1. Introduction
Medical diagnosis is a fundamental aspect of the clinical process, requiring doctors to perform step-by-step reasoning based on medical guidelines while integrating various patient information to analyze different indicators and reach accurate and well-informed conclusions [
57]. This process is time-consuming, and the growing demand for medical diagnosis is increasingly surpassing the capacity of existing healthcare resources, placing a significant burden on medical services. To alleviate this burden, efforts have been made to develop computer-aided diagnosis (CAD) techniques that assist with specific diagnostic tasks, such as tumor segmentation and cancer grading. However, these models function merely as tools, with the overall diagnostic process still relying heavily on human doctors. As a result, the development of AI systems capable of autonomously handling the entire diagnostic workflow has become a key research focus.
Recently, multi-modal large language models (MLLMs) [
1,
13,
21,
33,
42] have garnered significant attention for their strong interaction abilities. They can perform a variety of tasks based on user prompts, including Medical Vision Question Answering (MedVQA) [
51], Medical Report Generation (MRG), and clinical decision support [
9,
20]. These advancements underscore the potential of MLLMs in enhancing medical decision-making. However, despite these advancements, MLLMs still fall short of meeting the stringent standards required for disease diagnosis in clinical practice. Firstly, they often exhibit hallucinations and inconsistencies in reasoning, making it difficult to strictly adhere to established medical criteria. Moreover, many disease diagnoses clinically rely on quantitative evidence. For example, the vertical optic cup-to-disc ratio is crucial for glaucoma diagnosis, and left ventricular ejection fraction plays a key role in heart function evaluation. However, MLLMs struggle to perform quantitative analysis on images, thus hard to be applied in clinical practice. As a result, current MLLMs primarily function in one-hop medical visual question answering for diagnostic tasks, which falls short of the expert-driven, evidence-based analysis required in real-world medical diagnosis. These limitations raise significant safety concerns, highlighting the need for further advancements to improve their reliability and clinical applicability.
Figure 1.
Comparison of existing MLLMs and our MedAgent-Pro framework on two disease diagnoses. Red text highlights the limitations of MLLMs, while green text represents the evidence-based diagnosis provided by MedAgent-Pro. Our approach enhances diagnostic accuracy while offering comprehensive literature support and visual evidence.
Figure 1.
Comparison of existing MLLMs and our MedAgent-Pro framework on two disease diagnoses. Red text highlights the limitations of MLLMs, while green text represents the evidence-based diagnosis provided by MedAgent-Pro. Our approach enhances diagnostic accuracy while offering comprehensive literature support and visual evidence.
Due to the extensive computational resources and high-quality data required for fine-tuning [
33], enabling MLLMs with quantitative analysis capabilities and domain-specific expertise remains challenging, limiting their applicability in complex medical scenarios. Agentic systems [
12,
34,
56,
62,
72] offer a potential solution for assisting MLLMs in medical diagnosis. These systems, originally developed for general-purpose applications, integrate MLLMs with external tools to extend their capabilities beyond simple conversational interactions. Meanwhile, in the medical domain, specialized AI models such as classification [
74], grounding [
6], and segmentation [
45,
66,
78] have achieved great success while existing medical literature provides essential clinical criteria for disease diagnosis. By leveraging these resources, agentic systems can help compensate for the limitations of MLLMs, enhancing their effectiveness in medical applications. Several studies have explored the development of medical agentic systems [
16,
28,
32]. However, most existing approaches adapt general-purpose agentic frameworks to improve diagnostic accuracy without enhancing interpretability by providing supporting evidence. These frameworks incorporate debate or collaboration mechanisms between MLLMs to refine answers, but they still rely on empirical black-box decision-making and overlook the importance of expert-driven evidence, which is essential in modern medicine. In contrast, medical diagnosis requires structured reasoning based on guidelines, integration of multi-modal patient data, and evidence-based analysis to ensure reliability and safety. As a result, existing medical agentic systems often oversimplify diagnostic tasks, merely focusing on empiricism-based question answering rather than the in-depth, evidence-based analysis needed for real-world applications. These limitations highlight the need for approaches tailored to the stringent requirements of medical decision-making.
