure of the ancestral annelid brain. Front. Zoöl. 2019, 16, 6. [CrossRef]
- Fernández, I.; Pardos, F.; Benito, J.; Roldán, C. Ultrastructural observations on the phoronid nervous system. J. Morphol. 1996, 230, 265–281. [CrossRef]
- Temereva, E.; Wanninger, A. Development of the nervous system in Phoronopsis harmeri (Lophotrochozoa, Phoronida) reveals both deuterostome- and trochozoan-like features. BMC Evol. Biol. 2012, 12, 121–121. [CrossRef]
- Garlick, R.L.; Williams, B.J.; Riggs, A.F. The hemoglobins of Phoronopsis viridis, of the primitive invertebrate phylum phoronida: Characterization and subunit structure. Arch. Biochem. Biophys. 1979, 194, 13–23. [CrossRef]
- Ma, Z.; Wang, W.; Yang, X.; Rui, M.; Wang, S. Glial ferritin maintains neural stem cells via transporting iron required for self-renewal in Drosophila. eLife 2024, 13. [CrossRef]
- Tang, X.; Zhou, B. Ferritin is the key to dietary iron absorption and tissue iron detoxification inDrosophila melanogaster. FASEB J. 2013, 27, 288–298. [CrossRef]
- Wan, B.; Belghazi, M.; Lemauf, S.; Poirié, M.; Gatti, J.-L. Proteomics of purified lamellocytes from Drosophila melanogaster HopT identifies new membrane proteins and networks involved in their functions. Insect Biochem. Mol. Biol. 2021, 134, 103584. [CrossRef]
- Iatsenko, I.; Marra, A.; Boquete, J.-P.; Peña, J.; Lemaitre, B. Iron sequestration by transferrin 1 mediates nutritional immunity inDrosophila melanogaster. Proc. Natl. Acad. Sci. USA 2020, 117, 7317–7325. [CrossRef]
- Verkhratsky, A.; Arranz, A.M.; Ciuba, K.; Pękowska, A. Evolution of neuroglia. Ann. New York Acad. Sci. 2022, 1518, 120–130. [CrossRef]
- Collingwood, J.F.; Davidson, M.R. The role of iron in neurodegenerative disorders: insights and opportunities with synchrotron light. Front. Pharmacol. 2014, 5, 191. [CrossRef]
- Visanji, N.P.; Collingwood, J.F.; Finnegan, M.E.; Tandon, A.; House, E.; Hazrati, L.-N. Iron Deficiency in Parkinsonism: Region-Specific Iron Dysregulation in Parkinson's Disease and Multiple System Atrophy. J. Park. Dis. 2013, 3, 523–537. [CrossRef]
- Hackett, M.J.; McQuillan, J.A.; El-Assaad, F.; Aitken, J.B.; Levina, A.; Cohen, D.D.; Siegele, R.; Carter, E.A.; Grau, G.E.; Hunt, N.H.; et al. Chemical alterations to murine brain tissue induced by formalin fixation: implications for biospectroscopic imaging and mapping studies of disease pathogenesis. Anal. 2011, 136, 2941–2952. [CrossRef]
- Schrag, M.; Dickson, A.; Jiffry, A.; Kirsch, D.; Vinters, H.V.; Kirsch, W. The effect of formalin fixation on the levels of brain transition metals in archived samples. BioMetals 2010, 23, 1123–1127. [CrossRef]
- Perls, M. Nachweis von Eisenoxyd in gewissen Pigmenten. Archiv F Pathol Anat 1867, 39, 42–48. [CrossRef]
- Moos, T. Immunohistochemical localization of intraneuronal transferrin receptor immunoreactivity in the adult mouse central nervous system. J Comp Neurol 1996, 375, 675-692. [CrossRef]
- Moos, T.; Nielsen, T.R.; Skjørringe, T.; Morgan, E.H. Iron trafficking inside the brain. J. Neurochem. 2007, 103, 1730–1740. [CrossRef]
- McRae, R.; Bagchi, P.; Sumalekshmy, S.; Fahrni, C.J. In Situ Imaging of Metals in Cells and Tissues. Chem. Rev. 2009, 109, 4780–4827. [CrossRef]
- Brooks, J.; Everett, J.; Lermyte, F.; Tjhin, V.T.; Banerjee, S.; O'Connor, P.B.; Morris, C.M.; Sadler, P.J.; Telling, N.D.; Collingwood, J.F. Label-Free Nanoimaging of Neuromelanin in the Brain by Soft X-ray Spectromicroscopy. Angew. Chem. Int. Ed. Engl. 2020, 59, 11984–11991. [CrossRef]
- Crichton, R.R.; Dexter, D.T.; Ward, R.J. Brain iron metabolism and its perturbation in neurological diseases; Springer: 2012.
