Submitted:
25 March 2025
Posted:
26 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Bacterial Isolates
2.2. Antimicrobial Susceptibility Testing and Phenotypic Tests for β-lactamases
2.3. Molecular Detection of Resistance Genes
2.4. Inter-Array Genotyping CarbaResist Method
2.5. Whole Genome Sequencing (WGS)
2.6. Conjugation
2.7. Plasmid Analysis by PCR-Based Replicon Typing (PBRT)
2.8. Detection of Virulence Determinants
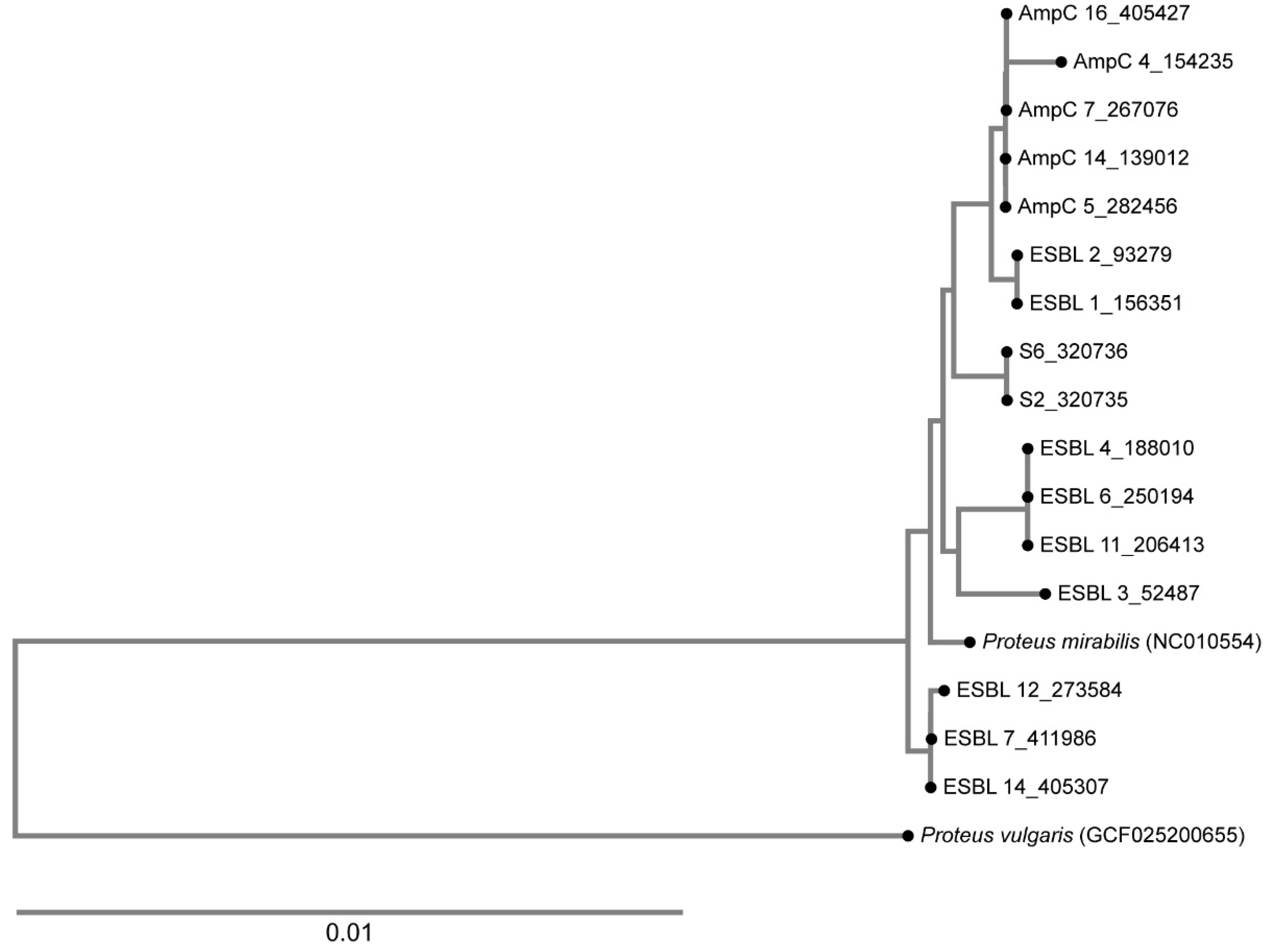
2.9. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. Antimicrobial Susceptibility Testing and Phenotypic Tests for β-lactamases
4.3. Molecular Detection of Resistance Genes
4.4. Detection of Resistance Genes by Inter-Array Kit CarbaResist
4.5. Whole Genome Sequencing (WGS)
4.6. Conjugation
4.7. Characterization of Plasmids
4.8. Detection of Virulence Determinants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sanches, M.S.; Silva, L.C.; Silva, C.R.D.; Montini, V.H.; Oliva, B.H.D.; Guidone, G.H.M.; Nogueira, M.C.L.; Menck-Costa, M.F.; Kobayashi, R.K.T.; Vespero, E.C.; Rocha, S.P.D. Prevalence of Antimicrobial Resistance and Clonal Relationship in ESBL/AmpC-Producing Proteus mirabilis Isolated from Meat Products and Community-Acquired Urinary Tract Infection (UTI-CA) in Southern Brazil. Antibiotics (Basel). 2023, 2023 10, 370. [Google Scholar] [CrossRef]
- Li, Y.; Yin, M.; Fang, C.; Fu, Y.; Dai, X.; Zeng, W.; Zhang, L. Genetic analysis of resistance and virulence characteristics of clinical multidrug-resistant Proteus mirabilis isolates. Front. Cell. Infect. Microbiol. 2023, 11, 1229194. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; Coque, T.M. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef]
- Coque, T.M.; Baquero, F.; Canton, R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Eurosurveillance 2008, 13, 19044. [Google Scholar] [PubMed]
- de Champs, C.; Bonnet, R.; Sirot, D.; Chanal, C.; Sirot, J. Clinical relevance of Proteus mirabilis in hospital patients: a two year survey. J Antimicrob Chemother. 2000, 45, 537–9. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC β-lactamases. J. Clin. Microbiol. 2009, 22, 161–182. [Google Scholar] [CrossRef]
- Migliavacca, R.; Migliavacca, A.; Nucleo, E.; Ciaponi, A.; Spalla, M.; De Luca, C.; Pagani, L. Molecular epidemiology of ESBL producing P. mirabilis strains from a long-term care and rehabilitation facility in Italy. New Microbiol. 2007, 30, 362–6. [Google Scholar]
- D’Andrea, M.M.; Literacka, E.; Zioga, A.; Giani, T.; Baraniak, A.; Fiett, J.; Sadowy, E.; Tassios, P.T.; Rossolini, G.M.; Gniadkowski, M.; Miriagou, V. Evolution and spread of a multidrug-resistant Proteus mirabilis clone with chromosomal AmpC-type cephalosporinases in Europe. Antimicrob Agents Chemother. 2011, 55, 2735–42. [Google Scholar] [CrossRef]
