Submitted:
10 April 2025
Posted:
10 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
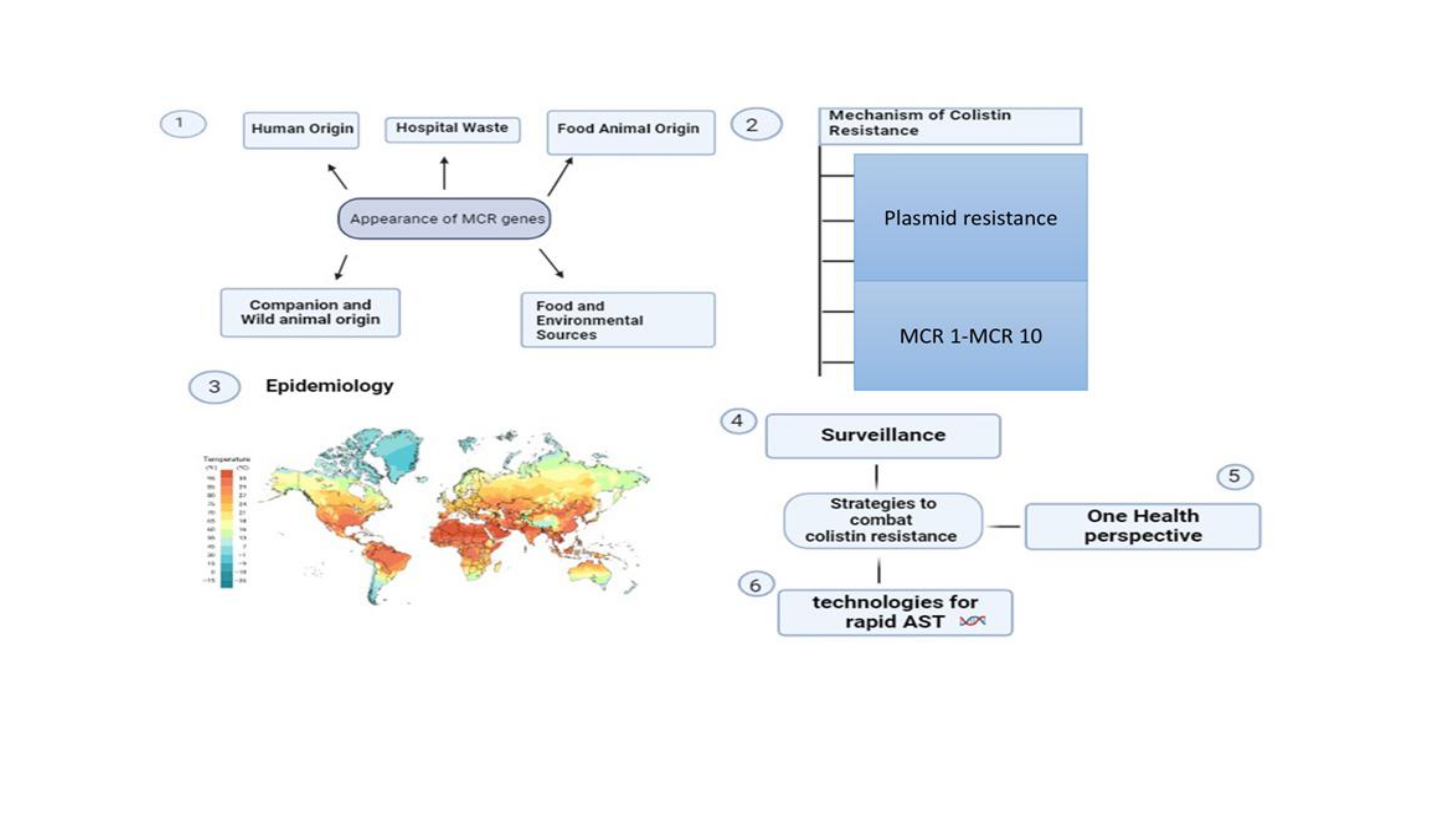
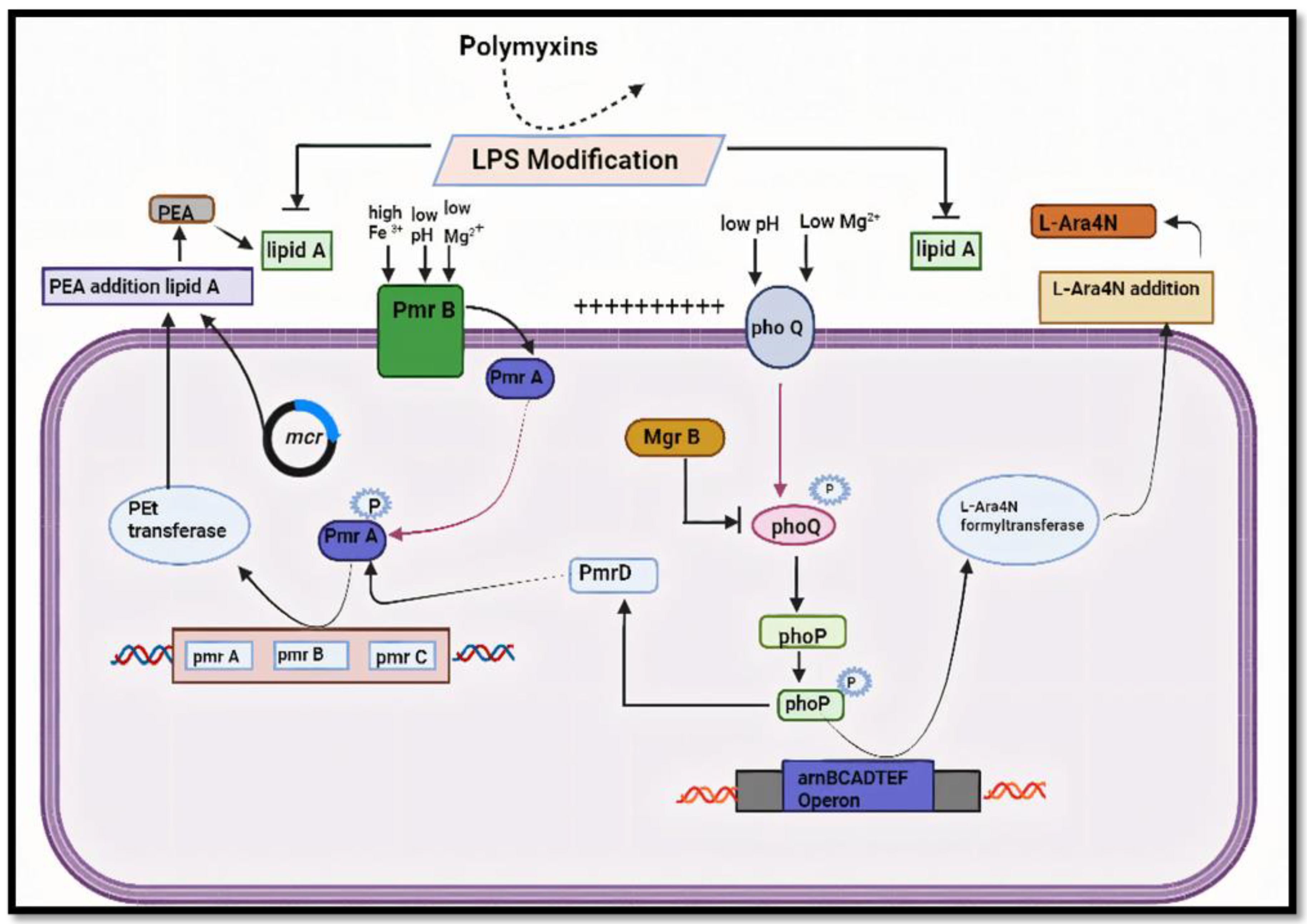
2. Mechanism of Colistin Resistance
2.1. Polymyxin Resistance Mediated by Plasmids
2.1.1. MCR-1 Gene
2.1.2. MCR-2 Gene
2.1.3. MCR-3 Gene
2.1.4. MCR-4 Gene
2.1.5. MCR-5 Gene
2.1.6. MCR-6 Gene
2.1.7. MCR-7 Gene
2.1.8. MCR-8 Gene
2.1.9. MCR-9 Gene
2.1.10. MCR-10 Gene
3. Clinical Implication
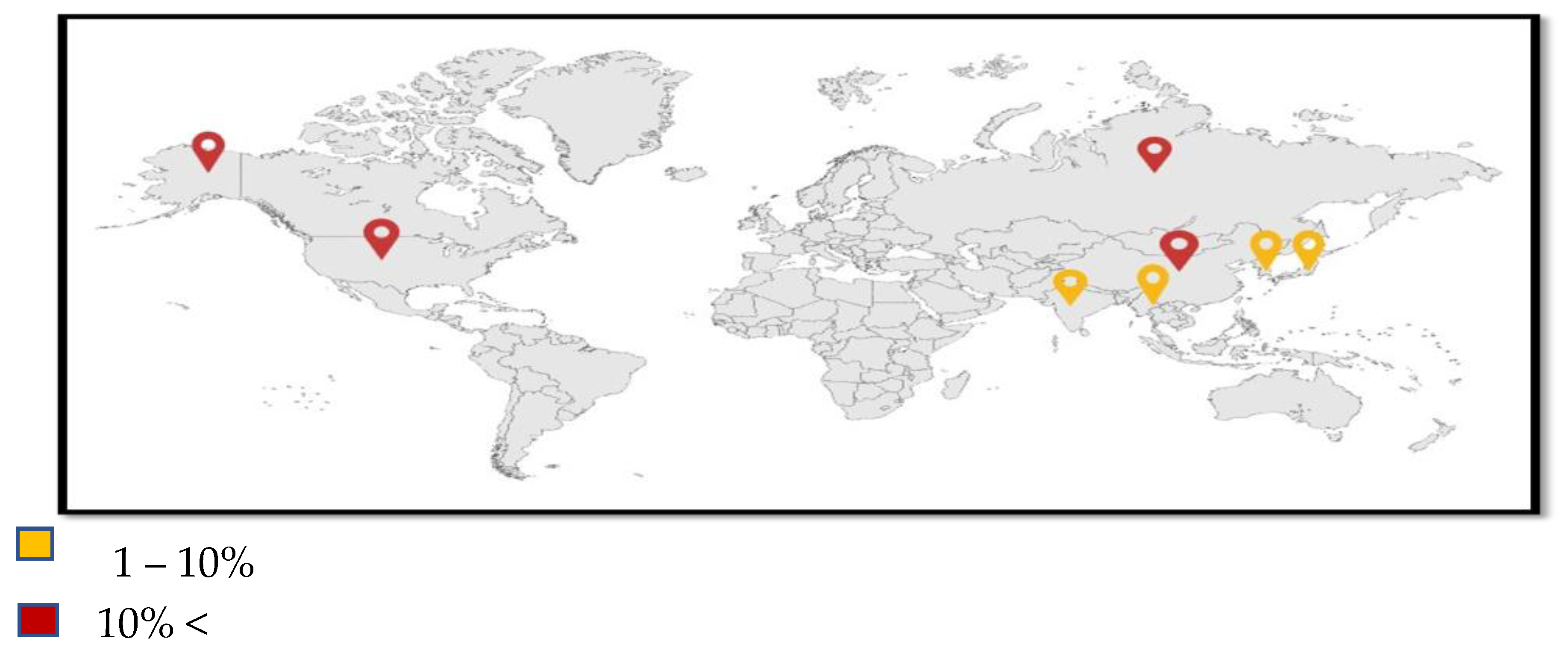
4. Epidemiology
4.1. American Countries
4.2. European Countries
4.3. African Countries
4.4. Asian Countries
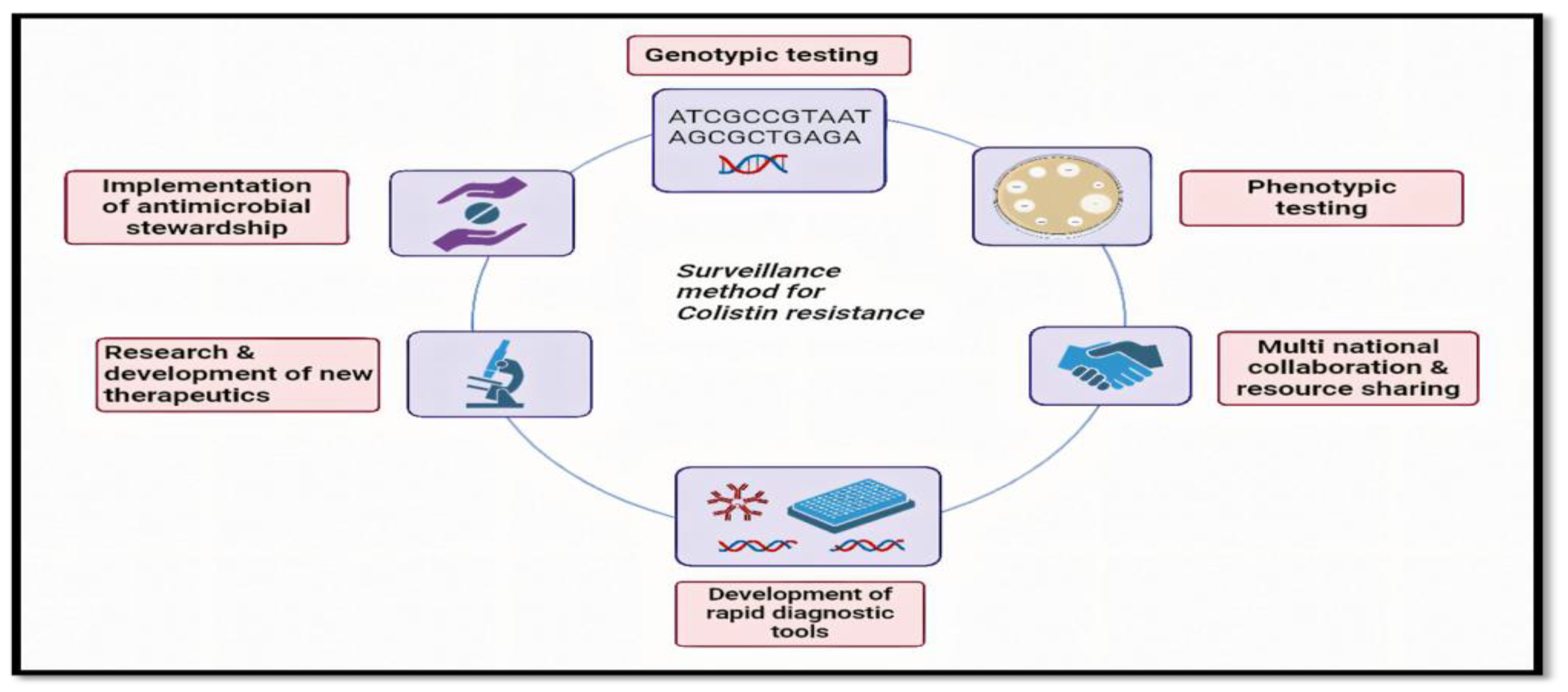
5. Surveillance
5.1. The initial Screening Process Using Selective Culture Medium
5.2. Automated AST, Antibiotic Gradient Testing, and Disk Diffusion Assays
5.3. Molecular Diagnostics
6. Combating
6.1. Colistin Combination Therapy
6.2. One Health Perspective
7. Future Perspective
7.1. Contemporary Instruments for Quick AMR Diagnosis
7.2. Available Technologies for Rapid AST
7.3. NAAT
7.4. Whole Genome Sequencing
8. Discussion
9. Conclusion
Acknowledgments
Credit Authorship statement
Funding
Availability of data and materials
Ethics approval and consent to participate:
Consent for publication
Competing interests
Author details:
Abbreviation
- GNB: Gram negative bacteria
- MCR: Mobilised colistin resistance
- MDR: Multidrug Resistance
- WHO: World Health Organization
- AMR: Antimicrobial resistance
- KPC: Klebsiella pneumoniae carbapenemase
- LOS: longer hospital stays
- IDSA: Infectious Diseases Society of America
- NDM: New Delhi metallo-β-lactamase
- AST: Antimicrobial susceptibility testing
- MIC: Minimum Inhibitory Concentration
- ICU: Intensive Care Unit
- MALDI-TOF MS: Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- LPS: Lipopolysaccharide
- EUCAST: European Committee on Antimicrobial Susceptibility Testing
- BMD: Broth microdilution
- GLASS: Global Antimicrobial Resistance Surveillance System
- FISH: Fluorescence In Situ Hybridization
- NAAT: Nucleic Acid Amplification Technology
- PDT: Photodynamic therapy
- CLSI: Clinical and laboratory standards institute
- WGS: Whole Genome Sequencing
- ABCR: A. baumanii resistant to colistin
- ABCS: A. baumanii sensitive to colistin
- MDR UTI: Multidrug-resistant Urinary tract infection
References
- Lim, L.M.; Ly, N.; Anderson, D.; Yang, J.C.; Macander, L.; Jarkowski III, A.; Forrest, A.; Bulitta, J.B.; Tsuji, B.T. Resurgence of colistin: a review of resistance, toxicity, pharmacodynamics, and dosing. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy 2010, 30, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Bialvaei, A.Z.; Samadi Kafil, H. Colistin, mechanisms and prevalence of resistance. Current medical research and opinion 2015, 31, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Hussein, N.H.; Al-Kadmy, I.M.; Taha, B.M.; Hussein, J.D. Mobilized colistin resistance (mcr) genes from 1 to 10: a comprehensive review. Molecular biology reports 2021, 48, 2897–2907. [Google Scholar] [CrossRef]
- Wozniak, T.M.; Barnsbee, L.; Lee, X.J.; Pacella, R.E. Using the best available data to estimate the cost of antimicrobial resistance: a systematic review. Antimicrobial Resistance & Infection Control 2019, 8, 1–12. [Google Scholar] [CrossRef]
- Kasiakou, S.K.; Michalopoulos, A.; Soteriades, E.S.; Samonis, G.; Sermaides, G.J.; Falagas, M.E. Combination therapy with intravenous colistin for management of infections due to multidrug-resistant Gram-negative bacteria in patients without cystic fibrosis. Antimicrobial agents and chemotherapy 2005, 49, 3136–3146. [Google Scholar] [CrossRef]
- America, I.D.S.o. Combating antimicrobial resistance: policy recommendations to save lives. Clinical Infectious Diseases 2011, 52, S397–S428. [Google Scholar] [CrossRef]
- Nation, R.L.; Velkov, T.; Li, J. Colistin and polymyxin B: peas in a pod, or chalk and cheese? Clinical infectious diseases 2014, 59, 88–94. [Google Scholar] [CrossRef]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure− activity relationships of polymyxin antibiotics. Journal of medicinal chemistry 2010, 53, 1898–1916. [Google Scholar] [CrossRef]
- Gelbicova, T.; Kolackova, I.; Krutova, M.; Karpiskova, R. The emergence of mcr-1-mediated colistin-resistant Escherichia coli and Klebsiella pneumoniae in domestic and imported turkey meat in the Czech Republic 2017–2018. Folia Microbiologica 2020, 65, 211–216. [Google Scholar] [CrossRef]
