Submitted:
21 April 2025
Posted:
27 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Determination of the GLP Content in G. lucidum Spore Powder and Fruiting Body Samples
2.2. Cloning of UGPase PCR Products
2.3. Sequencing and Bioinformatics Analysis
2.4. The PCR Amplification Efficiency and Standard Curve
2.5. Comparison of Relative Quantitative Contents of UGase mRNA
3. Discussion
4. Materials and Methods
4.1. Collection of Samples
4.2. Determination of GLP Contents in Spore Powder and Fruiting Bodies of G. lucidum
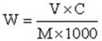 × 100%, where V = 500 mL, C denotes the concentration calculated according to the standard curve, and M represents the mass of the G. lucidum sample; W is the ratio of the GLP content of determined sample to standard contents (%).
× 100%, where V = 500 mL, C denotes the concentration calculated according to the standard curve, and M represents the mass of the G. lucidum sample; W is the ratio of the GLP content of determined sample to standard contents (%).4.3. Bioinformatics Analysis of G. lucidum Genome and Designing a Series of Primers
4.4. Total RNA Extraction and cDNA Synthesis
4.5. PCR Amplification,Cloning of UGPase Gene of G. Lucidum and Blasting
4.6. Establishment of Fluorescence Quantitative PCR Reaction System and Conditions
4.7. Preparation of the Fluorescence Quantitative Standard Curve and Counting of Relative Quantitative Contents of G. lucidum
4.8. Statistics Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China. China Medical Science Press. 2020 Vol, Beijing, p. 1.
- Seweryn, Z., Zia, A., Gamian, A. Health-Promoting of Polysaccharides Extracted from Ganoderma lucidum. Nutrients. 2021, 13 (8):2725.
- Wu, P., Zhang, C., Yin, Y., Zhang, X., Li, Q., Yuan, L., Sun, Y., Zhou, S., Ying, S., Wu, J. Bioactivities and industrial standardization status of Ganoderma lucidum: A comprehensive review. Heliyon. 2024, 10(19):e36987. [CrossRef]
- Zeng, P., Guo, Z., Zeng, X., Hao, C., Zhang, Y., Zhang, M. Molecular mechanisms of bioactive polysaccharides from Ganoderma lucidum (Lingzhi), a review. Int J Biol Macromol. 2020, 150: 765-774.
- Janse van Rensburg, H.C., Van den Ende, W. UDP-Glucose: A Potential Signaling Molecule in Plants?. Front Plant Sci. 2018, 9 (8):2230.
- Chivasa, S., Tomé, D., Slabas, A. UDP-glucose pyrophosphorylase is a novel plant cell death regulator. J Proteome Res. 2013, 12:1743-1753.
- Chambers, J.K., Macdonald, L.E., Sarau, H.M., Ames, R.S., Freeman, K., Foley, J.J.A G protein-coupled receptor for UDP-glucose. J Biol Chem. 2020, 275: 10767-10771.
- Decker, D., Aubert, J., Wilczynska, M., Kleczkowski, L.A. Exploring Redox Modulation of Plant UDP-Glucose Pyrophosphorylase. Int J Mol Sci. 2023,17;24(10):8914. [CrossRef]
- Zhang, J., Shi, X., Cheng, W. Comparison of the Anti-Inflammatory and Antioxidant Activities of Mycelial Polysaccharides from Different Strains of Lingzhi or Reishi Medicinal Mushroom, Ganoderma lucidum (Agaricomycetes). Int J Med Mushrooms.2020, 24(7):77-90.
- Yu, H. Z., Liu, Y. F. Zhou, S. Comparison of the polysaccharides from fruiting bodies, mycelia and spore powder of Ganoderma lingzhi. Mycosystema. 2016, 35 (2): 170-177(in Chinese).
- Guo, C., Guo, D., Fang, L., Sang, T., Wu, J., Guo, C. Ganoderma lucidum polysaccharide modulates gut microbiota and immune cell function to inhibit inflammation and tumorigenesis in colon. Carbohydr Polym. 2021, 67: 118231.