- Girlich, D.; Bonnin, R.A.; Dortet, L.; Naas, T. Genetics of Acquired Antibiotic Resistance Genes in Proteus spp. Front Microbiol. 2020, 21, 256. [Google Scholar] [CrossRef]
- Sardelić, S.; Bedenić, B.; Sijak, D.; Colinon, C.; Kalenić, S. Emergence of Proteus mirabilis isolates producing TEM-52 extended-spectrum β-lactamases in Croatia. Chemotherapy. 2010, 56, 208–13. [Google Scholar] [CrossRef] [PubMed]
- Tonkić, M.; Mohar, B.; Šiško-Kraljević, K.; Meško-Meglič, K.; Goić-Barišić, I.; Novak, A.; Kovačić, A.; Punda-Polić, V. High prevalence and molecular characterization of extended-spectrum β-lactamase-producing Proteus mirabilis strains in southern Croatia. J Med Microbiol. 2010, 59, 1185–1190. [Google Scholar] [CrossRef]
- Bedenić, B.; Firis, N.; Elveđi-Gašparović, V.; Krilanović, M.; Matanović, K.; Štimac, I.; Luxner, J.; Vraneš, J.; Meštrović, T.; Zarfel, G.; Grisold, A. Emergence of multidrug-resistant Proteus mirabilis in a long-term care facility in Croatia. Wien Klin Wochenschr. 2016, 128, 404–13. [Google Scholar] [CrossRef] [PubMed]
- Rubić, Z.; Soprek, S.; Jelić, M.; Novak, A.; Goić-Barisić, I.; Radić, M.; Tambić-Andrasević, A.; Tonkić, M. Molecular Characterization of β-Lactam Resistance and Antimicrobial Susceptibility to Possible Therapeutic Options of AmpC-Producing Multidrug-Resistant Proteus mirabilis in a University Hospital of Split, Croatia. Microb Drug Resist. 2021, 27, 162–169. [Google Scholar] [CrossRef]
- Shaaban, M.; Elshaer, S.L.; Abd El-Rahman, O.A. Prevalence of extended-spectrum β-lactamases, AmpC, and carbapenemases in Proteus mirabilis clinical isolates. BMC Microbiol. 2022, 11, 247. [Google Scholar] [CrossRef] [PubMed]
- Perillli, M.; Segatore, B.; Mugnaioli, C.; Celenza, G.; Rossolini, G.M. , Stefani, S.; Luzzaro, F.; Pini, B.; Amicosante, G. Persistence of TEM-52/TEM-92 and SHV-12 extended-spectrum β-lactamases in clinical isolates of Enterobacteriaceae in Italy. Microb Drug Resist. 2011, 17, 521–524. [Google Scholar] [CrossRef]
- Literacka, E.; Bedenic, B.; Baraniak, A.; Fiett, J.; Tonkic, M.; Jajic-Bencic, I.; Gniadkowski, M. blaCTX-M genes in Escherichia coli strains from Croatian Hospitals are located in new (blaCTX-M-3a) and widely spread (blaCTX-M-3a and blaCTX-M-15) genetic structures. Antimicrob Agents Chemother. 2009, 53, 1630–5. [Google Scholar] [CrossRef]
- Krilanović, M.; Tomić-Paradžik, M.; Meštrović, T.; Beader, N.; Herljević, Z.; Conzemius, R.; Barišić, I.; Vraneš, J.; Elveđi-Gašparović, V.; Bedenić, B. Extended-spectrum beta-lactamases and plasmid diversity in urinary isolates of Escherichia coli in Croatia: a nation-wide, multicentric, retrospective study. Folia Microbiol (Praha). 2020, 65, 649–667. [Google Scholar] [CrossRef]