- Ilbeigi, K.; Askari Badouei, M.; Vaezi, H.; Zaheri, H.; Aghasharif, S.; Kafshdouzan, K. Molecular survey of mcr1 and mcr2 plasmid mediated colistin resistance genes in Escherichia coli isolates of animal origin in Iran. BMC Research Notes 2021, 14, 1–5. [Google Scholar] [CrossRef]
- Xavier, B.B.; Lammens, C.; Ruhal, R.; Kumar-Singh, S.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Eurosurveillance 2016, 21, 30280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, L.; Wang, J.; Butaye, P.; Huang, K.; Qiu, H.; Zhang, X.; Gong, W.; Wang, C. Molecular detection of colistin resistance genes (mcr-1 to mcr-5) in human vaginal swabs. BMC Research Notes 2018, 11, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chen, H.; Zhang, H.; Ullah, S.; Hou, T.; Feng, Y. The MCR-3 inside linker appears as a facilitator of colistin resistance. Cell Reports 2021, 35. [Google Scholar] [CrossRef] [PubMed]
- Furlan, J.P.R.; dos Santos, L.D.R.; Ramos, M.S.; Gallo, I.F.L.; Stehling, E.G. Presence of colistin resistance mcr-4 gene and clinically relevant antimicrobial resistance genes in sand samples from a public beach. Water, Air, & Soil Pollution 2020, 231, 1–6. [Google Scholar] [CrossRef]
- Cherak, Z.; Loucif, L.; Ben Khedher, M.; Moussi, A.; Benbouza, A.; Baron, S.A.; Rolain, J.-M. MCR-5-producing colistin-resistant Cupriavidus gilardii strain from well water in Batna, Algeria. Msphere 2021, 6. [Google Scholar] [CrossRef]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. Journal of Antimicrobial Chemotherapy 2017, 72, 3317–3324. [Google Scholar] [CrossRef]
- Borowiak, M.; Baumann, B.; Fischer, J.; Thomas, K.; Deneke, C.; Hammerl, J.A.; Szabo, I.; Malorny, B. Development of a novel mcr-6 to mcr-9 multiplex PCR and assessment of mcr-1 to mcr-9 occurrence in colistin-resistant Salmonella enterica isolates from environment, feed, animals and food (2011–2018) in Germany. Frontiers in microbiology 2020, 11, 80. [Google Scholar] [CrossRef]
- Yang, Y.-Q.; Li, Y.-X.; Lei, C.-W.; Zhang, A.-Y.; Wang, H.-N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. Journal of Antimicrobial Chemotherapy 2018, 73, 1791–1795. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Zhou, Y.; Li, J.; Yin, W.; Wang, S.; Zhang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerging microbes & infections 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. MBio 2019, 10. [Google Scholar] [CrossRef]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerging microbes & infections 2020, 9, 508–516. [Google Scholar] [CrossRef]
- Kollef, M.H.; Golan, Y.; Micek, S.T.; Shorr, A.F.; Restrepo, M.I. Appraising contemporary strategies to combat multidrug resistant gram-negative bacterial infections–proceedings and data from the Gram-Negative Resistance Summit. Clinical infectious diseases 2011, 53, S33–S55. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-T.; Sun, J.-R.; Wang, Y.-C.; Chiu, C.-H.; Kuo, S.-C.; Chen, T.-L.; Yang, Y.-S.; Group, A.S. Multicentre study of risk factors for mortality in patients with Acinetobacter bacteraemia receiving colistin treatment. International journal of antimicrobial agents 2020, 55, 105956. [Google Scholar] [CrossRef]
- Balkhair, A.; Al Saadi, K.; Al Adawi, B. Epidemiology and mortality outcome of carbapenem-and colistin-resistant Klebsiella pneumoniae, Escherichia coli, Acinetobacter baumannii, and Pseudomonas aeruginosa bloodstream infections. IJID regions 2023, 7, 1–5. [Google Scholar] [CrossRef]
- Mantzarlis, K.; Makris, D.; Zakynthinos, E. Risk factors for the first episode of Acinetobacter baumannii resistant to colistin infection and outcome in critically ill patients. Journal of Medical Microbiology 2020, 69, 35–40. [Google Scholar] [CrossRef]
- Ejaz, H.; Younas, S.; Qamar, M.U.; Junaid, K.; Abdalla, A.E.; Abosalif, K.O.A.; Alameen, A.A.M.; Elamir, M.Y.M.; Ahmad, N.; Hamam, S.S.M. Molecular epidemiology of extensively drug-resistant mcr encoded colistin-resistant bacterial strains co-expressing multifarious β-lactamases. Antibiotics 2021, 10, 467. [Google Scholar] [CrossRef]
