Submitted:
28 March 2025
Posted:
31 March 2025
You are already at the latest version
Abstract
Keywords:
1. Background
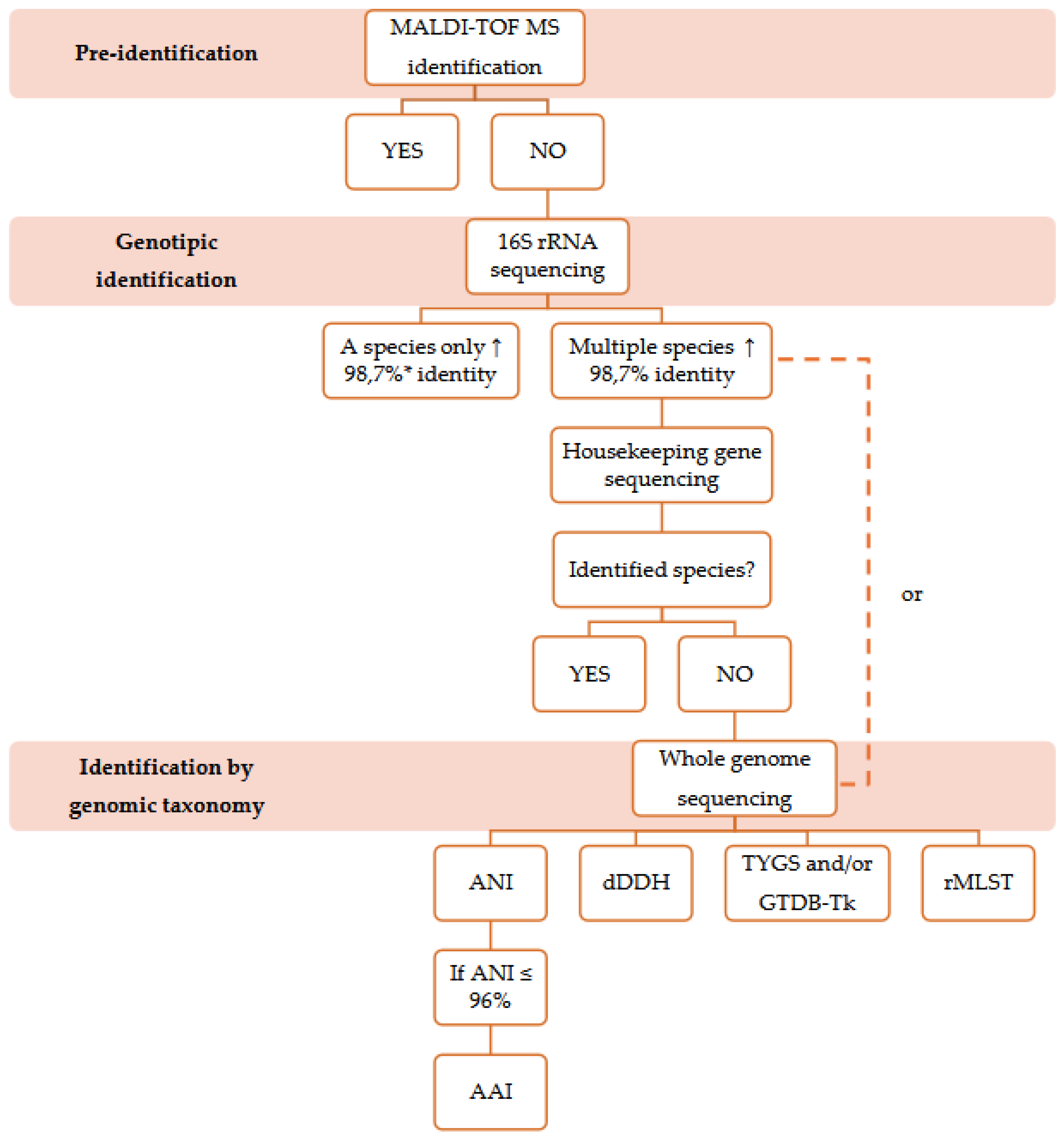
2. Phenotypic Identification
3. MALDI-TOF MS
4. Analysis of the 16S Ribosomal Gene Sequences
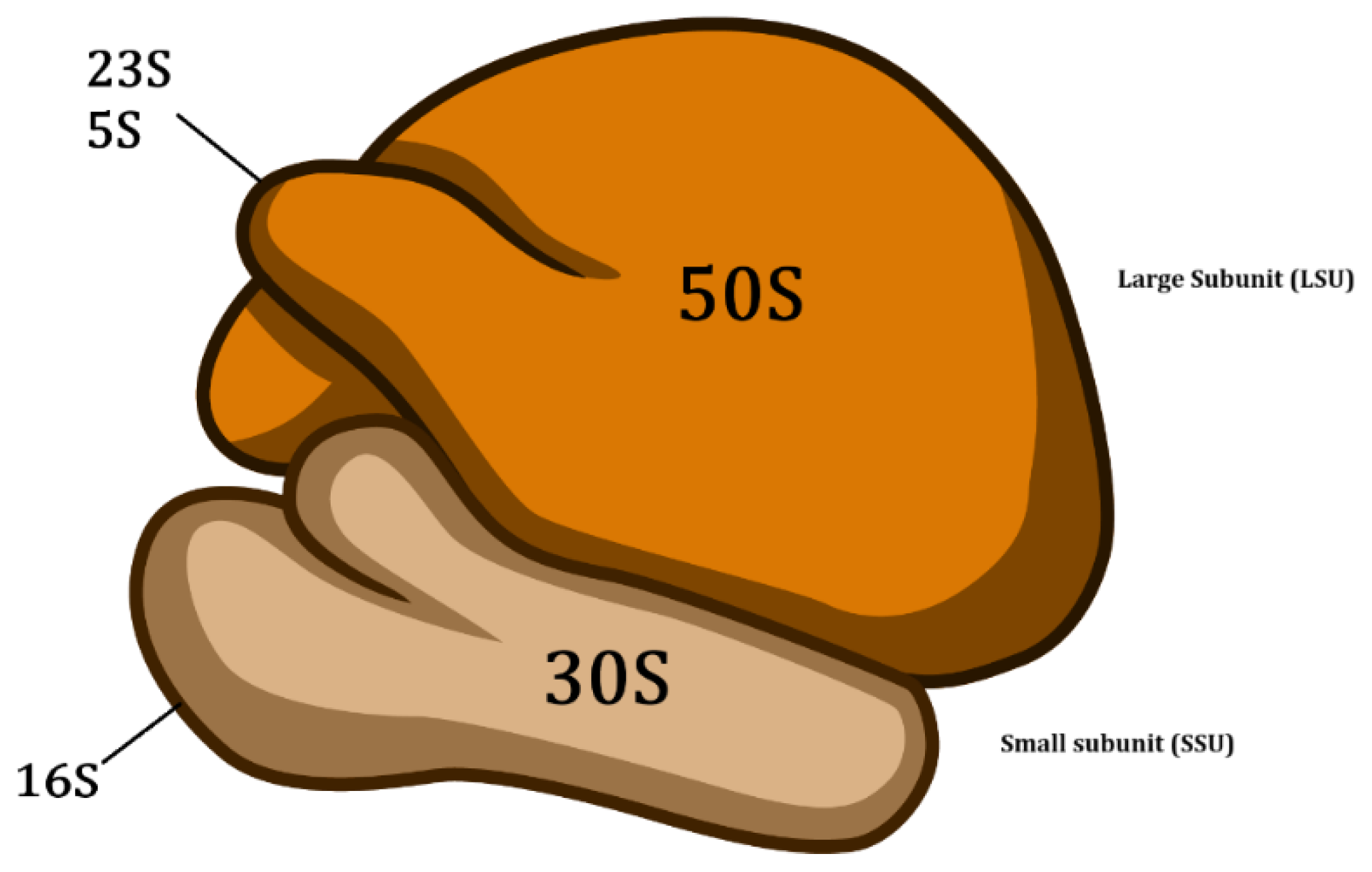
5. Analysis of the Housekeeping Gene Sequences
6. Genomic Taxonomy Tools
7. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AAI | Average Amino Acid Identity |
| ANI | Average Nucleotide Identity |
| ATCC | American Type Culture Collection |
| BacDive | Bacterial Diversity Metadatabase |
| BLAST | Basic Local Alignment Search tool |
| bp | Base pairs |
| DDBJ | DNA Data Bank of Japan |
| dDDH | digital DNA-DNA hybridization |
| DDH | Hibridization DNA-DNA |
| DSMZ | Deutsche Sammlung von Mikroorganismen und Zellkulturen |
| EMBL-EBI | European Molecular Biology Laboratory – European Bioinformatics Institute |
| G+C | Guanine and cytosine |
| GBDP | Genome BLAST Distance Phylogeny |
| GGDC | Genome-to-Genome Distance Calculator |
| GMPs | Good manufacturing practices |
| GTDB | Genome Taxonomy Database |