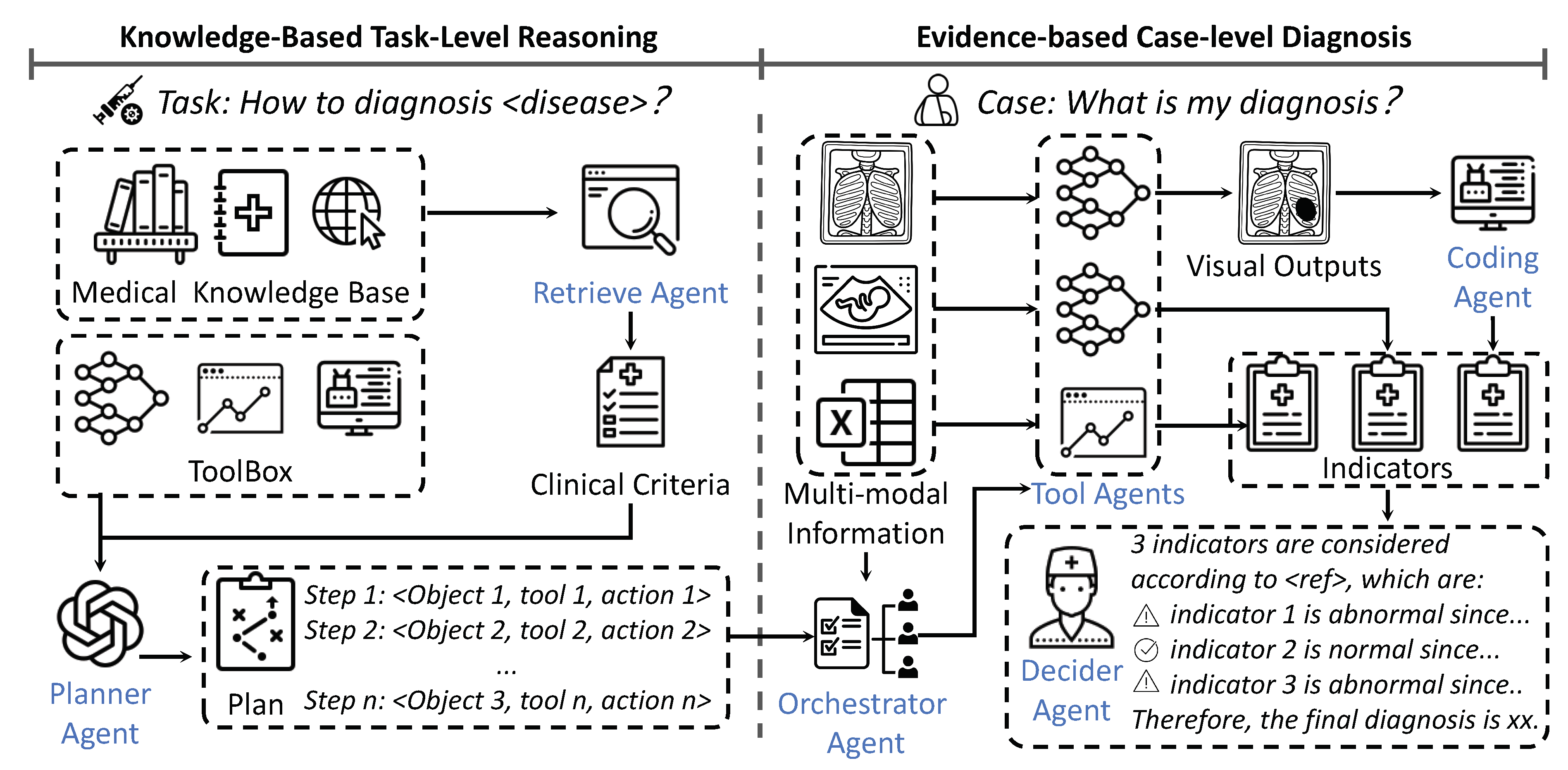
To align AI system with evidence-cased modern medical protocol, we propose a reasoning agentic workflow for evidence-based multi-modal Medical diagnosis, termed MedAgent-Pro, to enable comprehensive, evidence-based medical diagnosis. MedAgent-Pro achieves this by incentivizing the reasoning capabilities of MLLMs by integrating the retrieved medical guideline and medical expert tools. It adopts a hierarchical structure, where the task level formulates unified diagnostic plans for specific diseases, while the case level executes the plan and makes diagnostic decisions for suspected patients. At the task level, MLLMs act as planner agents, performing knowledge-based reasoning to generate reliable diagnostic plans by integrating retrieved clinical criteria. At the case level, medical expert models act as tool agents, analyzing both quantitative and qualitative indicators. Finally, a decider agent ensures a comprehensive and accurate diagnosis for each patient, supported by clinical literature and visual evidence. Our key contributions are listed below:
We propose MedAgent-Pro, a reasoning agentic workflow that can provide accurate and explainable medical diagnoses supported by visual evidence and clinical guidelines.
At the task level, MLLMs perform knowledge-based reasoning to generate well-founded diagnostic plans. Without fine-tuning, this is accomplished by integrating retrieved clinical criteria, ensuring greater reliability in diagnosis.
At the case level, various medical expert tools execute the corresponding steps in the plan to process multi-modal patient information, providing interpretable qualitative and quantitative analyses to support evidence-based decision-making.
We evaluate MedAgent-Pro on both 2D and 3D multi-modal medical diagnosis, where it achieves state-of-the-art performance, surpassing both general MLLMs and task-specific solutions. Additionally, case studies highlight MedAgent-Pro’s superior interpretability and reliability beyond quantitative results.