- Car, H.; Dobrić, M.; Pospišil, M.; Nađ, M.; Luxner, J.; Zarfel, G.; Grisold, A.; Nikić-Hecer, A.; Vraneš, J.; Bedenić, B. Comparison of Carbapenemases and Extended-Spectrum β-Lactamases and Resistance Phenotypes in Hospital- and Community-Acquired Isolates of Klebsiella pneumoniae from Croatia. Microorganisms. 2024, 2, 2224. [Google Scholar] [CrossRef]
- Bandić Pavlović, D.; Pospišil, M.; Nađ, M.; Vrbanović Mijatović, V.; Luxner, J.; Zarfel, G.; Grisold, A.; Tonković, D.; Dobrić, M.; Bedenić, B. Multidrug-Resistant Bacteria in Surgical Intensive Care Units: Antibiotic Susceptibility and β-Lactamase Characterization. Pathogens. 2024, 15, 11. [Google Scholar] [CrossRef]
- Bedenić, B.; Pešorda, L.; Krilanović, M.; Beader, N.; Veir, Z.; Schoenthaler, S.; Bandić-Pavlović, D.; Frančula-Zaninović, S.; Barišić, I. Evolution of Beta-Lactamases in Urinary Klebsiella pneumoniae Isolates from Croatia; from Extended-Spectrum Beta-Lactamases to Carbapenemases and Colistin Resistance. Curr Microbiol. 2022, 15, 355. [Google Scholar] [CrossRef]
- Seo, K.W.; Do, K.H.; Lee, W.K. Comparative Genetic Characterization of CTX-M-Producing Escherichia coli Isolated from Humans and Pigs with Diarrhea in Korea Using Next-Generation Sequencing. Microorganisms. 2023, 28, 1922. [Google Scholar] [CrossRef]
- Mahrouki, S.; Belhadj, O.; Chihi, H.; Mohamed, B.M.; Celenza, G.; Amicosante, G.; Perilli, M. Chromosomal blaCTX-M-₁₅ associated with ISEcp1 in Proteus mirabilis and Morganella morganii isolated at the Military Hospital of Tunis, Tunisia. J Med Microbiol. 2012, 61, 1286–1289. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Shan, G.; Yu, G.; Wei, J.; Zhang, Q.; Su, W.; Lin, Q.; Zheng, Z.; Wu, G.; Li, G.; Chang, Q.; Yuan, H.; He, Y.; Chen, Y.; Zhang, Y.; Huang, H.; Hu, W.; Song, R.; Weng, Y.; Li, X.; Liu, S. Whole genome sequencing of multidrug-resistant Proteus mirabilis strain PM1162 recovered from a urinary tract infection in China. J Glob Antimicrob Resist. 2023, 33, 44–50. [Google Scholar] [CrossRef]
- Tian, G.B.; Jiang, Y.Q.; Huang, Y.M.; Qin, Y.; Feng, L.Q.; Zhang, X.F.; Li, H.Y.; Zhong, L.L.; Zeng, K.J.; Patil, S.; Xing, Y.; Huang, X. Characterization of CTX-M-140, a Variant of CTX-M-14 Extended-Spectrum β-Lactamase with Decreased Cephalosporin Hydrolytic Activity, from Cephalosporin-Resistant Proteus mirabilis. Antimicrob Agents Chemother. 2016, 23, 6121–6. [Google Scholar] [CrossRef]
- Chao, C.M.; Lai, C.C.; Yu, W.L. Epidemiology of extended-spectrum β-lactamases in Enterobacterales in Taiwan for over two decades. Front Microbiol. 2023, 23, 1060050. [Google Scholar] [CrossRef]