- Landman, D.; Georgescu, C.; Martin, D.A.; Quale, J. Polymyxins revisited. Clinical microbiology reviews 2008, 21, 449–465. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, Y.; Shen, Y.; Shen, J.; Wu, C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. The Lancet infectious diseases 2016, 16, 293. [Google Scholar] [CrossRef]
- Bargiacchi, O.; Rossati, A.; Car, P.; Brustia, D.; Brondolo, R.; Rosa, F.; Garavelli, P.; De Rosa, F.G. Intrathecal/intraventricular colistin in external ventricular device-related infections by multi-drug resistant Gram negative bacteria: case reports and review. Infection 2014, 42, 801–809. [Google Scholar] [CrossRef]
- Coates, A.R.; Hu, Y.; Holt, J.; Yeh, P. Antibiotic combination therapy against resistant bacterial infections: synergy, rejuvenation and resistance reduction. Expert review of Anti-infective therapy 2020, 18, 5–15. [Google Scholar] [CrossRef]
- Sorlí, L.; Luque, S.; Li, J.; Campillo, N.; Danés, M.; Montero, M.; Segura, C.; Grau, S.; Horcajada, J.P. Colistin for the treatment of urinary tract infections caused by extremely drug-resistant Pseudomonas aeruginosa: dose is critical. Journal of Infection 2019, 79, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Aydın, M.; Ergönül, Ö.; Azap, A.; Bilgin, H.; Aydın, G.; Çavuş, S.; Demiroğlu, Y.; Alışkan, H.; Memikoğlu, O.; Menekşe, Ş. Rapid emergence of colistin resistance and its impact on fatality among healthcare-associated infections. Journal of Hospital Infection 2018, 98, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Yau, W.; Owen, R.J.; Poudyal, A.; Bell, J.M.; Turnidge, J.D.; Heidi, H.Y.; Nation, R.L.; Li, J. Colistin hetero-resistance in multidrug-resistant Acinetobacter baumannii clinical isolates from the Western Pacific region in the SENTRY antimicrobial surveillance programme. Journal of Infection 2009, 58, 138–144. [Google Scholar] [CrossRef]
- Band, V.I.; Satola, S.W.; Smith, R.D.; Hufnagel, D.A.; Bower, C.; Conley, A.B.; Rishishwar, L.; Dale, S.E.; Hardy, D.J.; Vargas, R.L. Colistin heteroresistance is largely undetected among carbapenem-resistant Enterobacterales in the United States. MBio 2021, 12. [Google Scholar] [CrossRef]
- Huang, H.; Dong, N.; Shu, L.; Lu, J.; Sun, Q.; Chan, E.W.-C.; Chen, S.; Zhang, R. Colistin-resistance gene mcr in clinical carbapenem-resistant Enterobacteriaceae strains in China, 2014–2019. Emerging microbes & infections 2020, 9, 237–245. [Google Scholar] [CrossRef]
- Sodhi, K.; Mittal, V.; Arya, M.; Kumar, M.; Phillips, A.; Kajla, B. Pattern of colistin resistance in Klebsiella isolates in an Intensive Care Unit of a tertiary care hospital in India. Journal of Infection and Public Health 2020, 13, 1018–1021. [Google Scholar] [CrossRef]
- Kawamoto, Y.; Kaku, N.; Akamatsu, N.; Sakamoto, K.; Kosai, K.; Morinaga, Y.; Ohmagari, N.; Izumikawa, K.; Yamamoto, Y.; Mikamo, H. The surveillance of colistin resistance and mobilized colistin resistance genes in multidrug-resistant Enterobacteriaceae isolated in Japan. International Journal of Antimicrobial Agents 2022, 59, 106480. [Google Scholar] [CrossRef]
- Luk-In, S.; Chatsuwan, T.; Kueakulpattana, N.; Rirerm, U.; Wannigama, D.L.; Plongla, R.; Lawung, R.; Pulsrikarn, C.; Chantaroj, S.; Chaichana, P. Occurrence of mcr-mediated colistin resistance in Salmonella clinical isolates in Thailand. Scientific Reports 2021, 11, 14170. [Google Scholar] [CrossRef]
- Cheong, H.S.; Kim, S.Y.; Seo, J.; Wi, Y.M.; Peck, K.R.; Ko, K.S. Colistin resistance and extensive genetic variations in PmrAB and PhoPQ in Klebsiella pneumoniae isolates from South Korea. Current microbiology 2020, 77, 2307–2311. [Google Scholar] [CrossRef]
- Makarov, D.A.; Ivanova, O.E.; Karabanov, S.Y.; Gergel, M.A.; Pomazkova, A.V. Antimicrobial resistance of commensal Escherichia coli from food-producing animals in Russia. Veterinary World 2020, 13, 2053. [Google Scholar] [CrossRef]
- Bir, R.; Gautam, H.; Arif, N.; Chakravarti, P.; Verma, J.; Banerjee, S.; Tyagi, S.; Mohapatra, S.; Sood, S.; Dhawan, B. Analysis of colistin resistance in carbapenem-resistant Enterobacterales and XDR Klebsiella pneumoniae. Therapeutic Advances in Infectious Disease 2022, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Manohar, P.; Shanthini, T.; Ayyanar, R.; Bozdogan, B.; Wilson, A.; Tamhankar, A.J.; Nachimuthu, R.; Lopes, B.S. The distribution of carbapenem-and colistin-resistance in Gram-negative bacteria from the Tamil Nadu region in India. Journal of medical microbiology 2017, 66, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.R.C.; Geetha, R.; Singh, M.; Rani, R.U.; Nekkanti, K.N. Mcr-1 expression in progression of colistin resistance gram negative bacilli of clinical specimens derived from Intensive Care Units, wards and hospital setting of Deccan Eco Region of Southern India. Journal of Pharmaceutical Negative Results 2022, 295–305. [Google Scholar] [CrossRef]