2. Related work
2.1. AI-Driven Multi-Modal Medical Diagnosis
Medical diagnosis is one of the most critical and complex tasks in clinical practice, demanding both expertise and precision. The development of artificial intelligence (AI) systems capable of assisting human clinicians in diagnostic decision-making has emerged as a key research objective [
5,
29,
47,
58]. However, multi-modal diagnosis presents significant challenges, as it requires the integration of both qualitative and quantitative assessments of patients’ visual examinations alongside a comprehensive analysis of diagnostic findings and patient-specific information. To address these challenges, prior research has primarily focused on developing visual models to support medical diagnosis, encompassing tasks such as classification [
2,
4,
14,
73,
74], detection [
7,
8,
15,
22,
61], and segmentation [
3,
25,
45,
55,
64,
66,
76]. These models extract critical diagnostic indicators, which are subsequently synthesized by medical professionals to formulate clinical decisions.
In parallel, efforts have been made to develop end-to-end AI architectures for medical diagnosis with minimal human intervention, particularly through multi-modal approaches like Vision Question Answering (VQA) [
27,
71,
75]. The emergence of MLLMs [
1,
13,
42,
69] has further expanded the potential of AI-driven diagnosis, as these models exhibit advanced reasoning capabilities approaching human-level performance across various domains. Several studies have explored fine-tuning general-purpose MLLMs for medical applications [
33,
52], achieving competitive performance in Medical VQA tasks. Despite these advancements, most existing Medical VQA datasets [
23,
31,
41] remain overly simplistic compared to real-world diagnostic scenarios. They primarily focus on classification tasks, lacking comprehensive analytical depth and explanatory reasoning, making them insufficient for revealing the interpretability limitations of MLLM-based diagnoses. While some methods improve explainability by linking text outputs to specific image regions [
30,
39,
40,
70], they still fall short of structured reasoning necessary for rigorous diagnosis. Consequently, further research is required to advance the application of MLLMs in real-world medical diagnosis, with particular emphasis on their reliability, interpretability, and clinical integration.
2.2. LLM-Based AI Agents
Developing reliable autonomous intelligent systems has long been regarded as a highly promising research avenue. With the advent of the agent concept, which refers to an entity capable of perceiving its environment and taking action, agent-based intelligent systems have garnered considerable attention in recent years. Large language models (LLMs) have shown remarkable potential in reasoning and planning, closely aligning with human expectations for agents that can perceive their surroundings, make informed decisions, and take actions within interactive environments. Driven by this potential, LLM-based agents have made significant advancements in navigating complex environments and tackling intricate tasks across diverse applications such as industrial engineering [
37,
49,
53,
67], scientific experimentation [
10,
19,
44], embodied agents [
11,
24,
65], gaming [
18,
36,
54], and societal simulation [
26,
46,
68].
With these advancements, LLM-based agents surpass traditional task-specific models by dynamically adapting to diverse applications without additional training. However, their capabilities remain prone to hallucinations, often leading to inconsistent answers for the same case. Moreover, LLMs excel at qualitative analysis but struggle with generating precise quantitative results. As a result, an agentic system relying solely on LLMs is insufficient for medical diagnosis [
35,
43,
59,
79]. Instead, a more structured, tool-based system is needed to ensure accuracy and reliability in clinical applications.
3. Methods
Our MedAgent-Pro framework consists of two main stages: task-level reasoning and case-level diagnosis, as shown in
Figure 2. At the task level, the planner agent utilizes retrieved medical pipelines to formulate a diagnostic plan that aligns with established medical principles for each disease. At the case level, specialized models follow the plan’s steps to analyze multi-modal patient data, extracting diverse biomarkers and indicators. Finally, the decider agent integrates all information to generate an explainable diagnosis supported by visual evidence. In the following sections, we introduce the agents and tools involved and provide a detailed explanation of the two-step process.
3.1. Agents Involved in MedAgent-Pro
In this section, we first introduce the various agents in our MedAgent-Pro framework and specify the models that function as each agent.
3.1.1. Agents in Task-Lavel Reasoning
Retrieve-Augumented Generation (RAG) Agent We utilize RAG to retrieve relevant medical documents, ensuring the development of reliable diagnostic processes that follow clinical criteria. For retrieval, we employ the built-in functionality of LangChain [
60], and retrieve from medical library [
48,
50].
Planner Agent The planner agent generates a diagnostic plan based on the retrieved guidelines and available tools. We employ GPT-4o [
1] as the planner agent due to its strong reasoning capabilities.
3.1.2. Agents in Case-Lavel Diagnosis
Orchestrator Agent responsible for conducting a preliminary analysis of the patient’s multi-modal information and determining which steps of the diagnostic plan will be executed. We employ GPT-4o [
1] since it can recognize different input effectively.