- Karpenko, A.; Shelenkov, A.; Petrova, L.; Gusarov, V.; Zamyatin, M.; Mikhaylova, Y.; Akimkin, V. Two multidrug-resistant Proteus mirabilis clones carrying extended spectrum beta-lactamases revealed in a single hospital department by whole genome sequencing. Heliyon. 2024, 2, e40821. [Google Scholar] [CrossRef]
- Biggel, M.; Boss, S.; Uea-Anuwong, T.; Lugsomya, K.; Magouras, I.; Stephan, R. Complete Genome Sequence of the Extensively Drug-Resistant Extended-Spectrum β-Lactamase-Producing Proteus mirabilis Isolate HK294, Obtained from Poultry Feces in Hong Kong. Microbiol. Resour. Announc. 2023, 20, e0022523. [Google Scholar] [CrossRef]
- Martínez-Álvarez, S.; Châtre, P.; François, P.; Zarazaga, M.; Madec, J.Y.; Haenni, M.; Torres, C. Comparative phylogenomics of extended-spectrum β-lactamase-producing Escherichia coli revealed a wide diversity of clones and plasmids in Spanish chicken meat. Int. J. Food Microbiol 2025, 2, 110900. [Google Scholar] [CrossRef]
- Lemlem, M.; Aklilu, E.; Mohamed, M.; Kamaruzzaman, N.F.; Devan, S.S.; Lawal, H.; Kanamma, A.A. Prevalence and molecular characterization of ESBL-producing Escherichia coli isolated from broiler chicken and their respective farms environment in Malaysia. BMC. Microbiol. 2024, 26, 499. [Google Scholar] [CrossRef]
- Martínez-Álvarez, S.; Châtre, P.; François, P.; Abdullahi, I.N.; Simón, C.; Zarazaga, M.; Madec, J.Y.; Haenni, M.; Torres, C. Unexpected role of pig nostrils in the clonal and plasmidic dissemination of extended-spectrum β-lactamase-producing Escherichia coli at farm level. Ecotoxicol. Environ. Saf. 2024, 15, 116145. [Google Scholar] [CrossRef]
- Nakano, R.; Nakano, A.; Abe, M.; Nagano, N.; Asahara, M.; Furukawa, T.; Ono, Y.; Yano, H.; Okamoto, R. Prevalence and mechanism of fluoroquinolone resistance in clinical isolates of Proteus mirabilis in Japan. Heliyon. 2019, 1, e01291. [Google Scholar] [CrossRef]
- Mahrouki, S.; Perilli, M.; Bourouis, A.; Chihi, H.; Ferjani, M.; Ben Moussa, M.; Amicosante, G.; Belhadj, O. Prevalence of quinolone resistance determinant qnrA6 among broad- and extended-spectrum beta-lactam-resistant Proteus mirabilis and Morganella morganii clinical isolates with sul1-type class 1 integron association in a Tunisian Hospital. Scand J Infect Dis. 2013, 45, 600–5. [Google Scholar] [CrossRef] [PubMed]
- Clinical Laboratory Standard Institution. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Approved Standard M100-S22; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- European Committee for Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12. 2022. Available online: http://www.eucast.org (accessed on 1st October 2023).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G. ; Harbarth, S; Hindler, J.F; Kahlmeter, G; Olsson-Liljequist, B; Paterson, D.L; Rice, L.B; Stelling J.; Struelens, M.J.; Vatopoulos, A.; Weber, J.T.; Monnet, D. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect 2002, 18, 268–281. [Google Scholar] [CrossRef]