- Everett, J.; Lermyte, F.; Brooks, J.; Tjendana-Tjhin, V.; Plascencia-Villa, G.; Hands-Portman, I.; Donnelly, J.M.; Billimoria, K.; Perry, G.; Zhu, X. Biogenic metallic elements in the human brain? Sci Adv 2021, 7, eabf6707.
- Pezacki, A.T.; Gao, J.; Chang, C.J. Designing small-molecule and macromolecule sensors for imaging redox-active transition metal signaling. Curr. Opin. Chem. Biol. 2024, 83, 102541. [CrossRef]
- Pezacki, A.T.; Gonciarz, R.L.; Okamura, T.; Matier, C.D.; Torrente, L.; Cheng, K.; Miller, S.G.; Ralle, M.; Ward, N.P.; DeNicola, G.M.; et al. A tandem activity-based sensing and labeling strategy reveals antioxidant response element regulation of labile iron pools. Proc. Natl. Acad. Sci.USA 2024, 121, e2401579121. [CrossRef]
- Wu, Y.; Torabi, S.-F.; Lake, R.J.; Hong, S.; Yu, Z.; Wu, P.; Yang, Z.; Nelson, K.; Guo, W.; Pawel, G.T.; et al. Simultaneous Fe 2+ /Fe 3+ imaging shows Fe 3+ over Fe 2+ enrichment in Alzheimer’s disease mouse brain. Sci. Adv. 2023, 9, eade7622. [CrossRef]
- Collingwood, J.F.; Adams, F. Chemical imaging analysis of the brain with X-ray methods. Spectrochim. Acta Part B: At. Spectrosc. 2017, 130, 101–118. [CrossRef]
- Brooks, J.; Everett, J.; Hill, E.; Billimoria, K.; Morris, C.M.; Sadler, P.J.; Telling, N.; Collingwood, J.F. Nanoscale synchrotron x-ray analysis of intranuclear iron in melanised neurons of Parkinson’s substantia nigra. Commun. Biol. 2024, 7, 1–7. [CrossRef]
- Telling, N.D.; Everett, J.; Collingwood, J.F.; Dobson, J.; van der Laan, G.; Gallagher, J.J.; Wang, J.; Hitchcock, A.P. Iron Biochemistry is Correlated with Amyloid Plaque Morphology in an Established Mouse Model of Alzheimer's Disease. Cell Chem. Biol. 2017, 24, 1205–1215.e3. [CrossRef]
- Lermyte, F.; Everett, J.; Brooks, J.; Bellingeri, F.; Billimoria, K.; Sadler, P.J.; O’connor, P.B.; Telling, N.D.; Collingwood, J.F. Emerging Approaches to Investigate the Influence of Transition Metals in the Proteinopathies. Cells 2019, 8, 1231. [CrossRef]
- Hayflick, S.J.; Kurian, M.A.; Hogarth, P. Neurodegeneration with brain iron accumulation. Handb Clin Neurol 2018, 147, 293-305.