-
Tool Agents We utilize various tool agents to complete different tasks in the diagnostic plans, including:
- -
Classification Models We use image classification models for analyzing some quantitative indicators. We use some general classification models like BioMedclip [
74], as well as some task-specific models like RetiZero [
63] for Ophthalmic.
- -
Segmentation Models We use the Medical SAM Adapter [
66] as the segmentation model due to its ability to achieve strong performance on the target task with only a small amount of data. To further optimize its effectiveness, we trained task-specific adapters for the target task like optic cup/disc segmentation.
- -
Vision Question Answering (VQA) Models We utilize VQA models to generate visual descriptions of medical images rather than performing complex reasoning. We employ general VQA models such as LLaVa-Med [
33], alongside task-specific models like VisionUnite [
38] for ophthalmic applications.
Coding Agent The coding module is designed to generate simple code for computing additional metrics from the raw outputs of vision models (i.e. segmentation masks). We use GPT-o1 for its strong coding ability.
Summary Agent: Since LLM outputs are often lengthy, we introduce a summary agent to refine the LLM decider’s response into a simple "yes" or "no" for accuracy evaluation. Additionally, the summary agent condenses the VQA tool’s output into "yes," "no," or "uncertain." We employ GPT-4o [
1] for its strong summarization capabilities.
Decider Agent: In charge of making the final diagnosis based on the indicators obtained from previous steps. The implementation includes two approaches, which will be introduced in the following sections.
3.2. Knowledge-Based Task-Level Reasoning
The goal of this phase is to devise a diagnostic process that aligns with medical guidelines for specific diseases. The planning process is generally divided into two steps. First, we gather information on the clinical diagnosis process for the disease. In previous studies, this step is generally handled by LLMs themselves. However, due to the inherent hallucination phenomenon and inconsistencies in reasoning, the information provided by these models is often highly unpredictable, which contradicts the structured nature of clinical diagnosis.
To address this issue, we introduce a Retrieval-Augmented Generation (RAG) agent to ensure the reliability of the information by retrieving medical resources, including clinical guidelines and hospital protocols. When processing queries about specific diseases, the agent consults medical texts, guidelines, and protocols to summarize relevant information [
48,
50], rather than relying on unsupported LLM-generated content. This approach enables evidence-based planning, enhancing the reliability and accuracy of the diagnostic process.
Once the diagnostic information is generated, it needs to be structured into an actionable diagnostic workflow. Specifically, the planning agent assigns tasks to different tool agents based on the available tools for the task. The result is a multi-step diagnostic process, where each step follows a triplet format: (object, tool, action). This format specifies which tools and operations should be applied to the input object or intermediate results of each modality at each stage. By structuring the process in this way, the system provides clear and practical diagnostic steps tailored to each specific case.
3.3. Evidence-Based Case-Level Diagnosis
Existing MLLMs approach medical diagnosis through end-to-end VQA. However, this method often lacks effective visual explanations and primarily relies on qualitative analysis without quantitative results to support the diagnosis. Although some methods enhance explainability by linking outputs to specific regions in input images, they merely highlight attended areas without providing underlying reasoning. In contrast, our proposed MedAgent-Pro system leverages advanced vision models as specialized tools, utilizing their fine-grained visual perception capabilities to perform detailed analyses rather than relying on the limited visual processing abilities of MLLMs.
Specifically, when given multi-modal patient inputs, the orchestrator agent automatically identifies the input modality, matches it with the
object field in the planned triplet, and executes the corresponding diagnostic steps. Based on these steps, various specialized models are involved as tool agents to conduct detailed analyses. Additionally, the coding agent generates code to process visual data for computing quantitative indicators or producing intermediate results as inputs for subsequent steps. Finally, the summary agent consolidates the analysis of each indicator into "yes," "no," or "uncertain" options, while the decider agent integrates all indicators, formulates the final diagnosis, and provides supporting evidence. In this paper, we explore two approaches for the final diagnosis. The first, LLM decider, involves feeding all obtained indicators into an LLM (GPT-4o in our case), allowing it to autonomously integrate the information and generate a diagnosis. The second approach, MOE (mixture-of-experts) decider, utilizes an LLM to assign weights to different indicators with a threshold-based
, and compute a final risk score
s for the decision-making process:
where
n is the total number of indicators,
represents the weight assigned to the
i-th indicator, and
encodes the indicator’s status. The final decision is determined by:
For example, diagnosing glaucoma requires a comprehensive analysis of a patient’s multi-modal data, including fundus images, OCT scans, and IOP measurements. When provided with only a fundus image, the orchestrator agent queries the diagnostic plan and identifies four indicators: The VQA tools first scan the entire image to detect potential disc hemorrhages, while the segmentation tools identify the optic cup and disc. The coding agent then calculates the vertical cup-to-disc ratio by calculating their vertical diameter, and then extracts the region around the optic disc for further analysis. The VQA module examines this region to identify signs of peripapillary atrophy, while the coding agent computes rim thickness in different directions. Similarly, when provided with 3D echocardiography and basic patient information (height, weight, etc.), the orchestrator agent queries the diagnostic plan and identifies four key indicators. The segmentation tools first detect the myocardium and left ventricle at both start-diastolic and end-diastolic phases. The coding agent then calculates the left ventricular ejection fraction, start-diastolic diameter, and end-diastolic diameter. By incorporating the patient’s height and weight, the left ventricular mass index is further derived. Once all indicators are obtained, the summary agent consolidates the findings, and the decider agent formulates the final diagnosis. A detailed implementation and the results of each agent are provided in the case study within the experimental section.