- Sharma, A.; Agrawal, M. Colistin Susceptibility for Carbapenem-Resistan t Gram-Negative Bacilli; Comparative Study of E-test and Vitek 2 Compact™ with Broth Microdilution. Galore International Journal of Health Sciences an d Research 2019, 4, 110–115, P-ISSN: 2456-9321. [Google Scholar]
- Das, S.; Roy, S.; Roy, S.; Goelv, G.; Sinha, S.; Mathur, P.; Walia, K.; Bhattacharya, S. Colistin susceptibility testing of gram-negative bacilli: Better performance of vitek2 system than E-test compared to broth microdilution method as the gold standard test. Indian journal of medical microbiology 2020, 38, 58–65. [Google Scholar] [CrossRef]
- Chakraverti, T.K.; Tripathi, P.C. Prevalence and Antibiotic Resistant Pattern of Pseudomonas aeruginosa at a Tertiary Care Centre of North India. Int. J. Curr. Microbiol. App. Sci 2018, 7, 1061–1069. [Google Scholar] [CrossRef]
- Choudhary, A.K. Isolation prevalence and antimicrobial resistance pattern of pseudomonas aeruginosa from urine samples in a tertiary care hospital udaipur, rajasthan. International Journal of Life Sciences, Biotechnology and Pharma Research, 2023, 12, 3, 2250-3137.
- Satpathy, M.; Sharma, N.; Kaur, P.; Arora, A. Detection of antimicrobial resistance genes in extended spectrum beta-lactamase-producing Escherichia coli from milk of indigenous Beetal goats of Punjab. Iranian Journal of Veterinary Research 2023, 24, 37. [Google Scholar] [CrossRef]
- Malik, S.; Rana, J.S.; Nehra, K. Prevalence and antibiotic susceptibility pattern of uropathogenic Escherichia coli strains in Sonipat region of Haryana in India. Biomedical and Biotechnology Research Journal (BBRJ) 2021, 5, 80–87. [Google Scholar] [CrossRef]
- Rudrapathy, P.; Samuel, S.; Murugesan, S. Microbiological Profile and Antibiotic Resistance of Bloodstream Infections among Cancer Patients at a Tertiary Care Cancer Centre in North Kerala, India. The Hospital 2022, 6, 16. [Google Scholar] [CrossRef]
- Kar, P.; Behera, B.; Mohanty, S.; Jena, J.; Mahapatra, A. Detection of colistin resistance in carbapenem resistant Enterobacteriaceae by reference broth microdilution and comparative evaluation of three other methods. Journal of Laboratory Physicians 2021, 13, 263–269. [Google Scholar] [CrossRef]
- Gupta, M.; Naik, A.K.; Singh, S.K. Bacteriological profile and antimicrobial resistance patterns of burn wound infections in a tertiary care hospital. Heliyon 2019, 5. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Akhtar, N.; Kesharwani, R.; Chandra, P.; Ayub, A. The Antimicrobial Resistance Pattern of pathogens isolated from patients with Health Care Associated Infections in a Tertiary Care Hospital of Chhattisgarh, India. European Journal of Molecular & Clinical Medicine, 2022 9, ISSN 2515-8260.
- Kishore, N.; Kakati, B.; Khanduri, S. Study of colistin and tigecycline sensitivity for carbapenem resistant isolates of klebsiella pneumoniae (crkp) patients in the intensive care unit (icu) of hims, dehradun uttarakhand. Int J Acad Med Pharm 2022, 4, 71–75. [Google Scholar]
- Chauhan, R.; Sharma, P. Phenotypic detection of Metallo-β-lactamase (MBL) producers among multidrug resistant (MDR) strains of P. aeruginosa in Himachal Pradesh. Indian J Basic App Med Res 2013, 3, 303–313. [Google Scholar]
- Samom, P.; Preeti, G.; Laifangbam, S.; Nahakpam, M. Bacteriological Profile Andantibiogram of ICU Isolates in A Tertiary Care Institute in Manipur–A Six-Year Study. BLOOD, 023, 554, 20.47. https://www.ncbi.nlm.nih.gov/nlmcatalog/474373.
- Biswas, U.; Biswas, R.; Samanta, I.; Das, S. Global emergence of mcr-mediated colistin resistance in gram-negative bacteria: focusing the current status in India under one health lens. 2022. [CrossRef]
- Keen III, E.F.; Robinson, B.J.; Hospenthal, D.R.; Aldous, W.K.; Wolf, S.E.; Chung, K.K.; Murray, C.K. Prevalence of multidrug-resistant organisms recovered at a military burn center. Burns 2010, 36, 819–825. [Google Scholar] [CrossRef]