- Sanodiya, B. S., Thakur, G. S., Baghel, R. K. Ganoderma lucidum: a potent pharmacological macrofungus. Curr. Pharm. Biotechnol. 2009, 10: 717-742.
- Fernandes, A., Nair, A., Kulkarni, N., Todewale, N., Jobby, R. Exploring Mushroom Polysaccharides for the Development of Novel Prebiotics: A Review. Int J Med Mushrooms. 2023, 25(2):1-10. [CrossRef]
- Cadar, E., Negreanu-Pirjol, T., Pascale, C., Sirbu, R., Prasacu, I., Negreanu-Pirjol, B.S., Tomescu, C.L., Ionescu, A.M. Natural Bio-Compounds from Ganoderma lucidum and Their Beneficial Biological Actions for Anticancer Application: A Review. Antioxidants (Basel). 2023, 25;12(11):1907. [CrossRef]
- Li, W., Zou, G., Bao, D., Wu, Y. Current Advances in the Functional Genes of Edible and Medicinal Fungi: Research Techniques, Functional Analysis, and Prospects. J Fungi (Basel). 2024, 25;10(5):311. [CrossRef]
- Ahmad, M.F., Ahmad, F.A., Khan, M.I., Alsayegh, A.A., Wahab, S., Alam, M.I., Ahmed, F. Ganoderma lucidum: A potential source to surmount viral infections through β-glucans immunomodulatory and triterpenoids antiviral properties. Int J Biol Macromol. 2021, 30;187:769-779. [CrossRef]
- Zhao, L.Y., Dong, Y. H., Chen, G.T. Extraction, purification, characterization and antitumor activity of polysaccharides from Ganoderma lucidum. Carbohydrate Polymers. 2010, 80(3): 783-789.
- Loyd, A.L., Richter, B.S., Jusino, M.A., Truong, C., Smith, M.E., Blanchette, R.A., Smith, J.A. Identifying the “Mushroom of Immortality”: Assessing the Ganoderma Species Composition in Commercial Reishi Products. Front Microbiol. 2018,16;9:1557. [CrossRef]
- Binns, C.W., Lee, M.K., Lee, A.H. Problems and prospects: public health regulation of dietary supplements. Annu. Rev. Public Health. 2017, 39: 403-420.
- Wang, Q., Xu, M., Zhao, L., Wang, F., Li, Y., Shi, G., Ding, Z. Transcriptome dynamics and metabolite analysis revealed the candidate genes and regulatory mechanism of ganoderic acid biosynthesis during liquid superficial-static culture of Ganoderma lucidum. Microb Biotechnol. 2021, 14(2):600-613. [CrossRef]
- Cortina-Escribano, M., Veteli, P., Wingfield, M.J., Wingfield, B.D., Coetzee, M.P.A., Vanhanen, H., Linnakoski, R. Phylogenetic analysis and morphological characteristics of laccate Ganoderma specimens in Finland. Mycologia. 2024 ,116(6):1046-1062. [CrossRef]
- Fryssouli, V., Zervakis, G.I., Polemis, E., Typas, M.A. A global meta-analysis of ITS rDNA sequences from material belonging to the genus Ganoderma (Basidiomycota, Polyporales) including new data from selected taxa. MycoKeys. 2020, 75:71-143. [CrossRef]
- Pavlik, M., Zhou, S., Zhang, J., Tang, Q., Feng, N., Kurjak, D., Pavlík, M. Jr., Kunca, A. Comparative Analysis of Triterpene Composition between Ganoderma lingzhi from China and G. lucidum from Slovakia under Different Growing Conditions. Int J Med Mushrooms. 2020, 22(8):793-802. [CrossRef]
- Cai, M., Tan, Z., Wu, X., Liang, X., Liu, Y., Xie, Y., Li, X., Xiao, C., Gao, X., Chen, S., Hu, H., Wu, Q. Comparative transcriptome analysis of genes and metabolic pathways involved in sporulation in Ganoderma lingzhi. G3 (Bethesda). 2022, 12(3):jkab448. [CrossRef]
- Liu, D., Sun, X., Diao, W., Qi, X., Bai, Y., Yu, X., Li, L., Fang, H., Chen, Z., Liu, Q., Liang, C. Comparative transcriptome analysis revealed candidate genes involved in fruiting body development and sporulation in Ganoderma lucidum. Arch Microbiol. 2022, 204(8):514. [CrossRef]
- Jia, T., Ge, Q., Zhang, S., Zhang, Z., Liu, A., Fan, S. UDP-Glucose Dehydrogenases: identification, expression, and function analyses in upland cotton (Gossypium Hirsutum). Front Genet. 2020, 11:597890.