4. Experiment
We evaluated the performance of MedAgent-Pro in two essential and complex diagnostic tasks: glaucoma and heart disease. Diagnosing both diseases requires a comprehensive evaluation of multiple factors, including both qualitative and quantitative analysis. While current MLLMs struggle in most cases without the aid of external tools.
4.1. Datasets and Evaluation Metrics
For glaucoma diagnosis, we use the REFUGE2 dataset [
17], which is a 2D retinal fundus image dataset containing 1200 RGB images at a resolution of
annotated by experts. Each image includes segmentation masks for the optic disc and optic cup, along with a classification label indicating whether the patient has glaucoma.
For heart disease diagnosis, we utilize the MITEA dataset [
77], a 3D echocardiography dataset comprising 536 images from 143 human subjects. Each image contains segmentation masks for the left ventricular myocardium and cavity, along with a classification label identifying the patient’s heart condition across seven categories: healthy, aortic regurgitation, dilated cardiomyopathy, amyloidosis, hypertrophic cardiomyopathy, hypertrophy, and transplant. Due to the limited number of samples for each specific heart condition, we simplify the task to a binary classification: determining whether the patient has heart disease.
While the greatest advantage of our MedAgent-Pro system lies in the interpretability of medical diagnosis, we also conducted quantitative experiments to validate the superiority of our approach. For evaluation, we take two common metrics for classification: mACC and F1 score. The mACC represents the average accuracy across all classes; the F1 score provides robustness against class imbalance; Throughout all experiments, the best results are highlighted in bold, while the second-best results are underlined.
4.2. Comparison with Multi-Modal Foundation Models
We conduct comprehensive experiments to demonstrate the superiority of our MedAgent-Pro framework. We have included comparisons with advanced multi-modal LLM methods such as BioMedClip [
74], GPT-4o [
1], LLaVa-Med [
33], and Janus [
13]. Notably, since existing MLLMs cannot process 3D images, we randomly select three slices from the 3D echocardiography as visual input and repeat this process ten times to calculate the mean value to minimize randomness.
When comparing with general LLMs, we use mACC and F1 scores as evaluation metrics and evaluate both ways of the decider agent. For comparison with the REFUGE2 challenge method, since only AUC results are available for these methods and LLMs provide binary classifications without probability scores like traditional methods, we only include the comparison with the MOE decider.
As shown in
Table 1, our MedAgent-Pro framework significantly outperforms the current state-of-the-art multi-modal foundation models across both diagnostic tasks. The mACC metric improves by 32.3 and 19.8%, and the F1 score increases by 55.1% and 14.8%, demonstrating the effectiveness of our design on multi-agentic workflow. When faced with complex diagnoses such as glaucoma and heart disease, LLMs struggle to make accurate judgments based solely on the general appearance of fundus or echocardiography images, as they lack the ability to analyze visual features effectively. Particularly, GPT-4o often refuses to provide a clear diagnosis, while LLaVa-Med, despite being fine-tuned with medical knowledge, tends to classify all patients as healthy. The proposed MedAgent-Pro framework integrates various specialized models as tool agents to provide both analysis of both qualitative and quantitative indicators, thus approach enables precise and comprehensive diagnosis.
It can also be observed that within our MedAgent-Pro framework, the MOE decider consistently outperforms the LLM decider, achieving improvements of 14.5% and 3.0% in the mAcc metric for glaucoma and heart disease diagnosis, respectively. This is because the final diagnosis relies on multiple indicators, making it difficult for a single LLM to effectively integrate and analyze all relevant information. As a result, the LLM decider often focuses on only a subset of indicators while overlooking others, leading to reduced diagnostic accuracy. Meanwhile, the MOE decider can alleviate this issue by assigning weight for various indicators, facilitating a comprehensive decision-making.