- Falagas, M.E.; Rafailidis, P.I.; Matthaiou, D.K. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug resistance updates 2010, 13, 132–138. [Google Scholar] [CrossRef]
- Li, J.; Nation, R.L.; Turnidge, J.D.; Milne, R.W.; Coulthard, K.; Rayner, C.R.; Paterson, D.L. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. The Lancet infectious diseases 2006, 6, 589–601. [Google Scholar] [CrossRef]
- Walkty, A.; DeCorby, M.; Nichol, K.; Karlowsky, J.; Hoban, D.; Zhanel, G. In vitro activity of colistin (polymyxin E) against 3,480 isolates of gram-negative bacilli obtained from patients in Canadian hospitals in the CANWARD study, 2007-2008. Antimicrobial agents and chemotherapy 2009, 53, 4924–4926. [Google Scholar] [CrossRef]
- Kontopoulou, K.; Protonotariou, E.; Vasilakos, K.; Kriti, M.; Koteli, A.; Antoniadou, E.; Sofianou, D. Hospital outbreak caused by Klebsiella pneumoniae producing KPC-2 β-lactamase resistant to colistin. Journal of Hospital Infection 2010, 76, 70–73. [Google Scholar] [CrossRef]
- Maalej, S.M.; Meziou, M.R.; Mahjoubi, F.; Hammami, A. Epidemiological study of Enterobacteriaceae resistance to colistin in Sfax (Tunisia). Médecine et maladies infectieuses 2012, 42, 256–263. [Google Scholar] [CrossRef]
- Igumbor, E.; Gwanzura, L.; Chirara, M.; Obi, C.; Muza, D. Antibiotic sensitivity and plasmid profiles of Pseudomonas aeruginosa. The Central African Journal of Medicine 2000, 46, 296–300. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Hu, F.; Zhang, X.; Xu, X.; Liu, Y.; Zhu, D.; Wang, H. Independent emergence of colistin-resistant Enterobacteriaceae clinical isolates without colistin treatment. Journal of clinical microbiology 2011, 49, 4022–4023. [Google Scholar] [CrossRef] [PubMed]
- Vakili, B.; Fazeli, H.; Shoaei, P.; Yaran, M.; Ataei, B.; Khorvash, F.; Khaleghi, M. Detection of colistin sensitivity in clinical isolates of Acinetobacter baumannii in Iran. Journal of research in medical sciences: the official journal of Isfahan University of Medical Sciences 2014, 19, S67, https://pubmed.ncbi.nlm.nih.gov/25002899/. [Google Scholar] [PubMed]
- Lin, L.; Ling, B.-D.; Li, X.-Z. Distribution of the multidrug efflux pump genes, adeABC, adeDE and adeIJK, and class 1 integron genes in multiple-antimicrobial-resistant clinical isolates of Acinetobacter baumannii–Acinetobacter calcoaceticus complex. International journal of antimicrobial agents 2009, 33, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, A.; Gull, S.; Imran, H.; Khan, Z. Mechanistic Insights of Colistin Resistance and Its Public Health Implications. Applied Biochemistry and Microbiology 2023, 59, 597–607. [Google Scholar] [CrossRef]
- Noster, J.; Thelen, P.; Hamprecht, A. Detection of multidrug-resistant Enterobacterales—from ESBLs to carbapenemases. Antibiotics 2021, 10, 1140. [Google Scholar] [CrossRef]
- Feng, J.; Zhuang, Y.; Luo, J.; Xiao, Q.; Wu, Y.; Chen, Y.; Chen, M.; Zhang, X. Prevalence of colistin-resistant mcr-1-positive Escherichia coli isolated from children patients with diarrhoea in Shanghai, 2016–2021. Journal of Global Antimicrobial Resistance 2023, 34, 166–175. [Google Scholar] [CrossRef]
- McGann, P.; Snesrud, E.; Maybank, R.; Corey, B.; Ong, A.C.; Clifford, R.; Hinkle, M.; Whitman, T.; Lesho, E.; Schaecher, K.E. Escherichia coli harboring mcr-1 and bla CTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrobial agents and chemotherapy 2016, 60, 4420–4421. [Google Scholar] [CrossRef]
- Nordmann, P.; Jayol, A.; Poirel, L. A universal culture medium for screening polymyxin-resistant Gram-negative isolates. Journal of clinical microbiology 2016, 54, 1395–1399. [Google Scholar] [CrossRef]
- Caniaux, I.; Van Belkum, A.; Zambardi, G.; Poirel, L.; Gros, M. MCR: modern colistin resistance. European Journal of Clinical Microbiology & Infectious Diseases 2017, 36, 415–420. [Google Scholar] [CrossRef]
- Nordmann, P.; Jayol, A.; Poirel, L. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerging infectious diseases 2016, 22, 1038. [Google Scholar] [CrossRef]
- Jayol, A.; Dubois, V.; Poirel, L.; Nordmann, P. Rapid detection of polymyxin-resistant Enterobacteriaceae from blood cultures. Journal of clinical microbiology 2016, 54, 2273–2277. [Google Scholar] [CrossRef] [PubMed]
