Submitted:
11 July 2025
Posted:
15 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
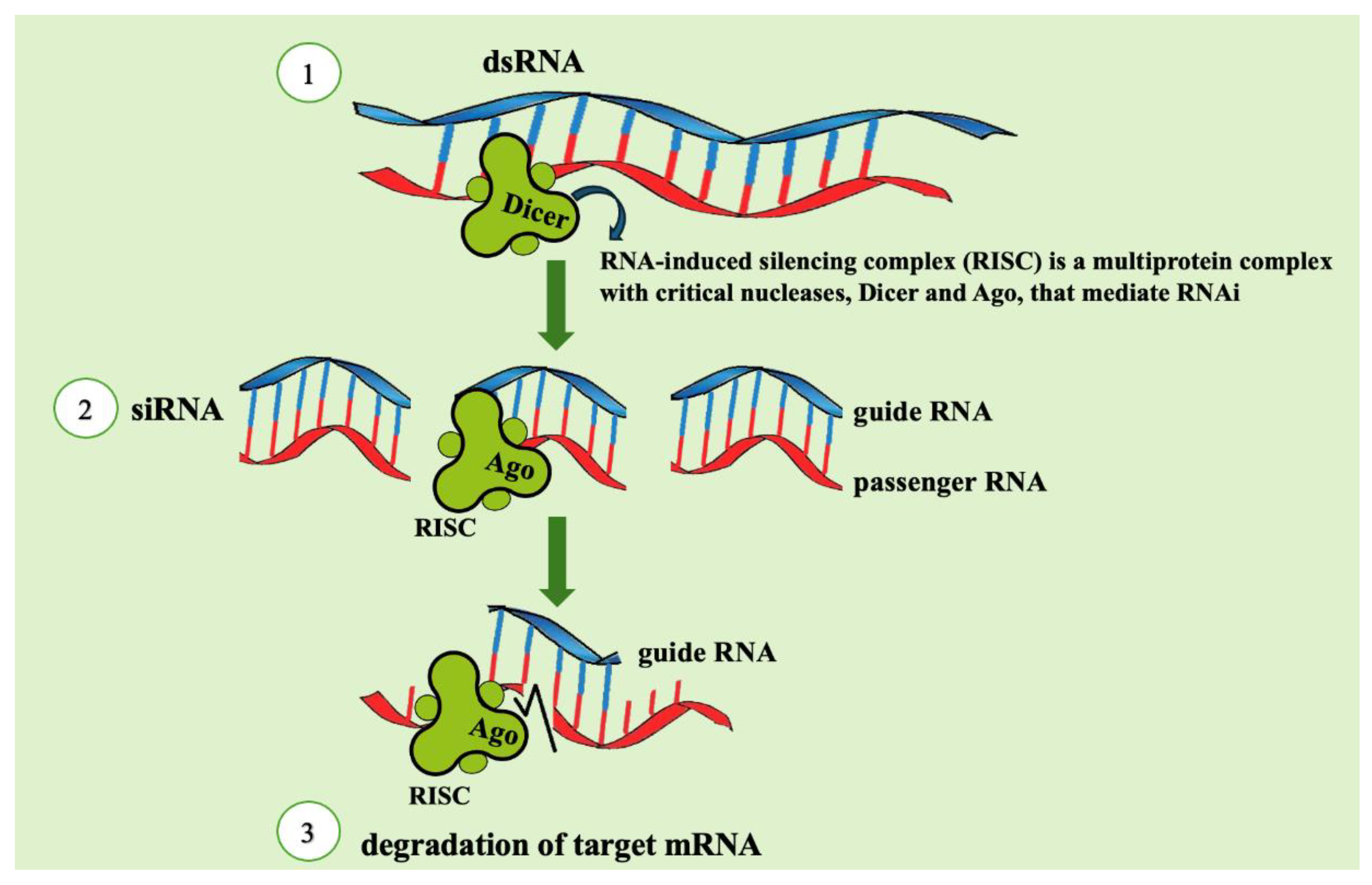
2. RNAi
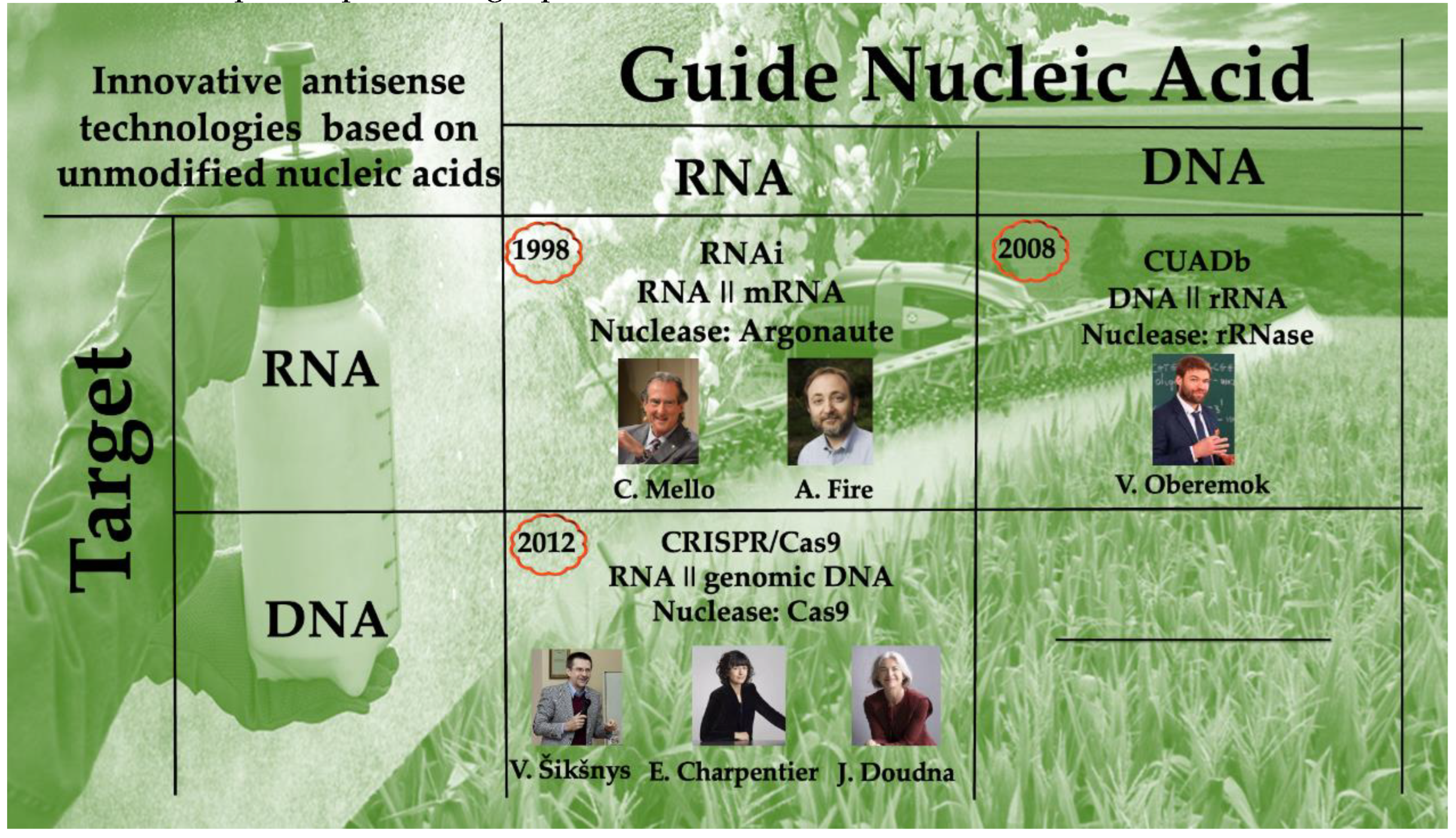
2.1. History of discovery
2.2. How RNAi works in insect pests
2.3. Perspectives and limitations of RNAi for insect pest control
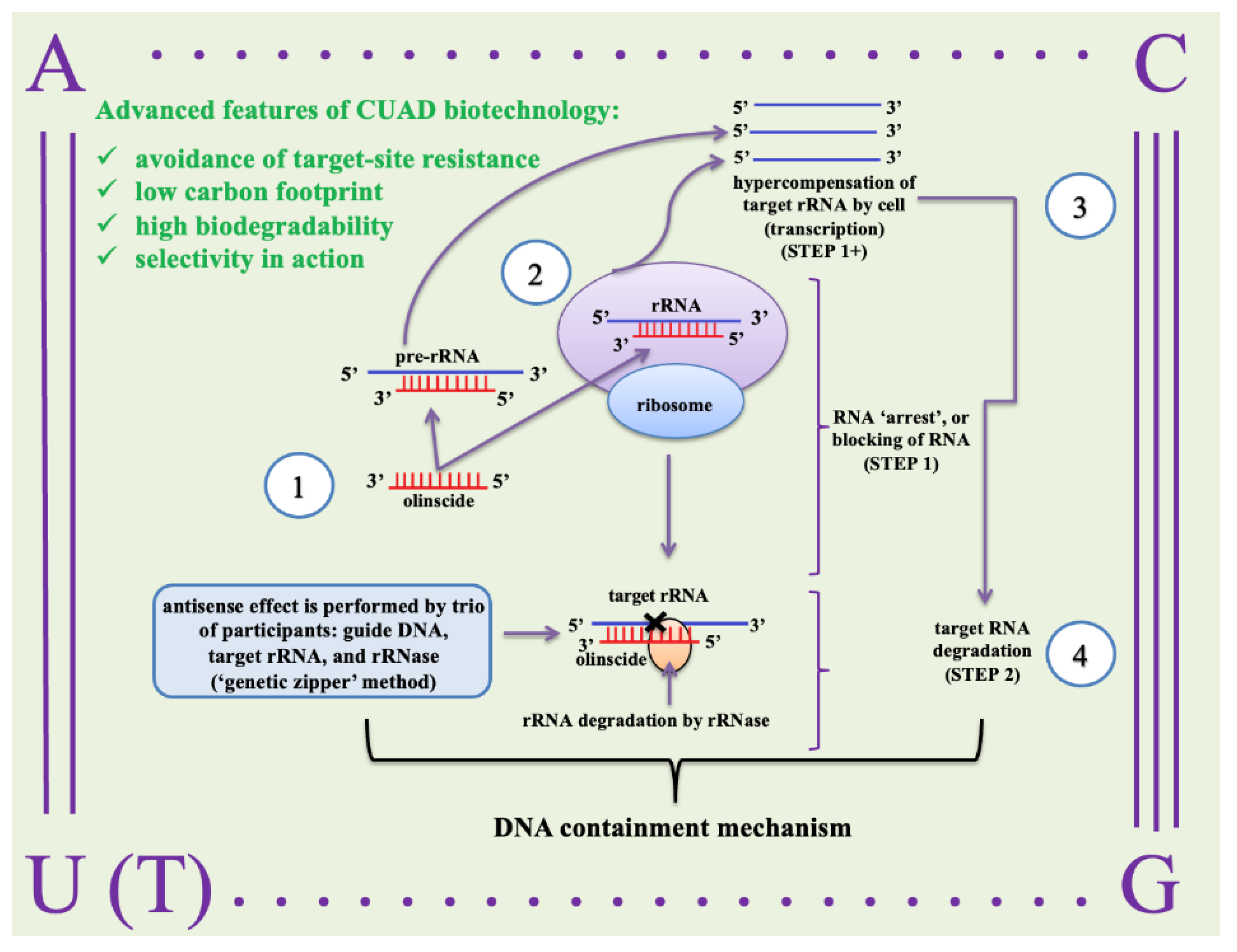
3. CUADb
3.1. History of discovery
3.2. How it works on insect pests
3.2. Perspectives and limitations of CUADb for insect pest control
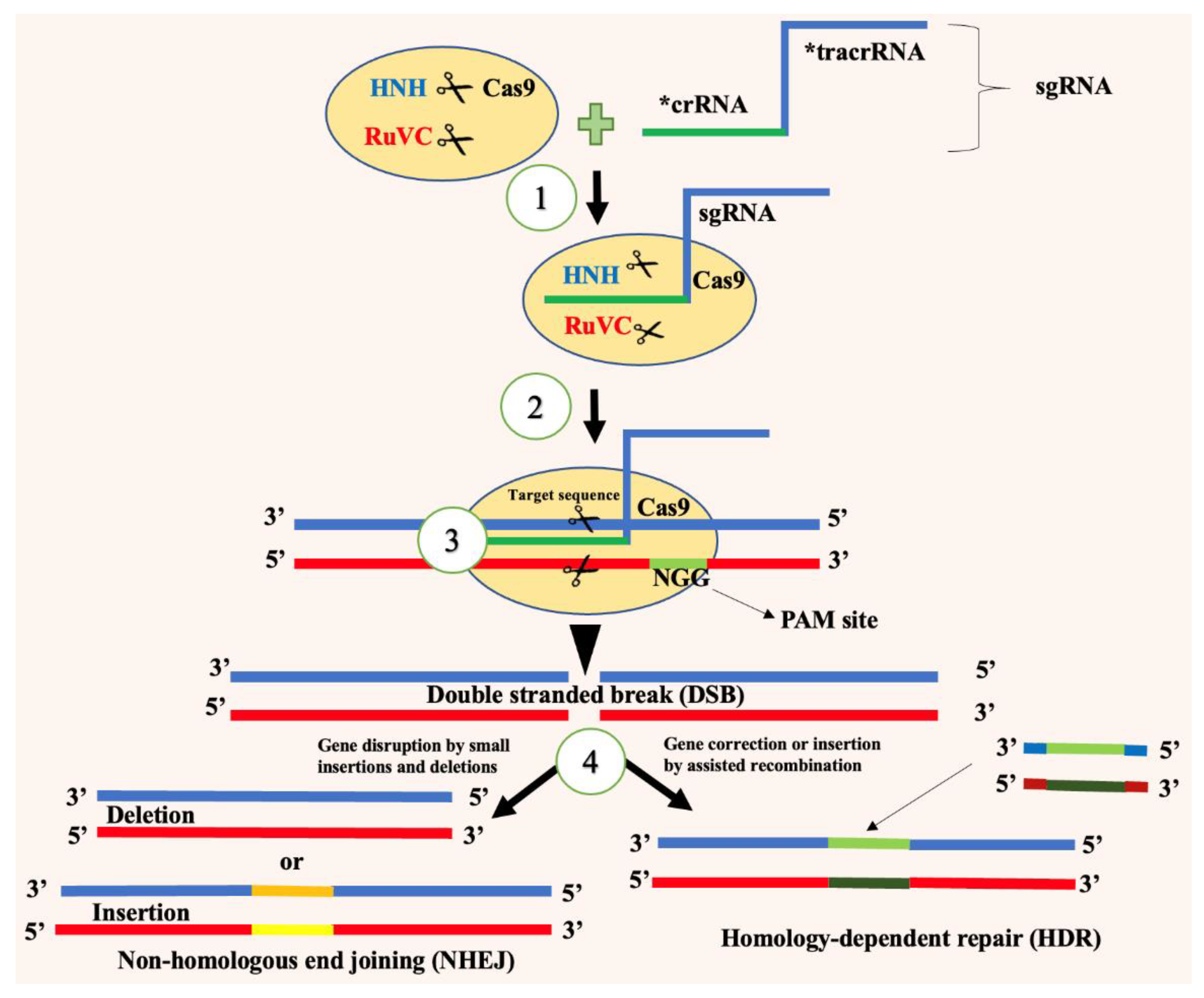
4. CRISPR/Cas
4.1. History of discovery
4.2. How it works on insect pests
4.3. Perspectives and limitations of CRISPR/Cas for insect pest control
5. Conclusion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Minchin, S.; Lodge, J. Understanding biochemistry: structure and function of nucleic acids. Essays Biochem. 2019, 63, 433–456. [Google Scholar] [CrossRef] [PubMed]
- Westhof, E.; Yusupov, M.; Yusupova, G. Recognition of Watson-Crick base pairs: constraints and limits due to geometric selection and tautomerism. F1000Prime Rep. 2014, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V. Method of Elimination of Phyllophagous Insects from Order Lepidoptera. Ukraine Patent UA. 2008, 445, 27. [Google Scholar]
- Gal'chinsky, N.V.; Yatskova, E.V.; Novikov, I.A.; Sharmagiy, A.K.; Plugatar, Y.V.; Oberemok, V.V. Mixed insect pest populations of Diaspididae species under control of oligonucleotide insecticides: 3′-end nucleotide matters. Pesticide Biochem. Physiol. 2024, 200, 105838. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V.; Laikova, K.V.; Gal’chinsky, N.V. Contact unmodified antisense DNA (CUAD) biotechnology: list of pest species successfully targeted by oligonucleotide insecticides. Front. Agron. 2024, 6, 1415314. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E.A. Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U S A. 2012, 109, E2579–E2586. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Rana, T.M. Molecular mechanisms of RNA-triggered gene silencing machineries. Acc Chem Res. 2012, 45, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V.; Laikova, K.V.; Gal'chinsky, N.V.; Useinov, R.Z.; Novikov, I.A.; Temirova, Z.Z.; Shumskykh, M.N.; Krasnodubets, A.M.; Repetskaya, A.I.; Dyadichev, V.V.; et al. DNA insecticide developed from the Lymantria dispar 5.8S ribosomal RNA gene provides a novel biotechnology for plant protection. Sci Rep. 2019, 9, 6197. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V.; Laikova, K.V.; Andreeva, O.A.; Gal'chinsky, N.V. Oligonucleotide insecticides and RNA-based insecticides: 16 years of experience in contact using of the next generation pest control agents. J. Plant. Dis. Prot. 2024, 131, 1837–1852. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Puzanova, Y.V.; Gal’chinsky, N.V. The 'genetic zipper' method offers a cost-effective solution for aphid control. Front. Insect Sci. 2024, 4, 1467221. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V.; Gal’chinsky, N.V.; Novikov, I.A.; Sharmagiy, A.K.; Yatskova, E.V.; Laikova, E.V.; Plugatar, Yu. rRNA-specific antisense DNA and dsDNA trigger rRNA biogenesis and cause potent insecticidal effect on insect pest Coccus hesperidum L. biorXiv 2024. biorXiv 2024. [Google Scholar]
- Wiles, M.V.; Qin, W.; Cheng, A.W.; Wang, H. CRISPR-Cas9-mediated genome editing and guide RNA design. Mamm Genome. [CrossRef]
- Li, T.; Yang, Y.; Qi, H.; Cui, W.; Zhang, L.; Fu, X.; He, X.; Liu, M.; Li, P-f. ; Yu T. CRISPR/Cas9 therapeutics: progress and prospects. Sig Transduct Target Ther. 2023, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zuo, Z.; Shao, W; Jin, Y. ; Meng, Y. The expanding roles of Argonautes: RNA interference, splicing and beyond. Brief Funct Genomics. 2018, 17, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Araújo, M.F.; Castanheira, E.M.S.; Sousa, S.F. The Buzz on Insecticides: A Review of Uses, Molecular Structures, Targets, Adverse Effects, and Alternatives. Molecules 2023, 28, 3641. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, J.A.; Fan, R.; Naz, H.; Bamisile, B.S.; Hafeez, M.; Ghani, M.I.; Wei, Y.; Xu, Y.; Chen, X. Insights into insecticide-resistance mechanisms in invasive species: Challenges and control strategies. Front. Physiol. 2023, 13, 1112278. [Google Scholar] [CrossRef] [PubMed]
- Gul, H.; Gadratagi, B.G.; Güncan, A.; Tyagi, S.; Ullah, F.; Desneux, N.; Liu, X. Fitness costs of resistance to insecticides in insects. Front. Physiol. 14, 1238111. https. [CrossRef]
- Hawkins, N.J.; Bass, C.; Dixon, A.; Neve, P. The evolutionary origins of pesticide resistance. Biol. Rev. Camb. Philos. Soc. 2019, 94, 135–155. [Google Scholar] [CrossRef] [PubMed]
- Belikova, A.; Zarytova, V.; Grineva, N. Synthesis of ribonucleosides and diribonucleoside phosphates containing 2-chloroethylamine and nitrogen mustard residues. Tetrahedron Lett. 1967, 37, 3557–3562. [Google Scholar] [CrossRef] [PubMed]
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA. 1978, 75, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S. The Heroes of CRISPR. Cell 2016, 164(1-2), 18-28. [CrossRef]
- Shmakova, A.A.; Shmakova, O.P.; Karpukhina, A.A.; Vassetzky, Y.S. CRISPR/Cas: History and Perspectives. Russ. J. Dev. Bio. 2022, 53, 272–282. [Google Scholar] [CrossRef]