- Meyer, E.; Kurian, M.A.; Hayflick, S.J. Neurodegeneration with Brain Iron Accumulation: Genetic Diversity and Pathophysiological Mechanisms. Annu. Rev. Genom. Hum. Genet. 2015, 16, 257–279. [CrossRef]
- Kolarova, H.; Tan, J.; Strom, T.M.; Meitinger, T.; Wagner, M.; Klopstock, T. Lifetime risk of autosomal recessive neurodegeneration with brain iron accumulation (NBIA) disorders calculated from genetic databases. EBioMedicine 2022, 77, 103869. [CrossRef]
- Svetel, M.; Dragasevic, N.; Petrovic, I.; Novakovic, I.; Tomic, A.; Kresojevic, N.; Stankovic, I.; Kostic, V. NBIA syndromes: a step forward from the previous knowledge. Neurol India 2021, 69, 1380-1388.
- Kumar, N.; Rizek, P.; Jog, M. Neuroferritinopathy: pathophysiology, presentation, differential diagnoses and management. Tremor Other Hyperkinet Mov (N Y) 2016, 6, 355.
- Levi, S.; Rovida, E. Neuroferritinopathy: From ferritin structure modification to pathogenetic mechanism. Neurobiol. Dis. 2015, 81, 134–143. [CrossRef]
- Chinnery, P.F.; Crompton, D.E.; Birchall, D.; Jackson, M.J.; Coulthard, A.; Lombès, A.; Quinn, N.; Wills, A.; Fletcher, N.; Mottershead, J.P.; et al. Clinical features and natural history of neuroferritinopathy caused by the FTL1 460InsA mutation. Brain 2006, 130, 110–119. [CrossRef]
- Kurzawa-Akanbi, M.; Keogh, M.; Tsefou, E.; Ramsay, L.; Johnson, M.; Keers, S.; Ochieng, L.W.; McNair, A.; Singh, P.; Khan, A.; et al. Neuropathological and biochemical investigation of Hereditary Ferritinopathy cases with ferritin light chain mutation: Prominent protein aggregation in the absence of major mitochondrial or oxidative stress. Neuropathol. Appl. Neurobiol. 2020, 47, 26–42. [CrossRef]
- Keita, M.; McIntyre, K.; Rodden, L.N.; Schadt, K.; Lynch, D.R. Friedreich Ataxia: Clinical Features and New Developments. Neurodegener. Dis. Manag. 2022, 12, 267–283. [CrossRef]
- Campuzano, V.; Montermini, L.; Moltò, M.D.; Pianese, L.; Cossée, M.; Cavalcanti, F.; Monros, E.; Rodius, F.; Duclos, F.; Monticelli, A.; et al. Friedreich’s Ataxia: Autosomal Recessive Disease Caused by an Intronic GAA Triplet Repeat Expansion. Science 1996, 271, 1423–1427. [CrossRef]
- Monfort, B.; Want, K.; Gervason, S.; D’autréaux, B. Recent Advances in the Elucidation of Frataxin Biochemical Function Open Novel Perspectives for the Treatment of Friedreich’s Ataxia. Front. Neurosci. 2022, 16, 838335. [CrossRef]
- Koutnikova, H.; Campuzano, V.; Foury, F.; Dollé, P.; Cazzalini, O.; Koenig, M. Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat. Genet. 1997, 16, 345–351. [CrossRef]
- Nair, M.; Adinolfi, S.; Pastore, C.; Kelly, G.; Temussi, P.; Pastore, A. Solution Structure of the Bacterial Frataxin Ortholog, CyaY: Mapping the iron binding sites. Structure 2004, 12, 2037–2048. [CrossRef]
- Koeppen, A.H.; Michael, S.C.; Knutson, M.D.; Haile, D.J.; Qian, J.; Levi, S.; Santambrogio, P.; Garrick, M.D.; Lamarche, J.B. The dentate nucleus in Friedreich’s ataxia: the role of iron-responsive proteins. Acta Neuropathol. 2007, 114, 163–173. [CrossRef]