- Bontron, S.; Poirel, L.; Nordmann, P. Real-time PCR for detection of plasmid-mediated polymyxin resistance (mcr-1) from cultured bacteria and stools. Journal of Antimicrobial Chemotherapy 2016, 71, 2318–2320. [Google Scholar] [CrossRef] [PubMed]
- Nijhuis, R.; Veldman, K.; Schelfaut, J.; Van Essen-Zandbergen, A.; Wessels, E.; Claas, E.; Gooskens, J. Detection of the plasmid-mediated colistin-resistance gene mcr-1 in clinical isolates and stool specimens obtained from hospitalized patients using a newly developed real-time PCR assay. Journal of antimicrobial chemotherapy 2016, 71, 2344–2346. [Google Scholar] [CrossRef]
- Pogue, J.M.; Mynatt, R.P.; Marchaim, D.; Zhao, J.J.; Barr, V.O.; Moshos, J.; Sunkara, B.; Chopra, T.; Chidurala, S.; Kaye, K.S. Automated alerts coupled with antimicrobial stewardship intervention lead to decreases in length of stay in patients with gram-negative bacteremia. Infection Control & Hospital Epidemiology 2014, 35, 132–138. [Google Scholar] [CrossRef]
- Sorlí, L.; Luque, S.; Grau, S.; Berenguer, N.; Segura, C.; Montero, M.M.; Álvarez-Lerma, F.; Knobel, H.; Benito, N.; Horcajada, J.P. Trough colistin plasma level is an independent risk factor for nephrotoxicity: a prospective observational cohort study. BMC infectious diseases 2013, 13, 1–9. [Google Scholar] [CrossRef]
- Gu, W.-J.; Wang, F.; Tang, L.; Bakker, J.; Liu, J.-C. Colistin for the treatment of ventilator-associated pneumonia caused by multidrug-resistant Gram-negative bacteria: a systematic review and meta-analysis. International journal of antimicrobial agents 2014, 44, 477–485. [Google Scholar] [CrossRef]
- Nagaoka, R.; Ikawa, K.; Onodera, M.; Koba, Y.; Hara, T.; Joichi, Y.; Yokozaki, M.; Ohge, H.; Morikawa, N. In vitro combined effects of double antibacterial drugs against multidrug-resistant Pseudomonas aeruginosa isolates: comparison among combinations of colistin, arbekacin, aztreonam, rifampicin and piperacillin. The Japanese Journal of Antibiotics 2014, 67, 167–174. [Google Scholar] [PubMed]
- Batirel, A.; Balkan, I.; Karabay, O.; Agalar, C.; Akalin, S.; Alici, O.; Alp, E.; Altay, F.A.; Altin, N.; Arslan, F. Comparison of colistin–carbapenem, colistin–sulbactam, and colistin plus other antibacterial agents for the treatment of extremely drug-resistant Acinetobacter baumannii bloodstream infections. European journal of clinical microbiology & infectious diseases 2014, 33, 1311–1322. [Google Scholar] [CrossRef]
- Petrosillo, N.; Taglietti, F.; Granata, G. Treatment options for colistin resistant Klebsiella pneumoniae: present and future. Journal of clinical medicine 2019, 8, 934. [Google Scholar] [CrossRef]
- Kempf, I.; Fleury, M.A.; Drider, D.; Bruneau, M.; Sanders, P.; Chauvin, C.; Madec, J.-Y.; Jouy, E. What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? International journal of antimicrobial agents 2013, 42, 379–383. [Google Scholar] [CrossRef]
- Tornimbene, B.; Eremin, S.; Escher, M.; Griskeviciene, J.; Manglani, S.; Pessoa-Silva, C.L. WHO global antimicrobial resistance surveillance system early implementation 2016–17. The Lancet Infectious Diseases 2018, 18, 241–242. [Google Scholar] [CrossRef] [PubMed]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q.M.R. Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Frontiers in medicine 2021, 8, 677720. [Google Scholar] [CrossRef] [PubMed]
- Claudia, S.-S.; Carmen, S.-S.; Andrés, D.; Marcela, M.-A.; Kerly, C.-A.; Bryan, B.M.; John, C.J.; José, G.-F. Risk factors associated with colistin resistance in carbapenemase-producing Enterobacterales: a multicenter study from a low-income country. Annals of Clinical Microbiology and Antimicrobials 2023, 22, 64. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Sharma, D.; Singh, A.; Sunita, K. Colistin resistance and management of drug resistant infections. Canadian Journal of Infectious Diseases and Medical Microbiology 2022, 2022, 4315030. [Google Scholar] [CrossRef]
- Novović, K.; Jovčić, B. Colistin resistance in Acinetobacter baumannii: molecular mechanisms and epidemiology. Antibiotics 2023, 12, 516. [Google Scholar] [CrossRef]
- Mudenda, S.; Chabalenge, B.; Daka, V.; Mfune, R.L.; Salachi, K.I.; Mohamed, S.; Mufwambi, W.; Kasanga, M.; Matafwali, S.K. Global strategies to combat antimicrobial resistance: a one health perspective. Pharmacology & Pharmacy 2023, 14, 271–328. [Google Scholar] [CrossRef]