| GTDB-Tk | Genome Taxonomy Database Toolkit |
| JGI | Joint Genome Institute |
| LPSN | List of Prokatiotic Names with Standing in Nomenclature |
| MALDI-TOF MS | Matrix-Assisted Laser Desorption Ionization–Time of Flight/Mass Spectrometry |
| MBP | multigene-based phylogenies |
| MLSA | multilocus sequence analysis |
| MLST | multilocus sequence typing |
| NCBI | National Center for Biotechnology Information |
| ORFs | open reading frames |
| RED | Relative Evolutionary Divergence |
| rMLST | Ribosomal Multilocus Sequence Typing |
| rRNA | Ribosomal RNA |
| TYGS | Type Strain Genome Server |
| WGS | Whole-genome sequencing |
References
- Costa, L.V. da; Miranda, R.V. da S. L. de; Reis, C.M.F. dos; Andrade, J.M. de; Cruz, F.V.; Frazão, A.M.; Fonseca, E.L. da; Ramos, J.N.; Brandão, M.L.L.; Vieira, V.V. MALDI-TOF MS Database Expansion for Identification of Bacillus and Related Genera Isolated from a Pharmaceutical Facility. J Microbiol Methods 2022, 203. [Google Scholar] [CrossRef]
- United States Pharmacopeial Convention. In The United States Pharmacopeia 43rd ed; The United States Pharmacopeia.
- Caldeira, N.G.S.; de Souza, M.L.S.; de Miranda, R.V.d.S. L.; da Costa, L.V.; Forsythe, S.J.; Zahner, V.; Brandão, M.L.L. Characterization by MALDI-TOF MS and 16S RRNA Gene Sequencing of Aerobic Endospore-Forming Bacteria Isolated from Pharmaceutical Facility in Rio de Janeiro, Brazil. Microorganisms 2024, 12, 724. [Google Scholar] [CrossRef]
- Food and Drug Administration - FDA. Guidance for Industry Sterile Drug Products Produced by Aseptic Processing —Current Good Manufacturing Practice,. ed by Aseptic Processing —Current Good Manufacturing Practice,.
- European Medicines Agency. The Rules Governing Medicinal Products in the European Union. Volume 4: European Union Guidelines for Good Manufacturing Practice for Medicinal Products for Human and Veterinary Use. Annex 1: Manufacture of Sterile Medicinal Products.. The Rules Governing Medicinal Products in the European Union. Volume 4: European Union Guidelines for Good Manufacturing Practice for Medicinal Products for Human and Veterinary Use. Annex 1: Manufacture of Sterile Medicinal Products.