- Todorich, B.; Zhang, X.; Connor, J.R. H-ferritin is the major source of iron for oligodendrocytes. Glia 2011, 59, 927–935. [CrossRef]
- Bartzokis, G. Age-related myelin breakdown: a developmental model of cognitive decline and Alzheimer’s disease. Neurobiol. Aging 2004, 25, 5–18. [CrossRef]
- Nasrabady, S.E.; Rizvi, B.; Goldman, J.E.; Brickman, A.M. White matter changes in Alzheimer’s disease: a focus on myelin and oligodendrocytes. Acta Neuropathol. Commun. 2018, 6, 1–10. [CrossRef]
- Honig, L.S.; Sun, Y.; Irizarry, M.C.; Swanson, C.J.; Dhadda, S.; Charil, A.; E Hart, D.; Noble, J.M.; Huey, E.D.; Teich, A.F. Neuropathological autopsy findings in an individual with Alzheimer’s disease who received long-term treatment with lecanemab (BAN2401). Alzheimer's Dement. 2022, 18. [CrossRef]
- Solopova, E.; Romero-Fernandez, W.; Harmsen, H.; Ventura-Antunes, L.; Wang, E.; Shostak, A.; Maldonado, J.; Donahue, M.J.; Schultz, D.; Coyne, T.M.; et al. Fatal iatrogenic cerebral β-amyloid-related arteritis in a woman treated with lecanemab for Alzheimer’s disease. Nat. Commun. 2023, 14, 8220. [CrossRef]
- Reish, N.J.; Jamshidi, P.; Stamm, B.; Flanagan, M.E.; Sugg, E.; Tang, M.; Donohue, K.L.; McCord, M.; Krumpelman, C.; Mesulam, M.-M.; et al. Multiple Cerebral Hemorrhages in a Patient Receiving Lecanemab and Treated with t-PA for Stroke. New Engl. J. Med. 2023, 388, 478–479. [CrossRef]
- Mascalchi, M.; Salvi, F.; Pirini, M.; D’errico, A.; Ferlini, A.; Lolli, F.; Plasmati, R.; Tessa, C.; Villari, N.; Tassinari, C. Transthyretin amyloidosis and superficial siderosis of the CNS. Neurology 1999, 53, 1498–1503. [CrossRef]
- Ziskin, J.L.; Greicius, M.D.; Zhu, W.; Okumu, A.N.; Adams, C.M.; Plowey, E.D. Neuropathologic analysis of Tyr69His TTR variant meningovascular amyloidosis with dementia. Acta Neuropathol. Commun. 2015, 3, 1–7. [CrossRef]
- Episkopou, V.; Maeda, S.; Nishiguchi, S.; Shimada, K.; A Gaitanaris, G.; E Gottesman, M.; Robertson, E.J. Disruption of the transthyretin gene results in mice with depressed levels of plasma retinol and thyroid hormone.. Proc. Natl. Acad. Sci. 1993, 90, 2375–2379. [CrossRef]
- Monk, J.A.; Sims, N.A.; Dziegielewska, K.M.; Weiss, R.E.; Ramsay, R.G.; Richardson, S.J. Delayed development of specific thyroid hormone-regulated events in transthyretin null mice. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E23–E31. [CrossRef]
- Richardson, S.J. Cell and molecular biology of transthyretin and thyroid hormones. Int Rev Cytol 2007, 258, 137-193. [CrossRef]
- Alshehri, B.; Pagnin, M.; Lee, J.Y.; Petratos, S.; Richardson, S.J. The Role of Transthyretin in Oligodendrocyte Development. Sci. Rep. 2020, 10, 4189. [CrossRef]