4.3. Case Study
Figure 3 illustrates the workflow of a typical case within our MedAgent-Pro framework. At the task level, the retrieval agent gathers relevant information to establish clinical criteria, while the planner agent formulates a diagnostic plan based on the given criteria and available tools. At the case level, the orchestrator agent selects the appropriate steps from the plan based on the input data, engaging various tool agents for indicator analysis. Finally, the summary agent consolidates the findings, and the decider agent delivers the final diagnosis.
4.4. Comparison with Task-specific Models
Additionally, we compare MedAgent-Pro with domain-specific approaches, including fine-tuned MLLMs for ophthalmology [
38,
63] and the top-performing methods from the REFUGE2 challenge leaderboard [
17]. Since the REFUGE2 leaderboard only provides the AUC metric, which measures the area under the ROC curve, and the LLM decider outputs only categorical decisions without probabilistic scores, we limit the comparison to the MOE decider.
As shown in
Table 2, MedAgent-Pro maintains its superiority in comparison with the REFUGE2 challenge winners and state-of-the-art MLLMs in ophthalmology. The AUC metric has improved by 6.8%, while the mACC and F1 scores have increased by 4.6% and 3.3%, respectively. The improvement is particularly impressive, given that the challenge winners employed numerous techniques and that expert MLLMs were trained on millions of ophthalmology data, whereas the MLLMs in our MedAgent-Pro framework maintain a zero-shot setting. This finding further demonstrates that leveraging MLLMs for a specific domain does not necessarily require fine-tuning but can instead be achieved through domain-specific tools, highlighting the potential of our MedAgent-Pro framework.
4.5. Ablation Study
In the diagnosis of glaucoma, four key indicators are analyzed in fundus images: vertical Cup-to-Disc Ratio (vCDR), Rim Thickness (RT), Peripapillary Atrophy (PPA), and Disc Hemorrhages (DH). To further analyze the accuracy of the indicators themselves and their impact on the final diagnosis, we conduct comprehensive ablation studies as shown in
Table 3.
Columns 5 and 6 of
Table 3 analyze the impact of individual indicators. Each indicator’s positive or negative result is directly taked as the final outcome without passing through the decider agent. The accuracy of indicators varies significantly, with vCDR and PPA being notably more accurate than the other two. On one hand, different indicators vary in their relevance to glaucoma. For example, vCDR is the gold standard for glaucoma diagnosis in clinical practice, whereas the others are not. On the other hand, some indicators are inherently more challenging to analyze. For example, when analyzing DH, the lack of effective detection tools necessitates the use of VQA and classification tools. However, these tools lack fine-grained perception of image details, thereby reducing accuracy.
When multiple indicators are combined for the final diagnosis, we can get the below findings: 1) The MOE decider effectively leverages multiple indicators, leading to a significant improvement: when using only one indicator, the highest mACC achieved is 81.7%, while the highest F1 score is 74.6%. However, with the MOE decider incorporating multiple indicators, the mACC increases to 90.4% and the F1 score reaches 81.5%. By compensating for the limitations of relying on a single indicator, the MOE decider achieves a more holistic and reliable diagnosis. 2) However, the LLM decider experiences a performance decline for multiple indicators, particularly in the absence of vCDR, where the F1 score drops to just 14.3% (7th row). This is because LLMs tend to adopt a conservative diagnostic approach—when critical information (vCDR) is missing, they often fail to make a definitive diagnosis. Furthermore, when multiple indicators are provided, LLMs may hesitate to confirm glaucoma even if only one appears normal and others are clearly abnormal. 3) Less accurate indicators can negatively impact the correctness of the final diagnosis. Comparing the last two rows, the F1 score drops by 5.1% and 16.5%, respectively. This decline is due to the low accuracy of DH, which has a high false positive rate, leading to a significant number of misdiagnoses.
5. Conclusion and Future Work
This paper introduces MedAgent-Pro, a reasoning-based agentic system designed to deliver accurate, evidence-based medical diagnoses. The system adopts a hierarchical structure: at the task level, knowledge-based reasoning generates reliable diagnostic plans by integrating retrieved clinical criteria, while at the case level, evidence-based diagnosis conducts step-by-step indicator analysis using multiple tool agents. Finally, a decider agent synthesizes all indicators and provides a final diagnosis supported by visual evidence. Comprehensive experiments show that MedAgent-Pro surpasses both general MLLMs and expert methods in diagnosing two diseases, demonstrating its exceptional accuracy and interpretability.
Future works will involve the following aspects: First, we aim to expand the datasets to further demonstrate the generalization capability of MedAgent-Pro. This includes increasing data scale, diversifying tasks, and incorporating more modalities to validate the framework’s performance in complex scenarios and multi-modal inputs. Additionally, human-in-the-loop validation with expert doctors will help assess clinical applicability and provide detailed analyzes to generate qualitative results, demonstrating the superior interpretability of our method compared to MLLMs. These advancements will further enhance the reliability and impact in computer-aided diagnosis.