- Thompson, C.C.; Vidal, L.; Salazar, V.; Swings, J.; Thompson, F.L. Microbial Genomic Taxonomy. In Trends in the systematics of bacteria and fungi; CABI: UK, 2021; pp. 168–178. [Google Scholar] [CrossRef]
- Moreira, F.M.; Pereira, P. de A.; Miranda, R.V. da S. L. de; Reis, C.M.F. dos; Braga, L.M.P. da S.; de Andrade, J.M.; do Nascimento, L.G.; Mattoso, J.M.V.; Forsythe, S.J.; da Costa, L.V.; et al. Evaluation of MALDI-TOF MS, Sequencing of D2 LSU RRNA and Internal Transcribed Spacer Regions (ITS) for the Identification of Filamentous Fungi Isolated from a Pharmaceutical Facility. J Pharm Biomed Anal 2023, 234, 115531. [Google Scholar] [CrossRef]
- Costa, L.V. da; Ramos, J.N.; Albuquerque, L. de S.; Miranda, R.V. da S. L. de; Valadão, T.B.; Veras, J.F.C.; Vieira, E.M.D.; Forsythe, S.; Brandão, M.L.L.; Vieira, V.V. Bacillus Lumedeiriae Sp. Nov., a Gram-Positive, Spore-Forming Rod Isolated from a Pharmaceutical Facility Production Environment and Added to the MALDI Biotyper® Database. Microorganisms 2024, 12, 2507. [Google Scholar] [CrossRef]
- Obasi, A.; Nwachukwu, S.; Ugoji, E.; Kohler, C.; Göhler, A.; Balau, V.; Pfeifer, Y.; Steinmetz, I. Extended-Spectrum β-Lactamase-Producing Klebsiella Pneumoniae from Pharmaceutical Wastewaters in South-Western Nigeria. Microbial Drug Resistance 2017, 23, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- da Silva Lage de Miranda, R.V.; da Costa, L.V.; de Sousa Albuquerque, L.; dos Reis, C.M.F.; da Silva Braga, L.M.P.; de Andrade, J.M.; Ramos, J.N.; Mattoso, J.M.V.; Forsythe, S.J.; Brandão, M.L.L. Identification of Sutcliffiella Horikoshii Strains in an Immunobiological Pharmaceutical Industry Facility. Lett Appl Microbiol 2023, 76. [Google Scholar] [CrossRef]
- Costa, L.V. da; Miranda, R.V. da S. L. de; Fonseca, E.L. da; Gonçalves, N.P.; Reis, C.M.F. dos; Frazão, A.M.; Cruz, F.V.; Brandão, M.L.L.; Ramos, J.N.; Vieira, V.V. Assessment of VITEK® 2, MALDI-TOF MS and Full Gene 16S RRNA Sequencing for Aerobic Endospore-Forming Bacteria Isolated from a Pharmaceutical Facility. J Microbiol Methods 2022, 194, 106419. [Google Scholar] [CrossRef]
- Sala-Comorera, L.; Vilaró, C.; Galofré, B.; Blanch, A.R.; García-Aljaro, C. Use of Matrix-Assisted Laser Desorption/Ionization–Time of Flight (MALDI–TOF) Mass Spectrometry for Bacterial Monitoring in Routine Analysis at a Drinking Water Treatment Plant. Int J Hyg Environ Health 2016, 219, 577–584. [Google Scholar] [CrossRef]
- Vithanage, N.R.; Yeager, T.R.; Jadhav, S.R.; Palombo, E.A.; Datta, N. Comparison of Identification Systems for Psychrotrophic Bacteria Isolated from Raw Bovine Milk. Int J Food Microbiol 2014, 189, 26–38. [Google Scholar] [CrossRef]
- Biomérieux. VITEK® 2. VITEK® 2. Fully integrated Identification and Antimicrobial Susceptibility Testing.