- Tomoyasu, Y.; Miller, S.C.; Tomita, S.; Schoppmeier, M.; Grossmann, D.; Bucher, G. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008, 9, R10. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, P. Key mechanistic principles and considerations concerning RNA interference. Front. Plant Sci. 2020, 11, 1237. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shu, R.; Liu, J. The development and improvement of ribonucleic acid therapy strategies. Mol. Ther. Nucleic Acids. 2021, 26, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.Y.; Palli, S.R. Mechanisms, applications, and challenges of insect RNA interference. Annual review of entomology 2020, 65, 293–311. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.M.; Silver, K.; Zhang, J.; Park, Y.; Zhu, K.Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest. Manag. Sci. 2019, 75, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.M.; Song, H.; Yu, Z.; Biondi, M.; Bai, J.; Shi, X.; Weerasekara, S.M.; Hua, D.H.; Silver, K.; Zhang, J.; et al. Comparison of strategies for enhancing RNA interference efficiency in Ostrinia nubilalis. Pest. Manag. Sci. 2021, 77, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T; Broeck, J. V. RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 2019, 9, 434563. [Google Scholar] [CrossRef] [PubMed]
- Girard, C.; Budin, K.; Boisnard, S.; Zhang, L.; Debuchy, R.; Zickler, D.; Espagne, E. RNAi-Related Dicer and Argonaute Proteins Play Critical Roles for Meiocyte Formation, Chromosome-Axes Lengths and Crossover Patterning in the Fungus Sordaria macrospora. Front. Cell Dev. Biol. 2021, 9, 684108. [Google Scholar] [CrossRef] [PubMed]
- Silver, K.; Cooper, A.M.; Zhu, K.Y. Strategies for enhancing the efficiency of RNA interference in insects. Pest. Manag. Sci. 2021, 77, 2645–2658. [Google Scholar] [CrossRef] [PubMed]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.L.; Barthel, A.; et al. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Lucena-Leandro, V.S.; Abreu, E.F.A.; Vidal, L.A.; Torres, C.R.; Junqueira, C.I.C.V.F.; Dantas, J.; Albuquerque, É. V.S. Current Scenario of Exogenously Induced RNAi for Lepidopteran Agricultural Pest Control: From dsRNA Design to Topical Application. Int. J. Mol. Sci. 2022, 23, 15836. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Tardajos, M.G.; Martinez Reyna, Z.L.; Dash, M.; Dubruel, P.; Smagghe, G. Increased RNAi Efficacy in via the Formulation of dsRNA With Guanylated Polymers. Front. Physiol. 2018, 9, 316. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.G.; Robinson, K.E.; Asgari, S.; Mitter, N. Current scenario of RNAi-based hemipteran control. Pest. Manag. Sci. 2021, 77, 2188–2196. [Google Scholar] [CrossRef] [PubMed]
- Kaplanoglu, E.; Kolotilin, I.; Menassa, R.; Donly, C. Plastid Transformation of Micro-Tom Tomato with a Hemipteran Double-Stranded RNA Results in RNA Interference in Multiple Insect Species. Int. J. Mol. Sci. 2022, 23, 3918. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Peng, H.; Yao, Q.; Chen, H.; Xie, Q.; Tang, B.; Zhang, W. Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PloS One 2009, 4, e6225. [Google Scholar] [CrossRef] [PubMed]
- Zha, W.; Peng, X.; Chen, R.; Du, B.; Zhu, L.; He, G. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PloS One 2011, 6, e20504. [Google Scholar] [CrossRef] [PubMed]
- Prentice, K.; Christiaens, O.; Pertry, I.; Bailey, A.; Niblett, C.; Ghislain, M.; Gheysen, G.; Smagghe, G. RNAi-based gene silencing through dsRNA injection or ingestion against the African sweet potato weevil Cylas puncticollis (Coleoptera: Brentidae). Pest. Manag. Sci. 2017, 73, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Camargo, R.A.; Barbosa, G.O.; Possignolo, I.P.; Peres, L.E.; Lam, E.; Lima, J.E.; Figueira, A.; Marques-Souza, H. RNA interference as a gene silencing tool to control Tuta absoluta in tomato (Solanum lycopersicum). PeerJ. 2016, 4, e2673. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Liu, J.; Chen, Y.; Zhang, J.; Zhong, G. RNAi-mediated knockdown of α-Spectrin depresses reproductive performance in female Bactrocera dorsalis. Pesticide Biochem. Physiol. 2023, 196, 105611. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.K.; Singh, S.; Upadhyay, S.K.; Tiwari, V.; Saxena, G.; Verma, P.C. RNAi-based gene silencing in Phenacoccus solenopsis and its validation by in planta expression of a double-stranded RNA. Pest. Manag. Sci. 2021, 77, 1796–1805. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Z.; Jiang, Y.X.; Li, M.W.; Li, J.W.; Zha, B.H.; Yang, G. Double-stranded RNA-degrading enzymes reduce the efficiency of RNA interference in Plutella xylostella. Insects 2021, 12, 712. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, X.; Li, J.; Hull, J.J.; Chen, L.; Liang, G. A spray-induced gene silencing strategy for Spodoptera frugiperda oviposition inhibition using nanomaterial-encapsulated dsEcR. International Journal of Biological Macromolecules 2024, 281, 136503. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.F.; Liu, T.T.; Zhao, Y.Q.; Luo, J.; Feng, H.Y.; Zhou Gong, L.L.; Zhang, M.Q.; He, Y.Y.; Hull, J.J.; Dewer, Y.; et al. RNA interference-screening of potentially lethal gene targets in the white-backed planthopper Sogatella furcifera via a spray-induced and nanocarrier-delivered gene silencing system. Journal of Agricultural and Food Chemistry 2024, 72, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, H.; Wang, Y.; Wei, L.; Lyu, J.; Shan, Z.; Zhang, X.; Fan, D. RNA Interference Reveals the Impacts of CYP6CY7 on Imidacloprid Resistance in Aphis glycines. Insects 2024, 15, 188. [Google Scholar] [CrossRef] [PubMed]