- Vancamp, P.; Gothié, J.-D.; Luongo, C.; Sébillot, A.; Le Blay, K.; Butruille, L.; Pagnin, M.; Richardson, S.J.; Demeneix, B.A.; Remaud, S. Gender-specific effects of transthyretin on neural stem cell fate in the subventricular zone of the adult mouse. Sci. Rep. 2019, 9, 19689. [CrossRef]
- Ahmed, Z.; Asi, Y.T.; Lees, A.J.; Revesz, T.; Holton, J.L. Identification and Quantification of Oligodendrocyte Precursor Cells in Multiple System Atrophy, Progressive Supranuclear Palsy and Parkinson's Disease. Brain Pathol. 2012, 23, 263–273. [CrossRef]
- Murthy, M.; Fodder, K.; Miki, Y.; Rambarack, N.; De Pablo Fernandez, E.; Pihlstrøm, L.; Mill, J.; Warner, T.T.; Lashley, T.; Bettencourt, C. DNA methylation patterns in the frontal lobe white matter of multiple system atrophy, Parkinson’s disease, and progressive supranuclear palsy: a cross-comparative investigation. Acta Neuropathol. 2024, 148, 1–20. [CrossRef]
- Miki, Y.; Tsushima, E.; Foti, S.C.; Strand, K.M.; Asi, Y.T.; Yamamoto, A.K.; Bettencourt, C.; Oliveira, M.C.B.; De Pablo-Fernández, E.; Jaunmuktane, Z.; et al. Identification of multiple system atrophy mimicking Parkinson’s disease or progressive supranuclear palsy. Brain 2021, 144, 1138–1151. [CrossRef]
- Ozawa, T.; Paviour, D.; Quinn, N.P.; Josephs, K.A.; Sangha, H.; Kilford, L.; Healy, D.G.; Wood, N.W.; Lees, A.J.; Holton, J.L.; et al. The spectrum of pathological involvement of the striatonigral and olivopontocerebellar systems in multiple system atrophy: clinicopathological correlations. Brain 2004, 127, 2657–2671. [CrossRef]
- Piras, I.S.; Bleul, C.; Schrauwen, I.; Talboom, J.; Llaci, L.; De Both, M.D.; Naymik, M.A.; Halliday, G.; Bettencourt, C.; Holton, J.L.; et al. Transcriptional profiling of multiple system atrophy cerebellar tissue highlights differences between the parkinsonian and cerebellar sub-types of the disease. Acta Neuropathol. Commun. 2020, 8, 1–20. [CrossRef]
- Kaindlstorfer, C.; Jellinger, K.A.; Eschlböck, S.; Stefanova, N.; Weiss, G.; Wenning, G.K. The Relevance of Iron in the Pathogenesis of Multiple System Atrophy: A Viewpoint. J. Alzheimer's Dis. 2018, 61, 1253–1273. [CrossRef]
- Fodder, K.; de Silva, R.; Warner, T.T.; Bettencourt, C. The contribution of DNA methylation to the (dys)function of oligodendroglia in neurodegeneration. Acta Neuropathol. Commun. 2023, 11, 1–15. [CrossRef]
- Bettencourt, C.; Foti, S.C.; Miki, Y.; Botia, J.; Chatterjee, A.; Warner, T.T.; Revesz, T.; Lashley, T.; Balazs, R.; Viré, E.; et al. White matter DNA methylation profiling reveals deregulation of HIP1, LMAN2, MOBP, and other loci in multiple system atrophy. Acta Neuropathol. 2019, 139, 135–156. [CrossRef]
- Bettencourt, C.; Miki, Y.; Piras, I.S.; de Silva, R.; Foti, S.C.; Talboom, J.S.; Revesz, T.; Lashley, T.; Balazs, R.; Viré, E.; et al. MOBP and HIP1 in multiple system atrophy: New α-synuclein partners in glial cytoplasmic inclusions implicated in the disease pathogenesis. Neuropathol. Appl. Neurobiol. 2021, 47, 640–652. [CrossRef]