- Bosshard, P.P.; Zbinden, R.; Abels, S.; Böddinghaus, B.; Altwegg, M.; Böttger, E.C. 16S RRNA Gene Sequencing versus the API 20 NE System and the VITEK 2 ID-GNB Card for Identification of Nonfermenting Gram-Negative Bacteria in the Clinical Laboratory. J Clin Microbiol 2006, 44, 1359–1366. [Google Scholar] [CrossRef]
- Seuylemezian, A.; Aronson, H.S.; Tan, J.; Lin, M.; Schubert, W.; Vaishampayan, P. Development of a Custom MALDI-TOF MS Database for Species-Level Identification of Bacterial Isolates Collected From Spacecraft and Associated Surfaces. Front Microbiol 2018, 9. [Google Scholar] [CrossRef]
- Zasada, A.A.; Mosiej, E. Contemporary Microbiology and Identification of Corynebacteria Spp. Causing Infections in Human. Lett Appl Microbiol 2018, 66, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Shah, H.N.; Shah, A.J.; Belgacem, O.; Ward, M.; Dekio, I.; Selami, L.; Duncan, L.; Bruce, K.; Xu, Z.; Mkrtchyan, H.V.; et al. MALDI-TOF MS and Currently Related Proteomic Technologies in Reconciling Bacterial Systematics. In Trends in the systematics of bacteria and fungi; CABI: UK, 2021; pp. 93–118. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Ebers, J. Taxonomic Parameters Revisited: Tarnished Gold Standards – ScienceOpen. Microbial Today. 2006, p. 152. Available online: https://www.scienceopen.com/document?vid=0cf4b084-5683-4ef4-a80c-c0df44a135dc (accessed on 17 March 2025).
- Lasch, P.; Stämmler, M.; Schneider, A. A MALDI-TOF Mass Spectrometry Database for Identification and Classification of Highly Pathogenic Microorganisms from the Robert Koch-Institute (RKI). [CrossRef]
- Caamaño-Antelo, S.; Fernández-No, I.C.; Böhme, K.; Ezzat-Alnakip, M.; Quintela-Baluja, M.; Barros-Velázquez, J.; Calo-Mata, P. Genetic Discrimination of Foodborne Pathogenic and Spoilage Bacillus Spp. Based on Three Housekeeping Genes. Food Microbiol 2015, 46, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Rajendhran, J.; Gunasekaran, P. Microbial Phylogeny and Diversity: Small Subunit Ribosomal RNA Sequence Analysis and Beyond. Microbiol Res 2011, 166, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Church, D.L.; Cerutti, L.; Gürtler, A.; Griener, T.; Zelazny, A.; Emler, S. Performance and Application of 16S rRNA Gene Cycle Sequencing for Routine Identification of Bacteria in the Clinical Microbiology Laboratory. Clin Microbiol Rev 2020, 33, e00053–19. [Google Scholar] [CrossRef]
- Madigan, M.; Martinko, J.; Bender, K.; Buckley, D.; Stahl, D. Brock Biology of Microorganisms, 14th ed.; Benjamin Cummings, 2015.
- Mahato, N.K.; Gupta, V.; Singh, P.; Kumari, R.; Verma, H.; Tripathi, C.; Rani, P.; Sharma, A.; Singhvi, N.; Sood, U.; et al. Microbial Taxonomy in the Era of OMICS: Application of DNA Sequences, Computational Tools and Techniques. Antonie Van Leeuwenhoek 2017, 110, 1357–1371. [Google Scholar] [CrossRef]
- D’Amore, R.; Ijaz, U.Z.; Schirmer, M.; Kenny, J.G.; Gregory, R.; Darby, A.C.; Shakya, M.; Podar, M.; Quince, C.; Hall, N. A Comprehensive Benchmarking Study of Protocols and Sequencing Platforms for 16S RRNA Community Profiling. BMC Genomics 2016, 17, 55. [Google Scholar] [CrossRef]
- Rodrigues, N.M.B.; Bronzato, G.F.; Santiago, G.S.; Botelho, L.A.B.; Moreira, B.M.; Coelho, I. da S.; Souza, M.M.S. de; Coelho, S. de M. de O. The Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry (MALDI-TOF MS) Identification versus Biochemical Tests: A Study with Enterobacteria from a Dairy Cattle Environment. Brazilian Journal of Microbiology 2016, 48, 132. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Mondotte, J.A.; Fazio, L.L.; Jetten, M. Authors Need to Be Prudent When Assigning Names to Microbial Isolates. Curr Microbiol 2021, 78, 4005–4008. [Google Scholar] [CrossRef]
- NIAID Visual & Medical Arts. Ribosome, NIAID NIH BIOART Source.