- Saberi, E.; Mondal, M.; Paredes-Montero, J.R.; Nawaz, K.; Brown, J.K.; Qureshi, J.A. Optimal dsRNA Concentration for RNA Interference in Asian Citrus Psyllid. Insects 2024, 15, 58. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.X.; Bao, H.Q.; Yan, Z.C.; Wang, J.; Wang, S.; Li, Y.X. Knockdown of vitellogenin receptor based on minute insect RNA interference methods affects the initial mature egg load in the pest natural enemy Trichogramma dendrolimi. Insect Science 2024. [CrossRef] [PubMed]
- Liu, J.; Yang, Y.; Yang, Q.; Lin, X.; Liu, Y.; Li, Z.; Swevers, L. Successful oral RNA interference efficiency in the silkworm Bombyx mori through nanoparticle-shielded dsRNA delivery. Journal of Insect Physiology 2025, 104749. [Google Scholar] [CrossRef] [PubMed]
- Darweesh, A.F.; Fahmy, I.; Ali, M.; Elwahy, A. RNA interference of cysteine protease genes for the management of whitefly (Bemisia tabaci) by oral route. Egyptian Journal of Botany 2025, 65, 43–56. [Google Scholar] [CrossRef]
- Shimomura, K.; Sakita, K.; Terajima, T.; Tomizawa, M. Gene Silencing of Olfactory Receptor Coreceptor by Systemic RNA Interference in Callosobruchus maculatus. Journal of Chemical Ecology 2025, 51, 5. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.P.; Yang, X.B.; Yang, H.; Zhou, C.; Long, G.Y.; Jin, D.C. Knockdown of the β-N-acetylhexosaminidase genes by RNA interference inhibited the molting and increased the mortality of the white-backed planthopper, Sogatella furcifera. Pesticide Biochem. Physiol. 2025, 207, 106216. [Google Scholar]
- Bera, P.; Suby, S.B.; Dixit, S.; Vijayan, V.; Kumar, N.; Sekhar, J.C.; Vadassery, J. Identification of novel target genes for RNAi mediated management of the pest, Fall Armyworm (Spodoptera frugiperda, JE Smith). Crop Protection 2025, 187, 106972. [Google Scholar] [CrossRef]
- Schellens, S.; Lenaerts, C.; Pérez Baca, M.D.R.; Cools, D.; Peeters, P.; Marchal, E.; Vanden Broeck, J. Knockdown of the Halloween genes Spook, Shadow and Shade influences oocyte development, egg shape, oviposition and hatching in the desert locust. Int. J. Mol. Sci. 2022, 23, 9232. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.; Young Noh, M.; Dittmer, N.T.; Muthukrishnan, S.; Kramer, K.J.; Kanost, M.R.; Arakane, Y. Cuticular protein with a low complexity sequence becomes cross-linked during insect cuticle sclerotization and is required for the adult molt. Sci. Rep. 2015, 5, 10484. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Dai, L.; Gao, H.; Sun, Y.; Liu, B.; Chen, H. Identification, expression patterns and RNA interference of aquaporins in Dendroctonus armandi (Coleoptera: Scolytinae) larvae during overwintering. Front. Physiol. 2019, 10, 967. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.M.; Ashfaq, M.; Khan, A.A.; Naseem, M.T.; Mansoor, S. Evaluation of potential RNA-interference-target genes to control cotton mealybug, Phenacoccus solenopsis (Hemiptera: Pseudococcuidae). Insect science 2018, 25, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Araujo, R.N.; Santos, A.; Pinto, F.S.; Gontijo, N.F.; Lehane, M.J.; Pereira, M.H. RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect biochemistry and molecular biology 2006, 36, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, D.; Yao, Q.; Zhang, J.; Dong, X.; Tian, H.; Chen, J.; Zhang, W. Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Molecular Biology 2010, 19, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.; Xiong, Y.; Xia, W.K.; Wei, D.D.; Wei, D.; Wang, J.J. Identification, characterization and functional analysis of a chitin synthase gene in the brown citrus aphid, Toxoptera citricida (Hemiptera, Aphididae). Insect Molecular Biology 2016, 25, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Shen, J. Target genes for RNAi in pest control: A comprehensive overview. Entomologia Generalis 2024, 44. [Google Scholar] [CrossRef]
- Pallis, S.; Alyokhin, A.; Manley, B.; Rodrigues, T.; Barnes, E.; Narva, K. Effects of Low Doses of a Novel dsRNA-based Biopesticide (Calantha) on the Colorado Potato Beetle. J. Econ. Entomol. 2023, 116, 456–461. [Google Scholar] [CrossRef] [PubMed]
- GreenLight Biosciences. Available online: https://www.greenlightbiosciences.com/international-crop-network-validates-ledprona-as-a-new-mode-of-action-group/ (accessed on 20 January 2025).
- Bachman, P.; Fischer, J.; Song, Z.; Urbanczyk-Wochniak, E.; Watson, G. Environmental fate and dissipation of applied dsRNA in soil, aquatic systems, and plants. Front. Plant Sci. 2020, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Sparks, T.C.; Nauen, R. IRAC: mode of action classification and insecticide resistance management. Pesticide Biochem. Physiol. 2015, 121, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Sparks, T.C.; Storer, N.; Porter, A.; Slater, R.; Nauen, R. Insecticide resistance management and industry: the origins and evolution of the insecticide resistance action committee (IRAC) and the mode of action classification scheme. Pest. Manag. Sci. 2021, 77, 2609–2619. [Google Scholar] [CrossRef] [PubMed]