- Vasala, A.; Hytönen, V.P.; Laitinen, O.H. Modern tools for rapid diagnostics of antimicrobial resistance. Frontiers in cellular and infection microbiology 2020, 10, 308. [Google Scholar] [CrossRef]
- Mashalla, Y.; Setlhare, V.; Massele, A.; Sepako, E.; Tiroyakgosi, C.; Kgatlwane, J.; Chuma, M.; Godman, B. Assessment of prescribing practices at the primary healthcare facilities in Botswana with an emphasis on antibiotics: findings and implications. International journal of clinical practice 2017, 71, e13042. [Google Scholar] [CrossRef]
- Van Belkum, A.; Bachmann, T.T.; Lüdke, G.; Lisby, J.G.; Kahlmeter, G.; Mohess, A.; Becker, K.; Hays, J.P.; Woodford, N.; Mitsakakis, K. Developmental roadmap for antimicrobial susceptibility testing systems. Nature Reviews Microbiology 2019, 17, 51–62. [Google Scholar] [CrossRef]
- Nijhuis, R.; Guerendiain, D.; Claas, E.; Templeton, K. Comparison of ePlex respiratory pathogen panel with laboratory-developed real-time PCR assays for detection of respiratory pathogens. Journal of clinical microbiology 2017, 55, 1938–1945. [Google Scholar] [CrossRef]
- Van Belkum, A.; Rochas, O. Laboratory-based and point-of-care testing for MSSA/MRSA detection in the age of whole genome sequencing. Frontiers in microbiology 2018, 9, 1437. [Google Scholar] [CrossRef] [PubMed]
- Ellington, M.; Ekelund, O.; Aarestrup, F.M.; Canton, R.; Doumith, M.; Giske, C.; Grundman, H.; Hasman, H.; Holden, M.; Hopkins, K.L. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clinical microbiology and infection 2017, 23, 2–22. [Google Scholar] [CrossRef] [PubMed]
- Mondal, A.H.; Khare, K.; Saxena, P.; Debnath, P.; Mukhopadhyay, K.; Yadav, D. A Review on Colistin Resistance: An Antibiotic of Last Resort. Microorganisms 2024, 12, 772. [Google Scholar] [CrossRef] [PubMed]
- Ajayi, A.O.; Odeyemi, A.T.; Akinjogunla, O.J.; Adeyeye, A.B.; Ayo-Ajayi, I. Review of antibiotic-resistant bacteria and antibiotic resistance genes within the one health framework. Infection Ecology & Epidemiology 2024, 14, 2312953. [Google Scholar] [CrossRef]
- Behera IC, Swain SK, Chandra M. Incidence of colistin-resistant Acinetobacter baumannii in an Indian tertiary care teaching hospital. Int J Adv Res (Indore), 2394.
- Mmatli, M.; Mbelle, N.M.; Osei Sekyere, J. Global epidemiology, genetic environment, risk factors and therapeutic prospects of mcr genes: A current and emerging update. Frontiers in Cellular and Infection Microbiology 2022, 12, 941358. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, T.; Wang, C.; Liang, G.; Lu, Q.; Wen, G.; Guo, Y.; Cheng, Y.; Wang, Z.; Shao, H. Prevalence of colistin resistance gene mcr-1 in Escherichia coli isolated from chickens in central China, 2014 to 2019. Journal of Global Antimicrobial Resistance 2022, 29, 241–246. [Google Scholar] [CrossRef]
- Hamame, A.; Davoust, B.; Cherak, Z.; Rolain, J.-M.; Diene, S.M. Mobile colistin resistance (mcr) genes in cats and dogs and their zoonotic transmission risks. Pathogens 2022, 11, 698. [Google Scholar] [CrossRef]
- Cohn, J.; Mendelson, M.; Kanj, S.S.; Shafiq, N.; Boszczowski, I.; Laxminarayan, R. Accelerating antibiotic access and stewardship: a new model to safeguard public health. The Lancet Infectious Diseases 2024. [Google Scholar] [CrossRef]
- Karakonstantis, S.; Kritsotakis, E.I.; Gikas, A. Treatment options for K. pneumoniae, P. aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: an approach based on the mechanisms of resistance to carbapenems. Infection 2020, 48, 835–851. [Google Scholar] [CrossRef]
- Protonotariou, E.; Meletis, G.; Pilalas, D.; Mantzana, P.; Tychala, A.; Kotzamanidis, C.; Papadopoulou, D.; Papadopoulos, T.; Polemis, M.; Metallidis, S. Polyclonal endemicity of carbapenemase-producing Klebsiella pneumoniae in ICUs of a Greek tertiary care hospital. Antibiotics 2022, 11, 149. [Google Scholar] [CrossRef]
- Luo, Q.; Wang, Y.; Xiao, Y. Prevalence and transmission of mobilized colistin resistance (mcr) gene in bacteria common to animals and humans. Biosafety and Health 2020, 2, 71–78. [Google Scholar] [CrossRef]
- Andrade, F.F.; Silva, D.; Rodrigues, A.; Pina-Vaz, C. Colistin update on its mechanism of action and resistance, present and future challenges. Microorganisms 2020, 8, 1716. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Humphries, R. Clinical and laboratory considerations for the rapid detection of carbapenem-resistant Enterobacteriaceae. Virulence 2017, 8, 427–439. [Google Scholar] [CrossRef]