- Davis, R.; Brown, P.D. Multiple Antibiotic Resistance Index, Fitness and Virulence Potential in 408 Respiratory Pseudomonas aeruginosa from Jamaica. J. Med. Microbiol. 2016, 65, 261–271. [Google Scholar] [PubMed]
- Jarlier, V.; Nicolas, M.H.; Fournier, G.; Philippon, A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis 1988, 10, 867–878. [Google Scholar] [CrossRef]
- Polsfuss, S.; Bloemberg, G.V.; Giger, J.; Meyer, V.; Böttger, E.C.; Hombach, M. Practical approach for reliable detection of AmpC β-lactamase-producing Enterobacteriaceae. J. Clin. Microbiol. 2011, 49, 2798–803. [Google Scholar] [CrossRef]
- Sanders, C.C.; Sanders, W.E. Jr; Goering, R.V. In vitro antagonism of beta-lactam antibiotics by cefoxitin. Antimicrob. Agents. Chemother. 1982, 21, 968–75. [Google Scholar] [CrossRef]
- Arlet, G.; Brami, G.; Decre, D.; Flippo, A.; Gaillot, O.; Lagrange, P.H.; Philippon, A. Molecular characterization by PCR restriction fragment polymorphism of TEM β-lactamases. FEMS Microbiol. Lett. 1995, 134, 203–208. [Google Scholar] [CrossRef]
- Nüesch-Inderbinen, M.T.; Hächler, H.; Kayser, F.H. Detection of genes coding for extended-spectrum SHV β-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur. J. Clin. Microbiol. Infect. Dis. 1996, 15, 398–402. [Google Scholar] [CrossRef]
- Woodford, N.; Ward, M.E.; Kaufmann, M.E.; Turton, J.; Fagan, E.J.; James, D.; Johnson, A.P.; Pike, R.; Warner, M.; Cheasty, T.; Pearson, A.; Harry, S.; Leach, J.B; Loughrey, A.; Lowes, J.A.; Warren, R.E.; Livermore, D.M. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 2004, 54, 735–743. [Google Scholar] [CrossRef]
- Robicsek, A.; Jacoby, G.A.; Hooper, D.C. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 2006, 6, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Fagan, E.J.; Ellington, M.J. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2006, 57, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Saladin, M.; Cao, V.T.B.; Lambert, T.; Donay, J.L.; Hermann, J.; Ould-Hocine, L. Diversity of CTX-M β-lactamases and Their Promoter Regions from Enterobacteriaceae Isolated in Three Parisian Hospitals. FEMS Microbiol. Lett. 2002, 209, 161–168. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012, 67, 2640–4. [Google Scholar] [CrossRef]