- Heidari, M.; Gerami, S.H.; Bassett, B.; Graham, R.M.; Chua, A.C.; Aryal, R.; House, M.J.; Collingwood, J.F.; Bettencourt, C.; Houlden, H.; et al. Pathological relationships involving iron and myelin may constitute a shared mechanism linking various rare and common brain diseases. Rare Dis. 2016, 4, e1198458. [CrossRef]
- Bettencourt, C.; Forabosco, P.; Wiethoff, S.; Heidari, M.; Johnstone, D.M.; Botía, J.A.; Collingwood, J.F.; Hardy, J.; Milward, E.A.; Ryten, M.; et al. Gene co-expression networks shed light into diseases of brain iron accumulation. Neurobiol. Dis. 2016, 87, 59–68. [CrossRef]
- Mifsud, G.; Zammit, C.; Muscat, R.; Di Giovanni, G.; Valentino, M. Oligodendrocyte Pathophysiology and Treatment Strategies in Cerebral Ischemia. CNS Neurosci. Ther. 2014, 20, 603–612. [CrossRef]
- Huang, S.; Ren, C.; Luo, Y.; Ding, Y.; Ji, X.; Li, S. New insights into the roles of oligodendrocytes regulation in ischemic stroke recovery. Neurobiol. Dis. 2023, 184, 106200. [CrossRef]
- Pantoni, L.; Garcia, J.H.; Gutierrez, J.A. Cerebral White Matter Is Highly Vulnerable to Ischemia. Stroke 1996, 27, 1641-1646; discussion 1647. [CrossRef]
- Sun, J.; Fang, Y.-Q.; Ren, H.; Chen, T.; Guo, J.-J.; Yan, J.; Song, S.; Zhang, L.-Y.; Liao, H. WIN55,212-2 protects oligodendrocyte precursor cells in stroke penumbra following permanent focal cerebral ischemia in rats. Acta Pharmacol. Sin. 2013, 34, 119–128. [CrossRef]
- Kishida, N.; Maki, T.; Takagi, Y.; Yasuda, K.; Kinoshita, H.; Ayaki, T.; Noro, T.; Kinoshita, Y.; Ono, Y.; Kataoka, H.; et al. Role of Perivascular Oligodendrocyte Precursor Cells in Angiogenesis After Brain Ischemia. J. Am. Hear. Assoc. 2019, 8, e011824. [CrossRef]
- Kyritsis, A.P.; Bondy, M.L.; Levin, V.A. Modulation of Glioma Risk and Progression by Dietary Nutrients and Antiinflammatory Agents. Nutr. Cancer 2011, 63, 174–184. [CrossRef]
- Torti, S.V.; Torti, F.M. Iron and cancer: more ore to be mined. Nat Rev Cancer 2013, 13, 342-355.
- Shenoy, G.; Connor, J.R. A closer look at the role of iron in glioblastoma. Neuro-Oncology 2023, 25, 2136–2149. [CrossRef]
- Troike, K.M.; Wang, S.Z.; Silver, D.J.; Lee, J.; E Mulkearns-Hubert, E.; Hajdari, N.; Ghosh, P.K.; E Kay, K.; Beilis, J.L.; E Mitchell, S.; et al. Homeostatic iron regulatory protein drives glioblastoma growth via tumor cell-intrinsic and sex-specific responses. Neuro-Oncology Adv. 2023, 6, vdad154. [CrossRef]
- Kerins, M.J.; Ooi, A. The Roles of NRF2 in Modulating Cellular Iron Homeostasis. Antioxid. Redox Signal. 2018, 29, 1756–1773. [CrossRef]
- Ma, Q.; Wang, X.; Li, J. LncRNA RP1-86C11.7 exacerbates the glioma progression and oncogenicity by hsa-miR-144-3p/TFRC signaling.. Transl. Oncol. 2021, 14, 101215. [CrossRef]
- Neilsen, B.K.; Sleightholm, R.; McComb, R.; Ramkissoon, S.H.; Ross, J.S.; Corona, R.J.; Miller, V.A.; Cooke, M.; Aizenberg, M.R. Comprehensive genetic alteration profiling in primary and recurrent glioblastoma. J Neurooncol 2019, 142, 111-118.