Acknowledgments
This work was supported by Ministry of Education Tier 1 Start up grant, NUS, Singapore (A-8001267-01-00); Ministry of Education Tier 1 grant, NUS, Singapore (A-8003261-00-00). Junde Wu is supported by the Engineering and Physical Sciences Research Council (EPSRC) under grant EP/S024093/1 and GE HealthCare.
References
- Achiam, J., Adler, S., Agarwal, S., Ahmad, L., Akkaya, I., Aleman, F.L., Almeida, D., Altenschmidt, J., Altman, S., Anadkat, S., et al.: Gpt-4 technical report. arXiv preprint arXiv:2303.08774 (2023).
- Association, A.D.: 2. classification and diagnosis of diabetes: standards of medical care in diabetes—2020. Diabetes care 43(Supplement_1), S14–S31 (2020).
- Aubreville, M., Stathonikos, N., Donovan, T.A., Klopfleisch, R., Ammeling, J., Ganz, J., Wilm, F., Veta, M., Jabari, S., Eckstein, M., et al.: Domain generalization across tumor types, laboratories, and species—insights from the 2022 edition of the mitosis domain generalization challenge. Medical Image Analysis 94, 103155 (2024).
- Azizi, S., Mustafa, B., Ryan, F., Beaver, Z., Freyberg, J., Deaton, J., Loh, A., Karthikesalingam, A., Kornblith, S., Chen, T., et al.: Big self-supervised models advance medical image classification. In: Proceedings of the IEEE/CVF international conference on computer vision. pp. 3478–3488 (2021).
- Bakator, M., Radosav, D.: Deep learning and medical diagnosis: A review of literature. Multimodal Technologies and Interaction 2(3), 47 (2018). [CrossRef]
- Bannur, S., Bouzid, K., Castro, D.C., Schwaighofer, A., Thieme, A., Bond-Taylor, S., Ilse, M., Pérez-García, F., Salvatelli, V., Sharma, H., et al.: Maira-2: Grounded radiology report generation. arXiv preprint arXiv:2406.04449 (2024).
- Baumgartner, M., Jäger, P.F., Isensee, F., Maier-Hein, K.H.: nndetection: a self-configuring method for medical object detection. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part V 24. pp. 530–539. Springer (2021).
- Bejnordi, B.E., Veta, M., Van Diest, P.J., Van Ginneken, B., Karssemeijer, N., Litjens, G., Van Der Laak, J.A., Hermsen, M., Manson, Q.F., Balkenhol, M., et al.: Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama 318(22), 2199–2210 (2017). [CrossRef]
- Benary, M., Wang, X.D., Schmidt, M., Soll, D., Hilfenhaus, G., Nassir, M., Sigler, C., Knödler, M., Keller, U., Beule, D., et al.: Leveraging large language models for decision support in personalized oncology. JAMA Network Open 6(11), e2343689–e2343689 (2023). [CrossRef]
- Boiko, D.A., MacKnight, R., Gomes, G.: Emergent autonomous scientific research capabilities of large language models. arXiv preprint arXiv:2304.05332 (2023).
- Brohan, A., Chebotar, Y., Finn, C., Hausman, K., Herzog, A., Ho, D., Ibarz, J., Irpan, A., Jang, E., Julian, R., et al.: Do as i can, not as i say: Grounding language in robotic affordances. In: Conference on robot learning. pp. 287–318. PMLR (2023).
- Chen, G., Dong, S., Shu, Y., Zhang, G., Sesay, J., Karlsson, B.F., Fu, J., Shi, Y.: Autoagents: A framework for automatic agent generation. arXiv preprint arXiv:2309.17288 (2023).
- Chen, X., Wu, Z., Liu, X., Pan, Z., Liu, W., Xie, Z., Yu, X., Ruan, C.: Janus-pro: Unified multimodal understanding and generation with data and model scaling. arXiv preprint arXiv:2501.17811 (2025).
- Coudray, N., Ocampo, P.S., Sakellaropoulos, T., Narula, N., Snuderl, M., Fenyö, D., Moreira, A.L., Razavian, N., Tsirigos, A.: Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nature medicine 24(10), 1559–1567 (2018). [CrossRef]
- Dou, Q., Chen, H., Yu, L., Zhao, L., Qin, J., Wang, D., Mok, V.C., Shi, L., Heng, P.A.: Automatic detection of cerebral microbleeds from mr images via 3d convolutional neural networks. IEEE transactions on medical imaging 35(5), 1182–1195 (2016). [CrossRef]
- Fallahpour, A., Ma, J., Munim, A., Lyu, H., Wang, B.: Medrax: Medical reasoning agent for chest x-ray. arXiv preprint arXiv:2502.02673 (2025).
- Fang, H., Li, F., Wu, J., Fu, H., Sun, X., Son, J., Yu, S., Zhang, M., Yuan, C., Bian, C., et al.: Refuge2 challenge: A treasure trove for multi-dimension analysis and evaluation in glaucoma screening. arXiv preprint arXiv:2202.08994 (2022).