- Khadbaatar, S., Bao, H., Gao, X., Huo, H. Study on Differences of Metabolites among Different Ganoderma Species with Comprehensive Metabolomics. J Fungi (Basel). 2024 Jul 27;10(8):524. [CrossRef]
- Brandt, W., Schulze, E., Liberman-Aloni, R., Bartelt, R., Pienkny, S., Carmeli-Weissberg, M., Frydman, A., Eyal, Y. Structural modeling of two plant UDP-dependent sugar-sugar glycosyltransferases reveals a conserved glutamic acid residue that is a hallmark for sugar acceptor recognition. J Struct Biol. 2021,213(3):107777. [CrossRef]
- Duan, S., Ai, J.X., Sun, L., Gao, L., Li, M., Chen, K., Li, D. Development and validation of a rapid kit for authenticity of murine meat in meat products with a species-specific PCR assay. Food Additives & Contaminants: Part A. 2020, 37(4): 552-560.
- Blundell, R., Camilleri, E., Baral, B., Karpiński, T.M., Neza, E., Atrooz, O.M. The Phytochemistry of Ganoderma Species and their Medicinal Potentials. Am J Chin Med. 2023, 51(4):859-882. [CrossRef]
- Rašeta, M., Kebert, M., Mišković, J., Kostić, S., Kaišarević, S., Stilinović N., Vukmirović, S., Karaman, M. Ganoderma pfeifferi Bres. and Ganoderma resinaceum Boud. as