- Elwell, L.P.; Falkow, S. The characterization of R plasmids and the detection of plasmid-specified genes. In Antibiotics in Laboratory Medicine, 2nd ed.; Lorian, V., Ed.; Williams and Wilkins: Baltimore, MD, USA, 1986; pp. 683–721. [Google Scholar]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threfall, E.J. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Carattoli, A.; Seiffert, S.N.; Schwendener, S.; Perreten, V.; Endimiani, A. Differentiation of IncL and IncM Plasmids Associated with the Spread of Clinically Relevant Antimicrobial Resistance. PLoS ONE 2015, 10, e0123063. [Google Scholar] [CrossRef]

| ESBL | AmpC | |||||||
| MIC range | MIC50 | MIC90 | Number and % of resistant isolates | MIC range | MIC50 | MIC90 | Number and % of resistant isolates | |
| amoxycillin-clavulanate | >128->128 | ≥128 | ≥128 | 17/17 (100%) | >128->128 | ≥128 | ≥128 | 22/22 (100%) |
| cefuroxime | >128->128 | ≥128 | ≥128 | 17/17 (100%) | >128->128 | ≥128 | ≥128 | 22/22 (100%) |
| piperacillin-tazobactam | 4-32 | 16 | 32 | 0/17 (0%) | 2-64 | 16 | 64 | 0/22 (0%) |
| ceftazidime | 2->128 | 16 | ≥128 | 10/17(58,8%) | 16->128 | >128 | >128 | 22/22 (95) |
| cefotaxime | 32->128 | ≥128 | ≥128 | 17/17 (100%) | >128->128 | ≥128 | ≥128 | 22/22 (100%) |
| ceftriaxone | 8->128 | 64 | ≥128 | 17/17 (100%) | 32->128 | ≥128 | ≥128 | 17/17 (100%) |
| cefepime | 4-64 | 32 | 64 | 16/17 (94%) | 4-32 | 8 | 32 | 6/22 (27%) |
| imipenem | 0,5-4 | 1 | 2 | 0/17 (0%) | 0,25-1 | 0,5 | 1 | 0/22 (0%) |
| meropenem | 0,06-0,25 | 0,06 | 0,12 | 0/17 (0%) | 0,06-0,25 | 0,12 | 0,25 | 0/22 (0%) |
| gentamicin | 0,25->128 | 32 | >128 | 13/17(76,4%) | 0,25->128 | 64 | >128 | 19/22 (86,3%) |
| amikacin | 8->128 | 32 | 128 | 6/17 (35,2%) | 32->128 | 128 | >128 | 20/22 (91%) |
| ciprofloxacin | 1->128 | 128 | >128 | 17/17 (100%) | 16_>128 | 128 | >128 | 22/22 (100%) |
| Isolate and Protocol Number | β-Lactam | AG | SUL | THR | |
| AmpC 1 (284989) |
blaCMY-16 blaTEM |
aac(6′) aac(3″)-Ia aac(6′)-Ib-cr aadA1 aadA2 |
Sul1 Sul2 |
dfrA1 | |
| ESBL 5 (156351) |
blaCTX-M-9 |
aac(6′) aac(6′)-Ib aac(3″)-IVa aadA1 aadA2 aphA |
Sul1 Sul2 |
dfrA5 |
|
Isolate and protocol number |
β-Lactam | Aminoglycosides | Sulphonamide | Trimethoprim | Chloramphenicol | Tetracycline | Fluoroquinolones | Plasmid Inc group |
| AmpC 4 (154235) |
blaCMY-16 (AJ781421) |
dfrA1 (X00926) |
||||||
| AmpC 5 (282456) |
blaCMY-16 AJ781421 blaTEM-1b AY458016 |
aac(6′)-Ib3 X60321 aadA1 JX185132 aph(6)-Id M28829 aph(3)-Ia X62115 |
Sul1 U12338 Sul2 HQ840942 |
dfrA1 X00926 |
CatA1 V00622 |
Tet (A) (AJ517790) |
||
| AmpC 7 (267076) |
blaCMY-16 (AJ781421) |
aadA1 (789/789) |
dfrA1 (X00926) |
CatA1 V00622 |
||||
| AmpC 14 (139012) |
blaCMY-16 (AJ781421) blaTEM-1b (AY458016) |
aac(6′)-Ib3 X60321 aadA1 (JX185132) aadA2 (JQ364967) aph(6)-Id (M28829) aph(3)-Ia (X62115) |
Sul1 U12338 Sul2 HQ840942 |
dfrA1 X00926 |
CatA1 V00622 |
Tet (J) (ACLE01000065) |
||
| AmpC 16 (405427) |
blaCMY-16 (AJ781421) blaTEM-1b (AY458016) |