- Khajuria, C.; Ivashuta, S.; Wiggins, E.; Flagel, L.; Moar, W.; Pleau, M.; Miller, K.; Zhang, Y.; Ramaseshadri, P.; Jiang, C. Development and characterization of the first dsRNA-resistant insect population from western corn rootworm, Diabrotica virgifera virgifera LeConte. PLoS One 2018, 13, e0197059. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Zhang, M.; Zhang, J. Characterization and potential mechanism of resistance to double-stranded RNA in willow leaf beetle, Plagiodera versicolora. Journal of Pest Science 2024, 97, 2217–2226. [Google Scholar] [CrossRef]
- Mishra, S.; Dee, J.; Moar, W.; Dufner-Beattie, J.; Baum, J.; Dias, N.P.; Alyokhin, A.; Buzza, A.; Rondon, S.I.; Clough, M.; et al. Selection for high levels of resistance to double-stranded RNA (dsRNA) in Colorado potato beetle (Leptinotarsa decemlineata Say) using non-transgenic foliar delivery. Scientific Reports 2021, 11, 6523. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Moar, W.; Jurat-Fuentes, J.L. Larvae of Colorado potato beetle (Leptinotarsa decemlineata Say) resistant to double stranded RNA (dsRNA) remain susceptible to small-molecule pesticides. Pest. Manag. Sci. 2024, 80, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Narva, K.; Toprak, U.; Alyokhin, A.; Groves, R.; Jurat-Fuentes, J.L.; Moar, W.; Nauen, R. Whipple S, Head G. Insecticide resistance management scenarios differ for RNA-based sprays and traits. Insect Mol Biol. [CrossRef]
- Christiaens, O.; Whyard, S.; Vélez, A.M.; Smagghe, G. Double-Stranded RNA Technology to Control Insect Pests: Current Status and Challenges. Front. Plant Sci. 2020, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Cedden, D.; Güney, G.; Rostás, M.; Bucher, G. Optimizing dsRNA sequences for RNAi in pest control and research with the dsRIP web platform. BMC Biol. 2025, 23, 114. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Peng, Y.; Zhang, H.; Wang, K.; Zhao, C.; Zhu, G.; Reddy Palli, S.; Han, Z. Off-target effects of RNAi correlate with the mismatch rate between dsRNA and non-target mRNA. RNA Biol. 2021, 18, 1747–1759. [Google Scholar] [CrossRef] [PubMed]
- Cedden, D.; Bucher, G. The quest for the best target genes for RNAi-mediated pest control. Insect Mol Biol. 2024. [CrossRef] [PubMed]
- Yang, J.; Cho, W.C.; Zheng, Y. Argonaute proteins: Structural features, functions and emerging roles. Journal of Advanced Research 2020, 24, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Höck, J.; Meister, G. The Argonaute protein family. Genome Biol. 2008, 9, 210. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Ishizuka, A.; Siomi, H.; Siomi, M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004, 18, 1655–1666. [Google Scholar] [CrossRef] [PubMed]
- Hoang BTL, Fletcher SJ, Brosnan CA, Ghodke AB, Manzie N, Mitter N. RNAi as a Foliar Spray: Efficiency and Challenges to Field Applications. Int. J. Mol. Sci. 2022, 23, 6639. [Google Scholar] [CrossRef] [PubMed]
- Socha, W.; Kwasnik, M.; Larska, M.; Rola, J.; Rozek, W. Vector-borne viral diseases as a current threat for human and animal health—One Health perspective. Journal of Clinical Medicine 2022, 11, 3026. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wang, B.; Lei, G.; Chen, G.; Liu, D. Advances in nanocarriers to improve the stability of dsRNA in the environment. Front. Bioeng. Biotechnol. 2022, 10, 974646. [Google Scholar] [CrossRef] [PubMed]
- Nitnavare, R.B.; Bhattacharya, J.; Singh, S. : Kour, A.; Hawkesford, M.J.; Arora, N. Next generation dsRNA-based insect control: Success so far and challenges. Front. Plant Sci. 2021, 12, 673576. [Google Scholar] [CrossRef] [PubMed]
- Narva, K.; Toprak, U.; Alyokhin, A.; Groves, R.; Jurat-Fuentes, J.L.; Moar, W.; Nauen, R.; Whipple, S.; Head, G. Insecticide resistance management scenarios differ for RNA-based sprays and traits. Insect Mol Biol. 2025. [Google Scholar] [CrossRef] [PubMed]
- Verdonckt, T.W.; Vanden Broeck, J. Methods for the Cost-Effective Production of Bacteria-Derived Double-Stranded RNA for in vitro Knockdown Studies. Front. Physiol. 2022, 13, 836106. [Google Scholar] [CrossRef] [PubMed]
- Palli, S.R. RNAi turns 25: contributions and challenges in insect science. Front. Insect Sci. 2023, 3, 1209478. [Google Scholar] [CrossRef] [PubMed]
- Egli, M.; Manoharan, M. Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 2023, 51, 2529–2573. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, K.; Fu, W.; Sheng, C.; Han, Z. Biochemical comparison of dsRNA degrading nucleases in four different insects. Front Physiol. 2018, 9, 624–10.3389. [Google Scholar] [CrossRef] [PubMed]
- Dias, N.; Stein, C.A. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. 2002, 1, 347–355. [Google Scholar] [PubMed]
- Bachellerie, J.P.; Cavaillé, J.; Hüttenhofer, A. The expanding snoRNA world. Biochimie. 2002. [Google Scholar] [CrossRef] [PubMed]
- Will, C.L. , Lührmann, R., 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. [CrossRef]
- Oberemok, V.V.; Laikova, K.V.; Zaitsev, A.S.; Shumskykh, M.N.; Kasich, I.N.; Gal'chinsky, N.V.; Bekirova, V.V.; Makarov, V.V.; Agranovsky, A.A.; Gushchin, V.A.; et al. Molecular Alliance of Lymantria dispar Multiple Nucleopolyhedrovirus and a Short Unmodified Antisense Oligonucleotide of Its Anti-Apoptotic IAP-3 Gene: A Novel Approach for Gypsy Moth Control. Int. J. Mol. Sci. 2017, 18, 2446. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V. DNA markers in investigation of interaction between Lymantria dispar multiple nucleopolyhedrovirus and its host gypsy moth. Dissertation, Taurida National VI Vernadsky University, 2011.