Potential Therapeutic Agents: A Comparative Study on Antiproliferative and Lipid-Lowering Properties. J Fungi (Basel). 2024,10(7):501. [CrossRef]
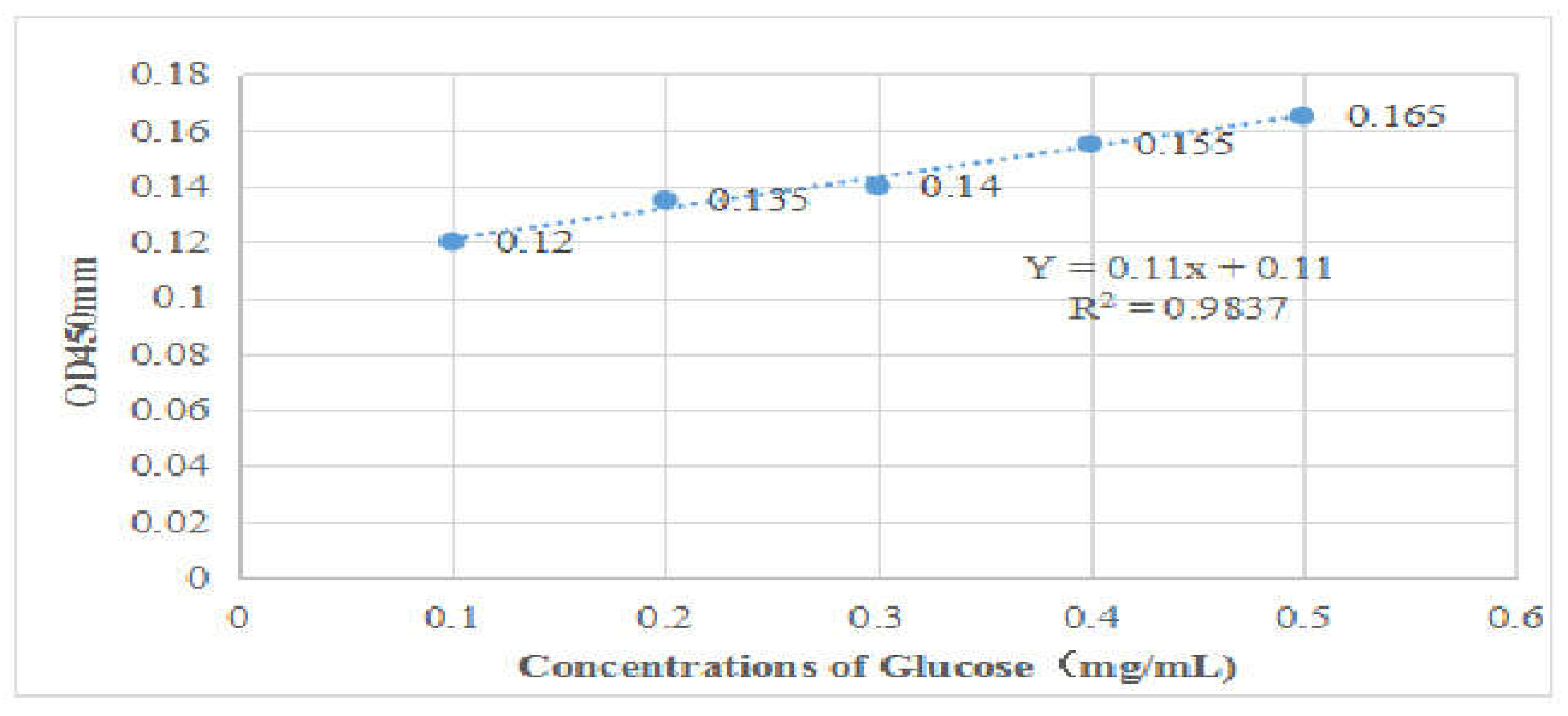
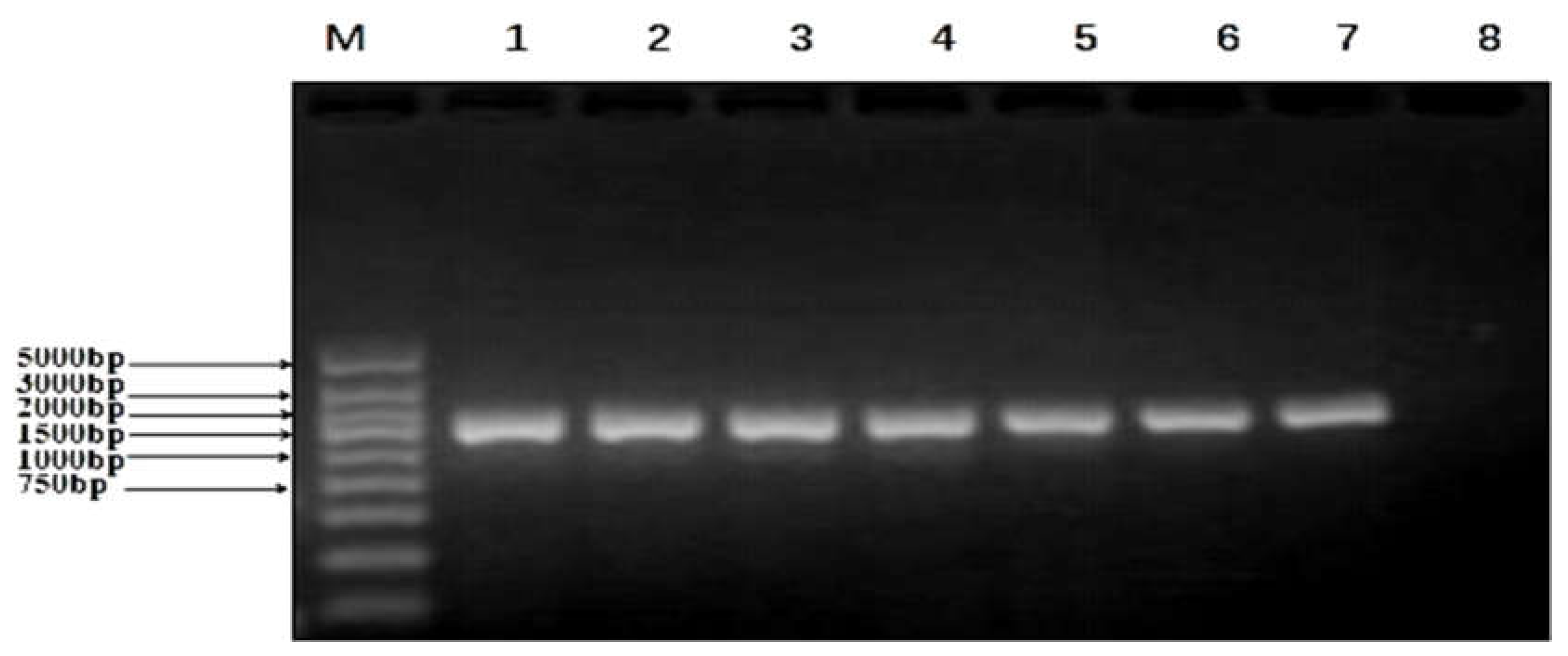
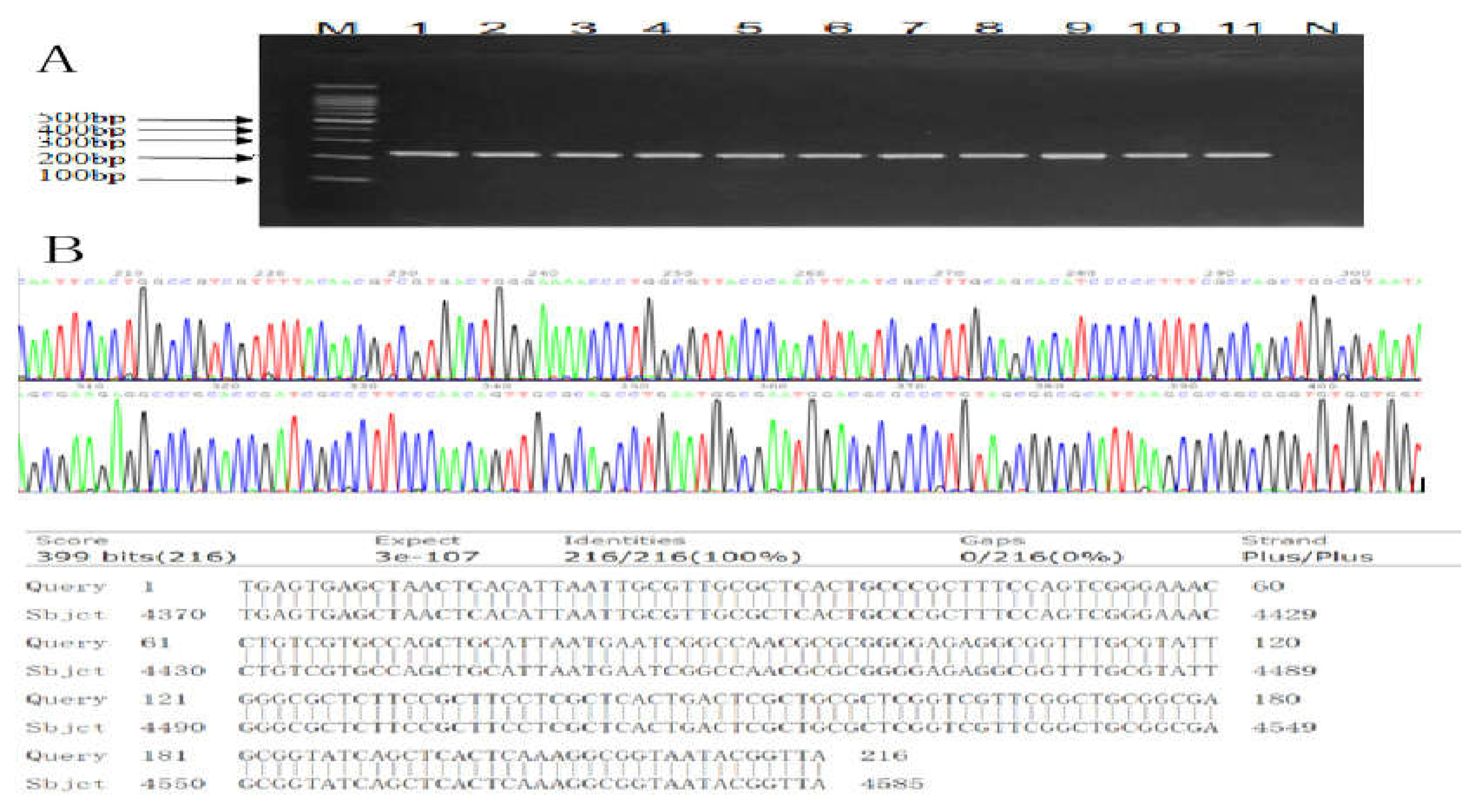
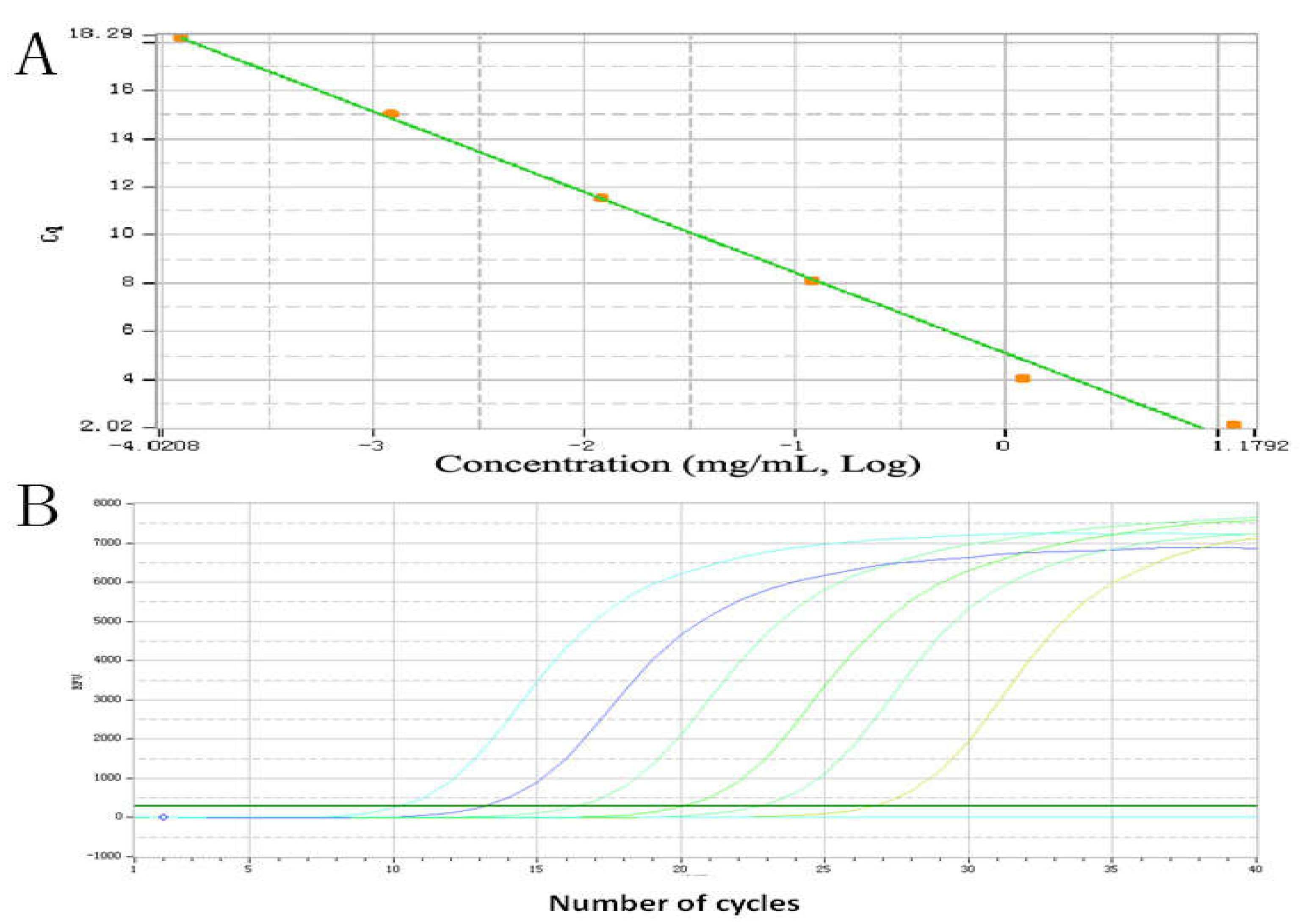
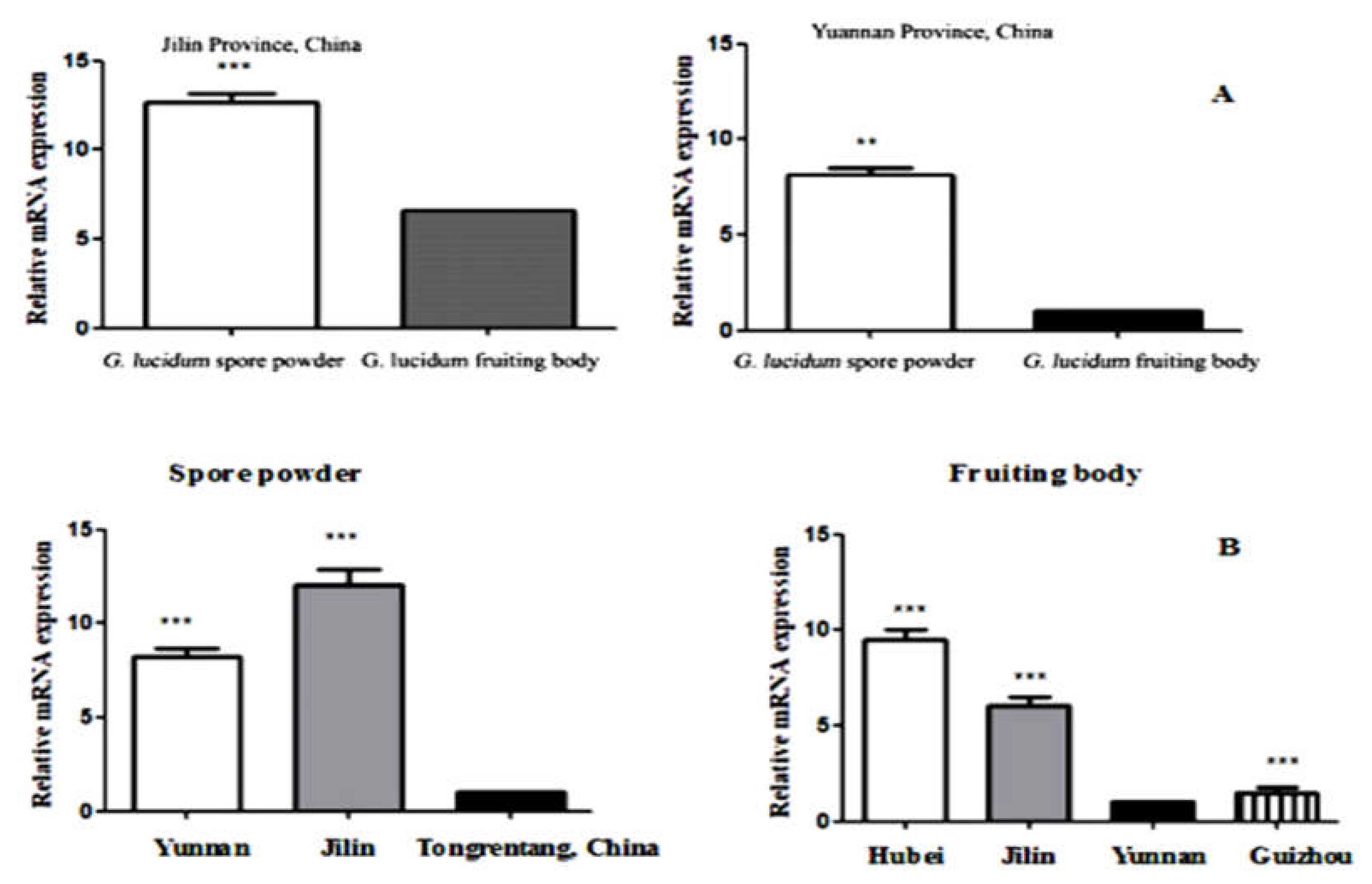
| Source | Serial No. | OD450 nm | Concentrations (mg/ml) | Relative GLP Contents (%) |
|---|---|---|---|---|
| GZsp | GZsp-1 | 1.589 | 13.445 | 3.361± 0.0046 |
| GZsp-2 | 1.590 | 13.454 | 3.364± 0.0045 | |
| GZsp-9 | 1.588 | 13.436 | 3.359± 0.0044 | |