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.J.; Ludwig, W.; Kämpfer, P. Notes on the Characterization of Prokaryote Strains for Taxonomic Purposes. Int J Syst Evol Microbiol 2010, 60 Pt 1, 249–266. [Google Scholar] [CrossRef]
- Sentausa, E.; Fournier, P.E. Advantages and Limitations of Genomics in Prokaryotic Taxonomy. Clinical Microbiology and Infection 2013, 19, 790–795. [Google Scholar] [CrossRef] [PubMed]
- Vlach, J.; Javůrková, B.; Karamonová, L.; Blažková, M.; Fukal, L. Novel PCR-RFLP System Based on RpoB Gene for Differentiation of Cronobacter Species. Food Microbiol 2017, 62, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Payne, G.W.; Vandamme, P.; Morgan, S.H.; LiPuma, J.J.; Coenye, T.; Weightman, A.J.; Jones, T.H.; Mahenthiralingam, E. Development of a RecA Gene-Based Identification Approach for the Entire Burkholderia Genus. Appl Environ Microbiol 2005, 71, 3917–3927. [Google Scholar] [CrossRef]
- Chun, J.; Rainey, F.A. Integrating Genomics into the Taxonomy and Systematics of the Bacteria and Archaea. Int J Syst Evol Microbiol 2014, 64, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Glaeser, S.P.; Kämpfer, P. Multilocus Sequence Analysis (MLSA) in Prokaryotic Taxonomy. Syst Appl Microbiol 2015, 38, 237–245. [Google Scholar] [CrossRef]
- Maiden, M.C.J.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russell, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.; Zurth, K.; Caugant, D.A.; et al. Multilocus Sequence Typing: A Portable Approach to the Identification of Clones within Populations of Pathogenic Microorganisms. Proc Natl Acad Sci U S A 1998, 95, 3140–3145. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Frederiksen, W.; Garrity, G.M.; Grimont, P.A.D.; Kämpfer, P.; Maiden, M.C.J.; Nesme, X.; Rosselló-Mora, R.; Swings, J.; Trüper, H.G.; et al. Report of the Ad Hoc Committee for the Re-Evaluation of the Species Definition in Bacteriology. Int J Syst Evol Microbiol 2002, 52, 1043–1047. [Google Scholar] [CrossRef]
- Hayashi Sant’Anna, F.; Bach, E.; Porto, R.Z.; Guella, F.; Hayashi Sant’Anna, E.; Passaglia, L.M.P. Genomic Metrics Made Easy: What to Do and Where to Go in the New Era of Bacterial Taxonomy. Crit Rev Microbiol 2019, 45, 182–200. [Google Scholar] [CrossRef]
- Thompson, C.C.; Chimetto, L.; Edwards, R.A.; Swings, J.; Stackebrandt, E.; Thompson, F.L. Microbial Genomic Taxonomy. BMC Genomics 2013, 14. [Google Scholar] [CrossRef]
- Land, M.; Hauser, L.; Jun, S.R.; Nookaew, I.; Leuze, M.R.; Ahn, T.H.; Karpinets, T.; Lund, O.; Kora, G.; Wassenaar, T.; et al. Insights from 20 Years of Bacterial Genome Sequencing. Funct Integr Genomics 2015, 15, 141. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.C.; Murray, R.G.E.; Stackebrandt, E.; et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int J Syst Evol Microbiol 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed Minimal Standards for the Use of Genome Data for the Taxonomy of Prokaryotes. Int J Syst Evol Microbiol 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Gosselin, S.; Fullmer, M.S.; Feng, Y.; Gogarten, J.P. Improving Phylogenies Based on Average Nucleotide Identity, Incorporating Saturation Correction and Nonparametric Bootstrap Support. Syst Biol 2022, 71, 396–409. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res 2022, 50, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; ran, Q.; Du, X.; Wu, S.; Wang, J.; Sheng, D.; Chen, Q.; Du, Z.; Li, Y. zhong. Two New Polyangium Species, P. Aurulentum Sp. Nov. and P. Jinanense Sp. Nov., Isolated from a Soil Sample. Syst Appl Microbiol 2021, 44. [Google Scholar] [CrossRef] [PubMed]