- Mathew, P.; Jaguga, C.; Mpundu, M.; Chandy, S.J. Building knowledge and evidence base on antimicrobial resistance in Africa, through ‘One Health’based surveillance. Clinical Epidemiology and Global Health 2020, 8, 313–317. [Google Scholar] [CrossRef]
- Allegranzi, B.; Pittet, D. Role of hand hygiene in healthcare-associated infection prevention. Journal of hospital infection 2009, 73, 305–315. [Google Scholar] [CrossRef]
- Giacobbe, D.R.; Mikulska, M.; Viscoli, C. Recent advances in the pharmacological management of infections due to multidrug-resistant Gram-negative bacteria. Expert review of clinical pharmacology 2018, 11, 1219–1236. [Google Scholar] [CrossRef]
- Talat, A.; Khan, F.; Khan, A.U. Genome analyses of colistin-resistant high-risk bla NDM-5 producing Klebsiella pneumoniae ST147 and Pseudomonas aeruginosa ST235 and ST357 in clinical settings. BMC microbiology 2024, 24, 174. [Google Scholar] [CrossRef]


| Mechanism | Description |
|---|---|
| Modification of LPS | Addition of positively charged molecules to lipopolysaccharide (LPS) structure, reducing colistin binding |
| Mutations in pmrAB | Alterations in regulatory genes controlling LPS modification |
| Mutations in mgrB | Loss of function mutations in the negative regulator of PhoPQ signaling pathway |
| Plasmid-mediated mechanisms | Acquisition of mobile genetic elements carrying resistance genes |
| Efflux pump overexpression | Increased expression of efflux pumps, reducing intracellular colistin concentration |
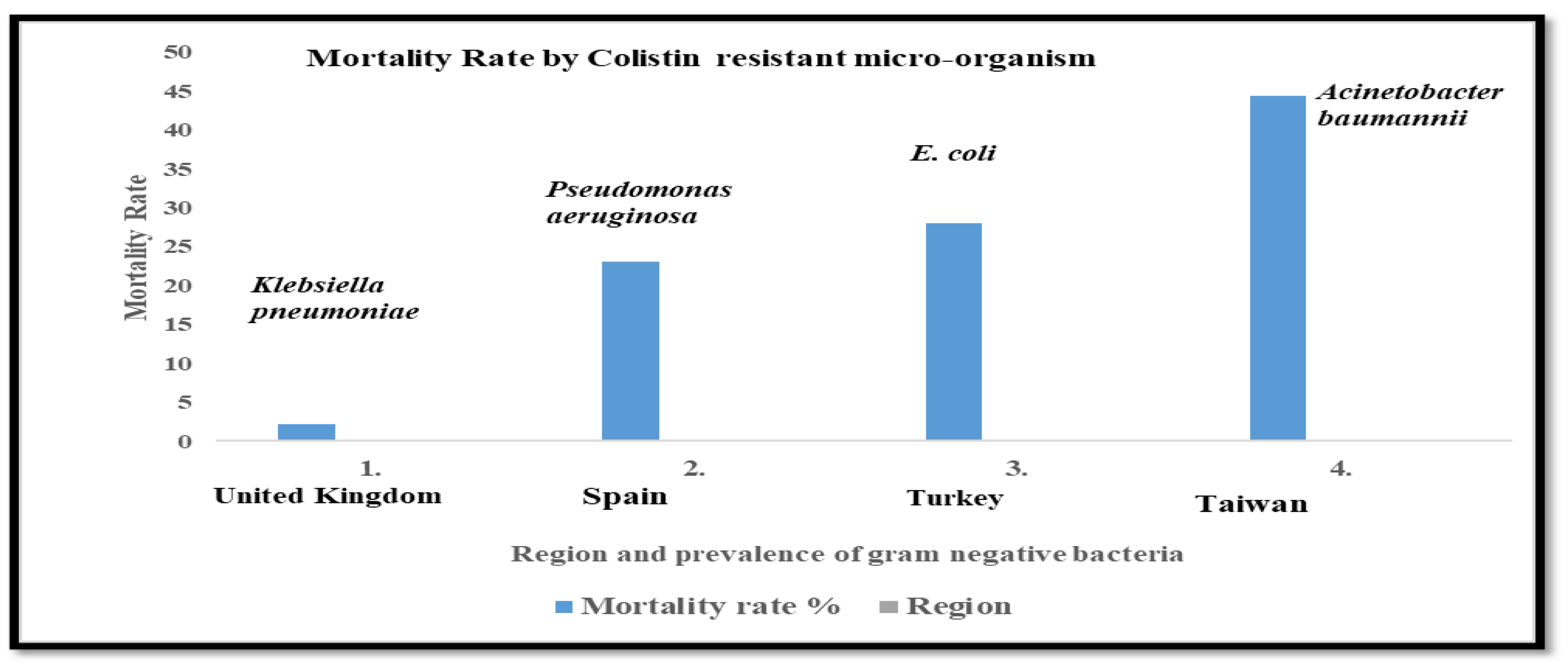
| S.No. | Mortality Rate % | Colistin Resistance Associated Infection (Micro-Organism Name) |
Region | Reference |
|---|---|---|---|---|
| 1. | 2.15 | Klebsiella pneumoniae | United Kingdom | [30] |
| 2. | 23.1 | Pseudomonas aeruginosa | Spain | [31] |
| 3. | 28 | E. coli | Turkey | [32] |
| 4. | 44.4 | Acinetobacter baumannii | Taiwan | [23] |
| Implication | Description |
|---|---|
| Limited Treatment Options | Reduced efficacy of colistin and polymyxins against resistant strains |
| Increased Morbidity/Mortality | Higher rates of treatment failure and patient mortality |
| Spread of Resistance | Potential for dissemination of resistant strains within healthcare settings |
| Need for Surveillance | Importance of monitoring colistin resistance rates for infection control |
| Development of Novel Therapies | Urgency for research into alternative treatment options |
| Region | Prevalence of Resistance (%) | Contributing Factors |
|---|---|---|
| North America | Low | Stringent antibiotic stewardship practices |
| Europe | Moderate | Increased use of colistin in agriculture |
| Asia | High | Widespread use of colistin in human medicine and agriculture |
| Africa | Varies | Limited surveillance and healthcare resources |
| South America | Varies | Variable regulatory oversight of antibiotic use |
| Country | Prevalence of Colistin Resistance (%) | References |
|---|---|---|
| United States | 69.2 % | [34] |
| China | 32.7 % | [35] |
| India | 1.28 % | [36] |
| Japan | 7.7 % | [37] |
| Thailand | 3.3 % | [38] |
| South Korea | 4.4 % | [39] |
| Russia | 30 % | [40] |
| State | Prevalence of Colistin Resistance (%) | Reference |
|---|---|---|
| Delhi | 15 | [41] |
| Tamil Nadu | 33 | [42] |
| Karnataka | 1.6 | [43] |
| Jaipur | 6.2 | [44] |
| West Bengal | 22.5 | [45] |
| Madhya Pradesh | 17 | [46] |
| Rajasthan | 17.4 | [47] |
| Punjab | 12.5 | [48] |
| Haryana | 3.8 | [49] |
| Kerala | 25 | [50] |
| Odisha | 13.5 | [51] |
| Jharkhand | 9.09 | [52] |
| Chhattisgarh | 0 | [53] |
| Uttarakhand | 8 | [54] |
| Himachal Pradesh | 11.76 | [55] |
| Manipur | 2 | [56] |
| Meghalaya | 2.5 | [57] |
| Method | Description |
|---|---|
| MIC Determination | Minimum inhibitory concentration testing to assess bacterial susceptibility to colistin |
| Whole-genome sequencing | Comprehensive analysis of bacterial genomes for resistance determinants |
| PCR-based assays | Polymerase chain reaction assays targeting specific resistance genes |
| Phenotypic Screening | High-throughput screening methods to detect resistant isolates |
| Epidemiological Surveillance | Monitoring of resistance trends in healthcare settings and communities |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