| HBsp | HBsp-1 | 1.165 | 9.591 | 2.398± 0.0045 |
| HBsp-2 | 1.162 | 9.564 | 2.391± 0.0046 | |
| HBsp-6 | 1.168 | 9.618 | 2.405± 0.0043 | |
| JLsp | JLsp-2 | 1.589 | 13.445 | 3.361± 0.0042 |
| JLsp-5 | 1.590 | 13.454 | 3.364± 0.0041 | |
| JLsp-6 | 1.588 | 13.436 | 3.359± 0.0045 | |
| TRsp | TRsp-3 | 1.165 | 9.591 | 2.398± 0.0045 |
| TRsp-4 | 1.162 | 9.564 | 2.391± 0.0044 | |
| TRsp-5 | 1.168 | 9.618 | 2.405± 0.0047 | |
| YNsp | YNsp-1 | 1.380 | 11.545 | 2.886± 0.0044 |
| YNsp-2 | 1.378 | 11.527 | 2.882± 0.0043 | |
| YNsp-5 | 1.381 | 11.554 | 2.889± 0.0044 |
| Source | Serial No. | OD450 nm | Concentrations (mg/ml) | Relative GLP Contents (%) |
|---|---|---|---|---|
| GZfb | GZfb-1 | 0.693 | 5.300 | 1.325± 0.0422 |
| GZfb-2 | 0.692 | 5.291 | 1.323± 0.04012 | |
| GZ-9 | 0.694 | 5.309 | 1.327± 0.0431 | |
| HBfb | HBfb-3 | 1.186 | 9.782 | 2.446 ± 0.0022 |
| HBfb-5 | 1.187 | 9.791 | 2.448 ± 0.0031 | |
| HBfb-6 | 1.186 | 9.782 | 2.446 ± 0.0031 | |
| JLfb | JLfb-5 | 0.904 | 7.217 | 1.804 ± 0.0042 |
| JLfb-7 | 0.905 | 7.227 | 1.807 ± 0.0041 | |
| JLfb-8 | 0.905 | 7.227 | 1.807 ± 0.0043 | |
| YNfb | YNfb-4 | 0.540 | 3.909 | 0.977± 0.0035 |
| YNfb-7 | 0.542 | 3.927 | 0.982± 0.0036 | |
| YNfb-8 | 0.539 | 3.900 | 0.975± 0.0037 |
| Serial No. | Source | Weight (g) |
|---|---|---|
| YNsp1-6 | Yuan Nan Province, China | 250 |
| YNfb1-9 | Yuan Nan Province, China | 500 |
| JLsp1-7 | Jilin Province, China | 250 |
| JLfb1-8 | Jilin Province, China | 500 |
| HBsp1-6 | Hubei Province, China | 250 |
| HBfb1-9 | Hubei Province, China | 500 |
| GZsp1-6 | Guizhou Province, China | 250 |
| GZfb1-9 | Guizhou Province, China | 500 |
| TRspc1-5 | Beijing, China | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