- Cuny, H.; Offret, C.; Boukerb, A.M.; Parizadeh, L.; Lesouhaitier, O.; Le Chevalier, P.; Jégou, C.; Bazire, A.; Brillet, B.; Fleury, Y. Pseudoalteromonas Ostreae Sp. Nov., a New Bacterial Species Harboured by the Flat Oyster Ostrea Edulis. Int J Syst Evol Microbiol 2021, 71. [Google Scholar] [CrossRef]
- Colston, S.M.; Fullmer, M.S.; Beka, L.; Lamy, B.; Peter Gogarten, J.; Graf, J. Bioinformatic Genome Comparisons for Taxonomic and Phylogenetic Assignments Using Aeromonas as a Test Case. mBio 2014, 5. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA Hybridization Values and Their Relationship to Whole-Genome Sequence Similarities. Int J Syst Evol Microbiol 2007, 57 Pt 1, 81–91. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic Insights That Advance the Species Definition for Prokaryotes. Proc Natl Acad Sci U S A 2005, 102, 2567–2572. [Google Scholar] [CrossRef]
- Qin, Q.L.; Xie, B. Bin; Zhang, X.Y.; Chen, X.L.; Zhou, B.C.; Zhou, J.; Oren, A.; Zhang, Y.Z. A Proposed Genus Boundary for the Prokaryotes Based on Genomic Insights. J Bacteriol 2014, 196, 2210. [Google Scholar] [CrossRef]
- Kim, D.; Park, S.; Chun, J. Introducing EzAAI: A Pipeline for High Throughput Calculations of Prokaryotic Average Amino Acid Identity. J Microbiol 2021, 59, 476–480. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Konstantinidis, K.T. Bypassing Cultivation To Identify Bacterial Species: Culture-Independent Genomic Approaches Identify Credibly Distinct Clusters, Avoid Cultivation Bias, and Provide True Insights into Microbial Species. Microbe Magazine 2014, 9, 111–118. [Google Scholar] [CrossRef]
- Nicholson, A.C.; Gulvik, C.A.; Whitney, A.M.; Humrighouse, B.W.; Bell, M.E.; Holmes, B.; Steigerwalt, A.G.; Villarma, A.; Sheth, M.; Batra, D.; et al. Division of the Genus Chryseobacterium: Observation of Discontinuities in Amino Acid Identity Values, a Possible Consequence of Major Extinction Events, Guides Transfer of Nine Species to the Genus Epilithonimonas, Eleven Species to the Genus Kaistella, and Three Species to the Genus Halpernia Gen. Nov., with Description of Kaistella Daneshvariae Sp. Nov. and Epilithonimonas Vandammei Sp. Nov. Derived from Clinical Specimens. Int J Syst Evol Microbiol 2020, 70, 4432–4450. [Google Scholar] [CrossRef]
- Walter, J.M.; Coutinho, F.H.; Dutilh, B.E.; Swings, J.; Thompson, F.L.; Thompson, C.C. Ecogenomics and Taxonomy of Cyanobacteria Phylum. Front Microbiol 2017, 8, 2132. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Riesco, R.; Trujillo, M.E. Update on the Proposed Minimal Standards for the Use of Genome Data for the Taxonomy of Prokaryotes. Int J Syst Evol Microbiol 2024, 74. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS Is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat Commun 2019, 10, 2182. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Rinke, C.; Mussig, A.J.; Chaumeil, P.A.; Hugenholtz, P. GTDB: An Ongoing Census of Bacterial and Archaeal Diversity through a Phylogenetically Consistent, Rank Normalized and Complete Genome-Based Taxonomy. Nucleic Acids Res 2022, 50, D785–D794. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bliss, C.M.; Bennett, J.S.; Bratcher, H.B.; Brehony, C.; Colles, F.M.; Wimalarathna, H.; Harrison, O.B.; Sheppard, S.K.; Cody, A.J.; et al. Ribosomal Multilocus Sequence Typing: Universal Characterization of Bacteria from Domain to Strain. Microbiology (Reading) 2012, 158 Pt 4, 1005–1015. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