aph(6)-Id (M28829) aph(3)-Ia X(62115) aadA1 (JX185132) aadA2 (JQ364967) aac(6′)-Ib-cr (DQ303918) aph(3″)-Ib (AF321551) armA (AY220558) |
Sul1 U12338 Sul2 HQ840942 |
dfrA1 (X00926) dfrA12 (AM040708) |
CatA1 (V00622) |
Tet (J) (ACLE01000065) Tet (A) (AJ517790) |
||
| ESBL 1 (156351) |
blaCTX-M-101 (HQ398214 |
aph(6)-Id (M28829) aac(3)-IId (EU022314) aadA1 (JX185132) aadA5 (AF137361) aph(3″)-Ia (X62115) aph(3″)-Ib (AF321551) |
Sul1 U12338 Sul2 (HQ840942) |
dfrA1 (X00926) |
cat (M11587) CatA1 (V00622) |
Tet (J) (ACLE01000065) |
||
| ESBL 2 (93279) |
blaCTX-M-15 (HQ398214) blaTEM-1d (AF188200) |
aph(6)-Id (M28829) aac(3)-IId (EU022314) aadA1 (JX185132) aadA5 (AF137361) aph(3)-Ia (X62115) aph(3″)-Ib (AF321551) aac(3)-IV (DQ241380) |
Sul1 U12338 Sul2 HQ840942 |
dfrA1 (X00926) dfrA17 (FJ460238) |
cat (M11587) CatA1 V00622 |
Tet (J) (ACLE01000065) |
||
| ESBL 3 (52487) |
blaCTX-M-32 (AJ557142) |
aph(6)-Id (M28829) aph(3)-Ia (X62115) aph(4)-Ia (V01499) aadA1 (JX185132) aph(3″)-Ib AF024602 |
Sul1 U12338 Sul2 HQ840942 |
dfrA1 (X00926) |
cat (M11587) |
Tet (J) (ACLE01000065) |
qnrD1 FJ228229 |
|
| ESBL 4 (188010) |
blaCTX-M-14b (DQ359215) |
aph(6)-Id (M28829 aac(3)-IIa (X51534) aadA1 (JX185132) aph(3″)-Ib (AF321551) |
dfrA1 (X00926) |
cat (M11587) |
Tet (J) (ACLE01000065) |
|||
| ESBL 6 (250194) |
blaCTX-M-14b (DQ359215) blaTEM-1d (AF188200) |
aac(3)-IIa (X51534) aadA1 (JX185132) aph(3″)-Ib (AF321551) |
Sul2 HQ840942 |
dfrA1 (X00926 |
cat (M11587) |
Tet (J) (ACLE01000065 |
||
| ESBL 7 (411986) |
blaCTX-M-65 (EF418608) blaOXA-1 (HQ170510) |
aph(6)-Id (M28829) aph(3′)-Ia (X62115) aph(4)-Ia (V01499) aac(6′)-Ib-cr (DQ303918) aph(3″)-Ib (AF321551) aac(3)-IV (DQ241380) aadA2b (D43625) |
Sul1 (U12338) Sul2 (HQ840942) |
dfrA32 (GU067642) |
catB3 (U13880) |
Tet (J) (ACLE01000065 tet(C) (AF055345) |
||
| ESBL 11 (206414) |
blaCTX-M-14b (DQ359215)¸ blaTEM-1d (AF188200 |
aph(6)-Id (M28829) aac(3)-IIa (X51534) aadA1 (JX185132) aph(3″)-Ib (AF321551) |
Sul2 (HQ840942) |
dfrA1 (X00926) |
cat (M11587) |
Tet (J) (ACLE01000065) |
||
| ESBL 12 (273584) |
blaCTX-M-65 (EF418608) blaOXA-1 (HQ170510) blaTEM-1A (HM749966) |
aph(6)-Id (M28829) aph(3′)-Ia (X62115) aph(4)-Ia (V01499) aph(3″)-Ib (AF321551) aac(6′)-Ib-cr (DQ303918) aac(3)-IV (DQ241380) aadA2b (D43625) |
Sul1 (U12338) Sul2 (HQ840942) |
dfrA32 (GU067642) |
catB3 (U13880) |
tet(C) (AF055345) tet(H) (Y15510) Tet (J) (ACLE01000065) |
||
| ESBL 14 (405307) |
blaCTX-M-65 (DQ359215) blaTEM-1d (AF188200) blaOXA-1 (HQ170510 |
aph(6)-Id aph(3′)-Ia (X62115) (M28829) aph(3″)-Ib (AF321551 aac(3)-IIa (X51534) aac(6′)-Ib-cr (DQ303918 aadA1 (JX185132) aadA2 (JQ364967) |
Sul1 (U12338) Sul2 (HQ840942 |
dfrA32 (GU067642) |
cat (M11587) catB3 (U13880) |
Tet (J) (ACLE01000065 tet(C) (AF055345) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