- Cerio, R.J.; Vandergaast, R.; Friesen, P.D. Host insect inhibitor-of-apoptosis SfIAP functionally replaces baculovirus IAP but is differentially regulated by its N-terminal leader. J. Virol. 2010, 84, 11448–11460. [Google Scholar] [CrossRef] [PubMed]
- Manju, M.; Nirosha, V.; Tullika, T.; Mankhanniang, G. DNA insecticides: an emerging tool in pest management. AGRIALLIS. 2022, 4. [Google Scholar]
- Oberemok, V.V.; Skorokhod, O.A. Single-stranded DNA fragments of insect-specific nuclear polyhedrosis virus act as selective DNA insecticides for gypsy moth control. Pestic. Biochem. Physiol. 2014, 113, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V.; Laikova, K.V.; Zaitsev, A.S.; Gushchin, V.A.; Skorokhod, O.A. The RING for gypsy moth control: Topical application of fragment of its nuclear polyhedrosis virus anti-apoptosis gene as insecticide. Pestic Biochem Physiol. 2016, 131, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, H.; Li, H.; Miao, X. Second-generation sequencing supply an effective way to screen RNAi targets in large scale for potential application in pest insect control. PloS One 2011, 6, e18644. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999, 24, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V.; Useinov, R.Z.; Skorokhod, O.A.; Gal’chinsky, N.V.; Novikov, I.A.; Makalish, T.P.; Yatskova, E.V.; Sharmagiy, A.K.; Golovkin, I.O.; Gninenko, Y.I.; et al. Oligonucleotide insecticides for green agriculture: regulatory role of contact DNA in plant- insect interactions. Int. J. Mol. Sci. 2022, 23, 15681. [Google Scholar] [CrossRef] [PubMed]
- Useinov, R.Z.; Galchinsky, N.; Yatskova, E.; Novikov, I.; Puzanova, Y.; Trikoz, N.; Sharmagiy, A.; Plugatar, Y; Laikova, K. ; Oberemok, V. To bee or not to bee: creating DNA insecticides to replace non-selective organophosphate insecticides for use against the soft scale insect Ceroplastes Japonicus green. J. Plant. Prot. Res. 2020, 60, 406–409. [Google Scholar]
- Gal’chinsky, N.; Useinov, R.; Yatskova, E.; Laikova, K.; Novikov, I.; Gorlov, M.; Trikoz, N.; Sharmagiy, A.; Plugatar, Y.; Oberemok, V. A breakthrough in the efficiency of contact DNA insecticides: rapid high mortality rates in the sap-sucking insects Dynaspidiotus britannicus Comstock and Unaspis euonymi Newstead. J. Plant. Prot. Res. 2020, 60, 220–223. [Google Scholar] [CrossRef]
- Gal’chinsky, N.V.; Yatskova, E.V.; Novikov, I.A.; Useinov, R.Z.; Kouakou, N.J.; Kouame, K.F.; Kra, K.D.; Sharmagiy, A.K.; Plugatar, Y.V.; Laikova, K.V.; et al. Icerya purchasi Maskell (Hemiptera: Monophlebidae) control using low carbon footprint oligonucleotide insecticides. Int. J. Mol. Sci. 2023, 24, 11650. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V.; Gal'chinsky, N.V.; Useinov, R.Z.; Novikov, I.A.; Puzanova, Y.V.; Filatov, R.I.; Kouakou, N.J.; Kouame, K.F.; Kra, K.D.; Laikova, K.V. Four Most Pathogenic Superfamilies of Insect Pests of Suborder Sternorrhyncha: Invisible Superplunderers of Plant Vitality. Insects 2023, 14, 462. [Google Scholar] [CrossRef] [PubMed]
- Novikov, A.; Yatskova, E.; Bily, A.; Puzanova, Y.; Sharmagiy, A.; Oberemok, V. Efficient control of the obscure mealybug Pseudococcus viburni with DNA insecticides. In vitro cellular & developmental biology-animal. Springer, New York, USA.
- Puzanova, Y.V.; Novikov, I.A.; Bilyk, A.I.; Sharmagiy, A.K.; Plugatar, Y.V.; Oberemok, V.V. Perfect complementarity mechanism for aphid control: oligonucleotide insecticide macsan-11 selectively causes high mortality rate for Macrosiphoniella sanborni Gillette. Int. J. Mol. Sci. 2023, 24, 11690. [Google Scholar] [CrossRef] [PubMed]
- Oberemok, V.V.; Novikov, I.A.; Yatskova, E.V.; Bilyk, A.I.; Sharmagiy, A.K.; Gal'chinsky, N.V. Potent and selective ‘genetic zipper’ method for DNA-programmable plant protection: innovative oligonucleotide insecticides against Trioza alacris Flor. Chem. Biol. Technol. Agric. 2024, 11, 144. [Google Scholar] [CrossRef]
- Gavrilova, D.; Grizanova, E.; Novikov, I.; Laikova, E.; Zenkova, A.; Oberemok, V.; Dubovskiy, I. Antisense DNA acaricide targeting pre-rRNA of two-spotted spider mite Tetranychus urticae as efficacy-enhancing agent of fungus Metarhizium robertsii. Journal of Invertebrate Pathology. 2025, 108297. [Google Scholar] [CrossRef] [PubMed]
- IZ. Available online: https://en.iz.ru/en/1870413/maria-neduk-denis-gricenko/agrarian-evolution-dna-drug-will-destroy-main-enemy-greenhouse-crops (accessed on 16 April 2025).