- Schonberg, D.L.; Lubelski, D.; Miller, T.E.; Rich, J.N. Brain tumor stem cells: Molecular characteristics and their impact on therapy. Mol. Asp. Med. 2014, 39, 82–101. [CrossRef]
- Schonberg, D.L.; Miller, T.E.; Wu, Q.; Flavahan, W.A.; Das, N.K.; Hale, J.S.; Hubert, C.G.; Mack, S.C.; Jarrar, A.M.; Karl, R.T.; et al. Preferential Iron Trafficking Characterizes Glioblastoma Stem-like Cells. Cancer Cell 2015, 28, 441–455. [CrossRef]
- Granholm, A.-C. Long-Term Effects of SARS-CoV-2 in the Brain: Clinical Consequences and Molecular Mechanisms. J. Clin. Med. 2023, 12, 3190. [CrossRef]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [CrossRef]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Author Correction: Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 408–408. [CrossRef]
- Reisbeck, L.; Linder, B.; Tascher, G.; Bozkurt, S.; Weber, K.J.; Herold-Mende, C.; van Wijk, S.J.L.; Marschalek, R.; Schaefer, L.; Münch, C.; et al. The iron chelator and OXPHOS inhibitor VLX600 induces mitophagy and an autophagy-dependent type of cell death in glioblastoma cells. Am. J. Physiol. Physiol. 2023, 325, C1451–C1469. [CrossRef]
- Jakobsson, A.W.; Kundu, S.; Guo, J.; Chowdhury, A.; Zhao, M.; Lindell, E.; Bergsten, P.; Swartling, F.J.; Sjöblom, T.; Zhang, X. Iron Chelator VLX600 Inhibits Mitochondrial Respiration and Promotes Sensitization of Neuroblastoma Cells in Nutrition-Restricted Conditions. Cancers 2022, 14, 3225. [CrossRef]
- Elincx-Benizri, S.; Glik, A.; Merkel, D.; Arad, M.; Freimark, D.; Kozlova, E.; Cabantchik, I.; Hassin-Baer, S. Clinical Experience With Deferiprone Treatment for Friedreich Ataxia. J. Child Neurol. 2016, 31, 1036–1040. [CrossRef]
- Pandolfo, M.; Arpa, J.; Delatycki, M.B.; Le Quan Sang, K.H.; Mariotti, C.; Munnich, A.; Sanz-Gallego, I.; Tai, G.; Tarnopolsky, M.A.; Taroni, F.; et al. Deferiprone in Friedreich ataxia: A 6-Month randomized controlled trial. Ann. Neurol. 2014, 76, 509–521. [CrossRef]
- Ayton, S.; Barton, D.; Brew, B.; Brodtmann, A.; Clarnette, R.; Desmond, P.; Devos, D.; Ellis, K.A.; Fazlollahi, A.; Fradette, C.; et al. Deferiprone in Alzheimer disease: a randomized clinical trial. JAMA Neurol 2025, 82, 11-18.
- Devos, D.; Labreuche, J.; Rascol, O.; Corvol, J.C.; Duhamel, A.; Guyon Delannoy, P.; Poewe, W.; Compta, Y.; Pavese, N.; Ruzicka, E.; et al. Trial of deferiprone in Parkinson’s disease. N Engl J Med 2022, 387, 2045-2055.
- Iankova, V.; Karin, I.; Klopstock, T.; Schneider, S.A. Emerging Disease-Modifying Therapies in Neurodegeneration With Brain Iron Accumulation (NBIA) Disorders. Front. Neurol. 2021, 12. [CrossRef]
- Klopstock, T.; Tricta, F.; Neumayr, L.; Karin, I.; Zorzi, G.; Fradette, C.; Kmieć, T.; Büchner, B.; E Steele, H.; Horvath, R.; et al. Safety and efficacy of deferiprone for pantothenate kinase-associated neurodegeneration: a randomised, double-blind, controlled trial and an open-label extension study. Lancet Neurol. 2019, 18, 631–642. [CrossRef]
- Wilson, R.B.; Lynch, D.R.; Fischbeck, K.H. Normal serum iron and ferritin concentrations in patients with Friedreich's ataxia. Ann. Neurol. 1998, 44, 132–134. [CrossRef]
- Currie, A. Science & Speculation. Erkenntnis 2021, 88, 597–619. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