- Oberemok, V.V.; Laikova, K.V.; Useinov, R.Z.; Gal’chinsky, N.V.; Novikov, I.A.; Gorlov, M.V.; Balykina, E.V.; Trikoz, N.N.; Yatskova, E.V.; Sharmagiy, A.K. High mortality of sap-sucking insects one week after topical application of DNA insecticides. In Vitro Cell. Dev. Biol. Anim. 2020, 56, 39. [Google Scholar]
- Plugatar, Y.V.; Chichkanova, E.S.; Yatskova, E.V.; Sharmagii, A.K.; Oberemok, V.V. An innovative method of Diaspis echinocacti Bouche control using DNA insecticide on Opuntia ficus-indica (L.) Mill. in the Nikitsky Botanical Garden, Crimea. South Russia: ecology Dev. 2021, 16, 119–128. [Google Scholar] [CrossRef]
- Oberemok, V.V.; Gal’chinsky, N.V. Oligonucleotide insecticides (contact unmodified antisense DNA biotechnology) and RNA biocontrols (double-stranded RNA technology): newly born fraternal twins in plant protection. bioRxiv 2024, 584797. [Google Scholar] [CrossRef]
- Oberemok, V.; Laikova, K.; Shumskykh, M.; Kenyo, I.; Kasich, I.; Deri, K.; Seidosmanova, E.; Krasnodubets, A.; Bekirova, V.; Gal'chinsky, N. A primary attempt of Leptinotarsa decemlineata control using contact DNA insecticide based on short antisense oligonucleotide of its CYP6B gene. J. Plant Prot. Res. 2018, 58, 106–108. [Google Scholar] [CrossRef]
- Du, Q.; Thonberg, H.; Wang, J.; Wahlestedt, C.; Liang, Z.A. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005, 33, 1671–1677. [Google Scholar] [CrossRef] [PubMed]
- Luige, O.; Karalė, K.; Bose, P.P.; Bollmark, M.; Tedebark, U.; Murtola, M.; Strömberg, R. Influence of sequence variation on the RNA cleavage activity of Zn2+-dimethyl-dppz-PNA-based artificial enzymes. RSC Adv. 2022, 12, 5398–5406. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Mojica, M.J.; Juez, G.; Rodríguez-Valera, F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol. Microbiol. 1993, 9, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.; Embden, J.D.A.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Aljabali, A.A.A.; El-Tanani, M.; Tambuwala, М.М. Principles of CRISPR-Cas9 technology: Advancements in genome editing and emerging trends in drug delivery. Journal of Drug Delivery Science and Technology 2024, 92, 105338. [Google Scholar] [CrossRef]
- Asokan, R.; Rai, A.; Dash, S.; Manamohan, M.; Ashok, K.; Bhargava, C.N.; Wishard, R.; Pradhan, S.K.; Parvathy, M.S. Application of genome editing in entomology. Indian Journal of Entomology 2022, 96–103. [Google Scholar]
- Khan, M.D.; Ahmad, B.; Ahmed, S.F.; Ijaz, M.; Abdin, Z.U.; Ghafoor, I.; Siddiqui, G.M.; Adrees, T.; Ali, S.; Shahzadi, N.; Rafa, H.U. CRISPR-Cas genome editing in crops: A promising frontier for climate-smart agriculture. Phytopathogenomics and Disease Control 2023, 2, 71–77. [Google Scholar] [CrossRef]
- Wang, J.Y.; Doudna, J.A. CRISPR technology: A decade of genome editing is only the beginning. Science 2023, 379, eadd8643. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Pausch, P.; Doudna, J.A. Structural biology of CRISPR–Cas immunity and genome editing enzymes. Nature Reviews Microbiology 2022, 20, 641–656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F. Development of CRISPR-Cas systems for genome editing and beyond. Quarterly Reviews of Biophysics. 2019, 52, e6. [Google Scholar] [CrossRef]
- Salum, Y.M.; Yin, A.; Zaheer, U.; Liu, Y.; Guo, Y.; He, W. CRISPR/Cas9-Based Genome Editing of Fall Armyworm (Spodoptera frugiperda): Progress and Prospects. Biomolecules 2024, 14, 1074. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Aumann, R.A.; Häcker, I.; Schetelig, M.F. CRISPR-based genetic control strategies for insect pests. J. Integr. Agric. 2023, 22, 651–668. [Google Scholar] [CrossRef]
- Ranian, K.; Zahoor, M.K.; Zulhussnain, M.; Ahmad, A. CRISPR/Cas9 mediated sex-ratio distortion by sex specific gene editing in Aedes aegypti. Saudi Journal of Biological Sciences 2022, 29, 3015–3022. [Google Scholar] [CrossRef] [PubMed]
- Zulhussnain, M.; Zahoor, M.K.; Ranian, K.; Ahmad, A.; Jabeen, F. CRISPR Cas9 mediated knockout of sex determination pathway genes in Aedes aegypti. Bulletin of Entomological Research. 2023, 113, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wang, M.; Zeng, J.; Sun, H.; Wei, X.; Jiang, H.; Shentu, X.; Sun, D. CRISPR/Cas Technology in Insect Insecticide Resistance. Insects 2025, 16, 345. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Handler, A.M. CRISPR/Cas9-mediated gene editing in an exogenous transgene and an endogenous sex determination gene in the Caribbean fruit fly, Anastrepha suspensa. Gene 2019, 691, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, X.e.; Liu, Z.; Xu, J.; Li, X.; Bi, H.; Andongma, A.A.; Niu, C.; Huang, Y. Mutation of doublesex induces sex-specific sterility of the diamondback moth Plutella xylostella. Insect Biochem. Mol. Biol. 2019, 112, 103180. [Google Scholar] [CrossRef] [PubMed]
- Nourani, L.; Mehrizi, A.A.; Pirahmadi, S.; Pourhashem, Z.; Asadollahi, E.; Jahangiri, B. CRISPR/Cas advancements for genome editing, diagnosis, therapeutics, and vaccine development for Plasmodium parasites, and genetic engineering of Anopheles mosquito vector. Infect Genet Evol. 2023, 109, 105419. [Google Scholar] [CrossRef] [PubMed]
- Zamanian, M.; Andersen, E.C. Prospects and challenges of CRISPR/Cas genome editing for the study and control of neglected vector-borne nematode diseases. FEBS J. 2016, 283, 3204–3221. [Google Scholar] [CrossRef] [PubMed]
- Bimal Kumar Sahoo, B.M.; Mahesh Pathak, M.; Sushruta Boruah, S.; Kasturi Sarmah, K. BIO-ENGINEERED DEFENDERS: CRISPR-CAS9 UNLEASHING DISEASE-RESISTANT INSECTS TO CURB VECTOR-BORNE DISEASES", Futuristic Trends in Agriculture Engineering & Food Sciences; IIP Series; 2024; Volume 3, Book 6, Volume 3, pp. 42–52. [CrossRef]
- Xu, J.; Xu, X.; Zhan, S.; Huang, Y. Genome editing in insects: current status and challenges. National Science Review 2019, 6, 399–401. [Google Scholar] [CrossRef] [PubMed]
- Courtier-Orgogozo, V.; Morizot, B.; Boëte, C. Using CRISPR-based gene drive for agriculture pest control. EMBO reports. 2017, 18, 1481–1481. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.H.; Du, J.; Chu, C.Y.; Madhav, M.; Hughes, G.L.; Champer, J. Symbionts and gene drive: two strategies to combat vector-borne disease. Trends in Genetics 2022, 38, 708–723. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, L.; Wei, L.; Wang, Y.; Han, Z. Advancements and Future Prospects of CRISPR-Cas-Based Population Replacement Strategies in Insect Pest Management. Insects 2024, 15, 653. [Google Scholar] [CrossRef] [PubMed]
- Meccariello, A.; Hou, S.; Davydova, S.; Fawcett, J.D.; Siddall, A.; Leftwich, P.T.; Krsticevic, F.; Papathanos, P.A.; Windbichler, N. Gene drive and genetic sex conversion in the global agricultural pest Ceratitis capitata. Nature Communications 2024, 15, 372. [Google Scholar] [CrossRef] [PubMed]
- Oye, K.A.; Esvelt, K.; Appleton, E.; Catteruccia, F.; Church, G.; Kuiken, T.; Lightfoot, S.B.Y.; McNamara, J.; Smidler, A.; Collins, J.P. Regulating gene drives. Science 2014, 345, 626–628. [Google Scholar] [CrossRef] [PubMed]
- Shukla, J.N.; Palli, S.R. Tribolium castaneum Transformer-2 regulates sex determination and development in both males and females. Insect biochemistry and molecular biology 2013, 43, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.J.; Gould, F.; Lorenzen, M.; Grubbs, N.; Edwards, O.; O’Brochta, D. Agricultural production: assessment of the potential use of Cas9-mediated gene drive systems for agricultural pest control. Journal of Responsible Innovation 2018, 5(sup1), S98–S120. [Google Scholar] [CrossRef]
- Tang, Y.H.; Zhang, X.; Dai, Z.C.; Li, H.; Yang, Y.; Zhao, T.J.; Yuan, D.Q.; Qian, W.L.; Cheng, D.J. CRISPR-Cas13-mediated RNA editing in the silkworm Bombyx mori. Zool Res. 2024, 45, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Bulle, M.; Sheri, V.; Aileni, M.; Zhang, B. Chloroplast Genome Engineering: A Plausible Approach to Combat Chili Thrips and Other Agronomic Insect Pests of Crops. Plants 2023, 12, 3448. [Google Scholar] [CrossRef] [PubMed]
- Han, W.K.; Yang, Y.L.; Si, Y.X.; Wei, Z.Q.; Liu, S.R.; Liu, X.L.; Yan, Q.; Dong, S.L. Involvement of GOBP2 in the perception of a sex pheromone component in both larval and adult Spodoptera litura revealed using CRISPR/Cas9 mutagenesis. Insect Biochemistry and Molecular Biology 2022, 141, 103719. [Google Scholar] [CrossRef] [PubMed]
- Gantz, V.M.; Jasinskiene, N.; Tatarenkova, O.; Fazekas, A.; Macias, V.M.; Bier, E.; James, A.A. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proceedings of the National Academy of Sciences 2015, 112, E6736–E6743. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.H.; Chereddy, S.C.; Howell, J.L.; Palli, S.R. Genome editing in the fall armyworm, Spodoptera frugiperda: Multiple sgRNA/Cas9 method for identification of knockouts in one generation. Insect biochemistry and molecular biology 2020, 122, 103373. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Wang, J.; Bi, H.; Li, X.; Merchant, A.; Zhang, P.; Zhang, Q.; Zhou, X. CRISPR/Cas9-mediated mutagenesis of sex-specific doublesex splicing variants leads to sterility in Spodoptera frugiperda, a global invasive pest. Cells 2022, 11, 3557. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhao, T.; Liu, X.; Li, T.; He, L.; Wang, Q.; Wang, L.; Zhou, L. CRISPR/Cas9-Mediated Mutagenesis of Antennapedia in Spodoptera frugiperda. Insects 2023, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Anu, C.N.; Ashok, K.; Bhargava, C.N.; Dhawane, Y.; Manamohan, M.; Jha, G.K.; Asokan, R. CRISPR/Cas9 mediated validation of spermatogenesis-related gene, tssk2 as a component of genetic pest management of fall armyworm, Spodoptera frugiperda (JE Smith) (Lepidoptera: Noctuidae). Archives of Insect Biochemistry and Physiology 2024, 116, e22121. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ma, Y.; Wang, F.; Yang, Y.; Wu, S.; Wu, Y. Disruption of nicotinic acetylcholine receptor α6 mediated by CRISPR/Cas9 confers resistance to spinosyns in Plutella xylostella. Pest. Manag. Sci. 2020, 76, 1618–1625. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Harvey-Samuel, T.; Yang, J.; You, M.; Alphey, L. CRISPR/Cas9-based functional characterization of the pigmentation gene ebony in Plutella xylostella. Insect Molecular Biology 2021, 30, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.Y.; Liang, L.Q.; Li, Q.F.; Zhu, Y.N.; Guo, Y.K.; Zheng, Q.L.; Lin, Y.; Yang, D.L.; Li, Z.G.; Su, S.K. CRISPR/Cas9 mediated knockout of Amyellow-y gene results in melanization defect of the cuticle in adult Apis mellifera. Journal of insect physiology 2021, 132, 104264. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Roy, M.C.; Al Baki, M.A.; Jung, J.K.; Lee, D.; Kim, Y. CRISPR/Cas9 mutagenesis against sex pheromone biosynthesis leads to loss of female attractiveness in Spodoptera exigua, an insect pest. PLoS One 2021, 16, e0259322. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.X.; Li, W.X.; Lyu, J.; Hu, Y.T.; Huang, G.; Zhang, W.Q. CRISPR/Cas9-mediated knockout of the NlCSAD gene results in darker cuticle pigmentation and a reduction in female fecundity in Nilaparvata lugens (Hemiptera: Delphacidae). Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 2021, 256, 110921. [Google Scholar]
- Fu, X.; Li, R.; Qiu, Q.; Wang, M.; Zhao, T.; Zhou, L. Study on the function of Helicoverpa armigera Wnt1 gene using CRISPR/Cas9 system. Journal of Asia-Pacific Entomology 2022, 25, 101869. [Google Scholar] [CrossRef]
- Bi, H.; Merchant, A.; Gu, J.; Li, X.; Zhou, X.; Zhang, Q. CRISPR/Cas9-mediated mutagenesis of abdominal-A and ultrabithorax in the Asian corn borer, Ostrinia furnacalis. Insects 2022, 13, 384. [Google Scholar] [CrossRef] [PubMed]
- Konu, M.; Kulmuni, J.; Viljakainen, L. Genetic modification of the ant Lasius niger using CRISPR-Cas9 technology. Insect Molecular Biology 2023, 32, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Xu, X.; Li, X.; Wang, Y.; Zhou, S.; Huang, Y. CRISPR/Cas9-mediated Serine protease 2 disruption induces male sterility in Spodoptera litura. Front. Physiol. 2022, 13, 931824. [Google Scholar] [CrossRef] [PubMed]
- Shirk, B.D.; Shirk, P.D.; Furlong, R.B.; Scully, E.D.; Wu, K.; Siegfried, B.D. Gene editing of the ABC Transporter/White locus using CRISPR/Cas9-mediated mutagenesis in the Indian Meal Moth. Journal of Insect Physiology 2023, 145, 104471. [Google Scholar] [CrossRef] [PubMed]
- Ashok, K.; Bhargava, C.N.; Babu, K.P.; Rohan, W.; Manamohan, M.; Rai, A.; Sanjay, K.P.; Parvathy, M.S.; Kennedy, J.S.; Asokan, R. First report on CRISPR/Cas9 mediated editing of the eye colour gene, tryptophan 2, 3-dioxygenase in egg plant shoot and fruit borer Leucinodes orbonalis Guenée (Lepidoptera: Crambidae). Journal of Asia-Pacific Entomology 2023, 26, 102031. [Google Scholar] [CrossRef]
- Ashok, K.; Bhargava, C.N.; Venkatesh, R.; Balasubramani, V.; Murugan, M.; Geethalakshmi, V.; Manamohan, M.; Jha, G.K.; Asokan, R. Molecular characterization and CRISPR/Cas9 validation of the precursor of egg yolk protein gene, vitellogenin of Leucinodes orbonalis Guenée (Lepidoptera: Crambidae). Gene 2025, 933, 148925. [Google Scholar] [CrossRef] [PubMed]
- Ashok, K.; Bhargava, C.N.; Asokan, R.; Pradeep, C.; Kennedy, J.S.; Rai, A.; Manamohan, M. First report on the utility of pupal case for early determination of CRISPR/Cas9 ribonucleoprotein mediated genomic edits in the oriental fruit fly, Bactrocera dorsalis (Hendel) (Tephritidae: Diptera). Archives of Insect Biochemistry and Physiology 2023, 113, e22024. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, C.N.; Ashok, K.; Asokan, R.; Prasad Babu, K.; Parvathy, M.S.; Yogi, D.; Shashikala, T.; Chiranth, R.K.; Ashok, U.; Harsha, C.G. CRISPR/Cas9 Mediated Editing of the white (wh) locus Affects Body Size and Reproduction of the Oriental Fruit Fly, Bactocera dorsalis (Hendel). Agricultural Research 2024, 1–8. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Karuppannasamy, A.; Sujatha, P.M.; Nagaraja, B.C.; Narayanappa, A.C.; Chalapathi, P.; Dhawane, Y.; Bynakal, S.; Riegler, M.; Maligeppagol, M.; et al. Embryonic microinjection of ribonucleoprotein complex (Cas9+ sgRNA) of white gene in melon fly, Zeugodacus cucurbitae (Coquillett) (Diptera: Tephritidae) produced white eye phenotype. Archives of Insect Biochemistry and Physiology 2023, 114, e22059. [Google Scholar] [CrossRef] [PubMed]
- Sujatha, P.M.; Karuppannasamy, A.; Chalapathi, P.; Dhawane, Y.; Narasimhappa, N.S.; Narayanappa, A.C.; Munikrishnappa, V.K.T.; Chikmagalur Nagaraja, B.; Thalooru, S.; Kesavan, S.; et al. CRISPR/Cas9 Ribo nucleoprotein complex-mediated editing of the OBP13 gene affected the response of male Bactrocera dorsalis (Diptera: Tephritidae) to methyl eugenol. Journal of Applied Entomology 2024. [CrossRef]
- Yadav, A.K.; Butler, C.; Yamamoto, A.; Patil, A.A.; Lloyd, A.L.; Scott, M.J. CRISPR/Cas9-based split homing gene drive targeting doublesex for population suppression of the global fruit pest Drosophila suzukii. Proceedings of the National Academy of Sciences 2023, 120, e2301525120. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; James, B.; Williams, M.; Bachler, A.; Tay, W.T.; Walsh, T.; Frese, M. Cry1 resistance in a CRISPR/Cas9-mediated HaCad1 gene knockout strain of the Australian cotton bollworm Helicoverpa armigera conferta (Lepidoptera: Noctuidae). Pest. Manag. Sci. 2025, 81, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, Y.; Nakamura, T.; Watanabe, T.; Barnett, A.A.; Tomonari, S.; Ylla, G.; Whittle, C.A.; Noji, S.; Mito, T.; Extavour, C.G. Establishment of CRISPR/Cas9-based knock-in in a hemimetabolous insect: targeted gene tagging in the cricket Gryllus bimaculatus. Development 2025, 152. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Bauer, D.E.; Chiarle, R. Assessing and advancing the safety of CRISPR-Cas tools: from DNA to RNA editing. Nat Commun. 2023, 14, 212. [Google Scholar] [CrossRef] [PubMed]
- Richardson, C.; Kelsh, R.N.; J Richardson, R. New advances in CRISPR/Cas-mediated precise gene-editing techniques. Dis. Model. Mech. 2023, 16, dmm049874. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Munawar, N.; Khan, Z.; Qusmani, A.T.; Khan, S.H.; Jamil, A.; Ashraf, S.; Ghouri, M.Z.; Aslam, S.; Mubarik, M.S.; et al. An Outlook on Global Regulatory Landscape for Genome-Edited Crops. Int. J. Mol. Sci. 2021, 22, 11753. [Google Scholar] [CrossRef] [PubMed]
- Asad, M.; Chang, Y.; Liao, J.; Yang, G. CRISPR/Cas9 Genome Editing in the Diamondback Moth: Current Progress, Challenges, and Prospects. Int. J. Mol. Sci. 2025, 26, 1515. [Google Scholar] [CrossRef] [PubMed]
- Komal, J.; Desai, H.R.; Samal, I.; Mastinu, A.; Patel, R.D.; Kumar, P.V.D.; Majhi, P.K.; Mahanta, D.K.; Bhoi, T.K. Unveiling the Genetic Symphony: Harnessing CRISPR-Cas Genome Editing for Effective Insect Pest Management. Plants (Basel) 2023, 12, 3961. [Google Scholar] [CrossRef] [PubMed]
- Moon, T.T.; Maliha, I.J.; Khan, A.A.M.; Chakraborty, M.; Uddin, M.S.; Amin, M.R.; Islam, T. CRISPR-Cas Genome Editing for Insect Pest Stress Management in Crop Plants. Stresses 2022, 2, 493–514. [Google Scholar] [CrossRef]
| Sl. No. | Names of model insects | Targeted gene(s) | Affected processes | References |
|---|---|---|---|---|
| 1 | Beet armyworm, Spodoptera exigua (Lepidoptera) | Chitin synthase gene A | Chitin synthesis | [40] |
| 2 | Brown planthopper, Nilaparvata lugens (Hemiptera) | NlHT1, Nlcar, Nltry | Digestive system | [41] |
| 3 | African sweet potato weevil, Cylas puncticollis (Coleoptera) | Snf7 | Digestive system | [42] |
| 4 | Tomato pinworm, Tuta absoluta (Lepidoptera) | Vacuolar ATPase-A and Arginine kinase | High mortality | [43] |
| 5 | Oriental fruit fly, Bactrocera dorsalis (Diptera) | α-Spectrin | Oviposition and ovary size | [44] |
| 6 | Cotton mealybug, Phenacoccus solenopsis (Hemiptera) | Krüppel homologue-1, ADP-ATP/Translocase, IDGF-1 | Not specified | [45] |
| 7 | Diamondback moth, Plutella xylostella (Lepidoptera) | PxCht | Chitin synthesis | [46] |
| 8 | Fall armyworm, Spodoptera frugiperda (Lepidoptera) | Met, EcR, USP | Reproductive system, fertility | [47] |
| 9 | White-backed planthopper, Sogatella furcifera (Hemiptera) | hsc70-3, PP-α | Insect metamorphosis | [48] |
| 10 | Soybean aphid, Aphis glycines (Hemiptera) | Cytochrome P450 monooxygenases (CYP450s) | Insect resistance | [49] |
| 11 | Asian citrus psyllid, Diaphorina citri (Hemiptera) | CHC, vATPase-A, Snf7 | Transmembrane system | [50] |
| 12 | Trichogramma dendrolimi (Lepidoptera) | Vitellogenin receptor (VgR) | Female reproductive system | [51] |
| 13 | Domestic silk moth, Bombyx mori (Lepidoptera) | BmToll9-2 | Chitin synthesis | [52] |
| 14 | Silverleaf whitefly, Bemisia tabaci (Hemiptera) | Cysteine protease | Digestive system | [53] |
| 15 | Cowpea weevil, Callosobruchus maculatus (Coleoptera) | Olfactory receptor coreceptor (Cmac\Orco) | Insect sensory system | [53] |
| 16 | White-backed planthopper, S. furcifera (Hemiptera) | β-N-acetylhexosaminidase genes | Insect metamorphosis | [55] |
| 17 | Fall armyworm, S. frugiperda (Lepidoptera) | COPIα, COPIβ, GSTU1 | Insect reproduction | [56] |
| 18 | Desert locust, Schistocerca gregaria (Orthoptera) | Cytochrome P450 | Ecdysteroid pathway | [57] |
| 19 | Red flour beetle, Tribolium castaneum (Coleoptera) | CPAPs | Cuticular proteins | [58] |
| 20 | Chinese white pine beetle, Dendroctonus armandi (Coleoptera) | Aquaporin | Osmoregulation | [59] |
| 21 | Cotton mealybug, P. solenopsis (Hemiptera) | Bursicon, V-ATPase | Cuticle hardening and V-ATPases act as proton pumps | [60] |
| 22 | Kissing bug, Rhodnius prolixus (Hemiptera) | Nitrophorin 2 (NP2) | Anticoagulant and apyrase activities in saliva | [61] |
| 23 | Brown plant hopper, N. lugens (Hemiptera) | NlTPS | Enzymatic activity | [62] |
| 24 | Citrus aphid, Toxoptera citricida (Hemiptera) | TCiCHS | Chitin synthesis | [63] |
| 25 | Potato psyllid, Bactericera cockerelli (Hemiptera) | SUC1, ST4 | Osmoregulatory | [64] |
| Sl. No. | Insects name | Targeted gene(s) | Affected system | References |
|---|---|---|---|---|
| 1 | Euonymous scale, Unaspis euonymi (Hemiptera) | 28S rRNA | Protein biosynthesis | [104,112] |
| 2 | Holly scale, Dynaspidiotus Britannicus (Hemiptera) | 28S rRNA | Protein biosynthesis | [5,104] |
| 3 | Japanese wax scale, Ceroplastes japonicus (Hemiptera) | 28S rRNA | Protein biosynthesis | [103] |
| 4 | Cactus scale, Diaspis echinocacti (Hemiptera) | 28S rRNA | Protein biosynthesis | [113] |
| 5 | Bay sucker, Trioza alacris (Hemiptera) | ITS2 of pre-rRNA and 28S rRNA | Protein biosynthesis | [109] |
| 6 | Cottony cushion scale, Icerya purchase (Hemiptera) | 28S rRNA | Protein biosynthesis | [105] |
| 7 | Chrysanthemum aphid, Macrosiphoniella sanborni (Hemiptera) | ITS2 of pre-rRNA | Protein biosynthesis | [108] |
| 8 | Mealybug, Pseudococcus viburni (Hemiptera) | 5.8S, 18S and 28S rRNA | Protein biosynthesis | [107] |
| 9 | Laureal scale, Aonidia lauri (Hemiptera) | 28S rRNA | Protein biosynthesis | [5] |
| 10 | Soft scale, Coccus hesperidum (Hemiptera) | 28S rRNA | Protein biosynthesis | [102] |
| 11 | Two-spotted spider mite, Tetranychus urticae (Trombidiformes) | ITS2 of pre-rRNA | Protein biosynthesis | [110] |
| 12 | Grey pine aphid, Schizolachnus pineti (Hemiptera) | ITS2 of pre-rRNA | Protein biosynthesis | [13] |
| 13 | Large pine aphid, Cinara pinea (Hemiptera) |
ITS2 of pre-rRNA | Protein biosynthesis | [13] |
| 14 | Pine needle aphid, Eulachnus rileyi (Hemiptera) | ITS2 of pre-rRNA | Protein biosynthesis | [13] |
| Sl. No. | Insect name | Target gene(s) | Affected system | Reference |
|---|---|---|---|---|
| 1 | Mosquito, Anopheles stephensi (Diptera) | Kynurenine hydroxylase | Parasite-resistance | [151] |
| 2 | Fall armyworm, S. frugiperda (Lepidoptera) |
Ebony Doublesex (dsx) (Sfdsx) Antennapedia (Antp) Spermatogenesis-related, tssk2 |
Melanin biosynthesis Sex differentiation Insect thorax and wing development Male reproductive system |
[152-155] |
| 3 | Diamondback moth, P. xylostella (Lepidoptera) |
Yellow Ebony LW-opsin |
Body pigmentation Body pigmentation Efficiency of phototaxis |
[46,156,157] |
| 4 | European bee, Apis mellifera (Hymenoptera) | Amyellow-y | Melanization in cuticle | [158] |
| 5 | Beet armyworm, S. exigua (Lepidoptera) | Desaturase (SexiDES5) | Sex pheromone biosynthesis | [159] |
| 6 | Brown planthopper, N. lugens (Hemiptera) | Cysteine sulfinic acid decarboxylase (CSAD) | Melanin metabolism | [160] |
| 7 | Chickpea pod borer, H. armigera (Lepidoptera) | Wnt1 | Segmentation, appendage development, and pigmentation | [161] |
| 8 | Asian corn borer, Ostrinia furnacalis (Lepidoptera) | Abdominal-A (Abd-A) and Ultrabithorax (Ubx) | Anatomical structure formation | [162] |
| 9 | Black garden ant, Lasius niger (Hymenoptera) | Cinnabar | Eye pigmentation | [163] |
| 10 | Common cutworm, S. litura (Lepidoptera) |
Serine protease 2 Odorant-binding proteins |
Male sterility Perception of a sex pheromone |
[150, 164] |
| 11 | Indian meal moth, Plodia interpunctella (Lepidoptera) | ATP binding cassette (ABC) proteins | Eye pigmentation | [165] |
| 12 | Eggplant shoot and fruit borer, Leucinodes orbonalis (Lepidoptera) |
Tryptophan 2, 3-dioxygenase Vitellogenin (Vg) |
Eye pigmentation Female reproductive system |
[166,167] |
| 13 | Mango fruit fly, B. dorsalis (Diptera) |
White White locus OBP13 |
Eye pigmentation Eye pigmentation Methyl eugenol |
[168-171] |
| 14 | Pomace fly, Drosophila suzukii (Diptera) | Doublesex | Population suppression | [172] |
| 15 | Australian cotton bollworm, H. armigera conferta (Lepidoptera) | Cadherin | Cry1Ac resistance | [173] |
| 16 | Cricket, Gryllus bimaculatus (Orthoptera) | Laccase 2 (Gb-lac2) | Cuticle system pigmentation | [174] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