- Gallotta, R., Todd, G., Zammit, M., Earle, S., Liapis, A., Togelius, J., Yannakakis, G.N.: Large language models and games: A survey and roadmap. arXiv preprint arXiv:2402.18659 (2024). [CrossRef]
- Ghafarollahi, A., Buehler, M.J.: Protagents: protein discovery via large language model multi-agent collaborations combining physics and machine learning. Digital Discovery (2024). [CrossRef]
- Ghezloo, F., Seyfioglu, M.S., Soraki, R., Ikezogwo, W.O., Li, B., Vivekanandan, T., Elmore, J.G., Krishna, R., Shapiro, L.: Pathfinder: A multi-modal multi-agent system for medical diagnostic decision-making applied to histopathology. arXiv preprint arXiv:2502.08916 (2025).
- Guo, D., Yang, D., Zhang, H., Song, J., Zhang, R., Xu, R., Zhu, Q., Ma, S., Wang, P., Bi, X., et al.: Deepseek-r1: Incentivizing reasoning capability in llms via reinforcement learning. arXiv preprint arXiv:2501.12948 (2025).
- Habli, Z., AlChamaa, W., Saab, R., Kadara, H., Khraiche, M.L.: Circulating tumor cell detection technologies and clinical utility: Challenges and opportunities. Cancers 12(7), 1930 (2020). [CrossRef]
- He, X., Zhang, Y., Mou, L., Xing, E., Xie, P.: Pathvqa: 30000+ questions for medical visual question answering. arXiv preprint arXiv:2003.10286 (2020).
- Huang, W., Xia, F., Xiao, T., Chan, H., Liang, J., Florence, P., Zeng, A., Tompson, J., Mordatch, I., Chebotar, Y., et al.: Inner monologue: Embodied reasoning through planning with language models. arXiv preprint arXiv:2207.05608 (2022).
- Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods 18(2), 203–211 (2021). [CrossRef]
- Jinxin, S., Jiabao, Z., Yilei, W., Xingjiao, W., Jiawen, L., Liang, H.: Cgmi: Configurable general multi-agent interaction framework. arXiv preprint arXiv:2308.12503 (2023).
- Khare, Y., Bagal, V., Mathew, M., Devi, A., Priyakumar, U.D., Jawahar, C.: Mmbert: Multimodal bert pretraining for improved medical vqa. In: 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI). pp. 1033–1036. IEEE (2021).
- Kim, Y., Park, C., Jeong, H., Chan, Y.S., Xu, X., McDuff, D., Lee, H., Ghassemi, M., Breazeal, C., Park, H.W.: Mdagents: An adaptive collaboration of llms for medical decision-making. In: The Thirty-eighth Annual Conference on Neural Information Processing Systems (2024).
- Kononenko, I.: Machine learning for medical diagnosis: history, state of the art and perspective. Artificial Intelligence in medicine 23(1), 89–109 (2001).
- Lai, X., Tian, Z., Chen, Y., Yang, S., Peng, X., Jia, J.: Step-dpo: Step-wise preference optimization for long-chain reasoning of llms. arXiv preprint arXiv:2406.18629 (2024).
- Lau, J.J., Gayen, S., Ben Abacha, A., Demner-Fushman, D.: A dataset of clinically generated visual questions and answers about radiology images. Scientific data 5(1), 1–10 (2018). [CrossRef]
- Li, B., Yan, T., Pan, Y., Luo, J., Ji, R., Ding, J., Xu, Z., Liu, S., Dong, H., Lin, Z., et al.: Mmedagent: Learning to use medical tools with multi-modal agent. arXiv preprint arXiv:2407.02483 (2024).
- Li, C., Wong, C., Zhang, S., Usuyama, N., Liu, H., Yang, J., Naumann, T., Poon, H., Gao, J.: Llava-med: Training a large language-and-vision assistant for biomedicine in one day. Advances in Neural Information Processing Systems 36 (2024).
- Li, G., Hammoud, H., Itani, H., Khizbullin, D., Ghanem, B.: Camel: Communicative agents for" mind" exploration of large language model society. Advances in Neural Information Processing Systems 36, 51991–52008 (2023).
- Li, J., Lai, Y., Li, W., Ren, J., Zhang, M., Kang, X., Wang, S., Li, P., Zhang, Y.Q., Ma, W., et al.: Agent hospital: A simulacrum of hospital with evolvable medical agents. arXiv preprint arXiv:2405.02957 (2024).
- Li, K., Hopkins, A.K., Bau, D., Viégas, F., Pfister, H., Wattenberg, M.: Emergent world representations: Exploring a sequence model trained on a synthetic task. arXiv preprint arXiv:2210.13382 (2022).
- Li, R., Zhang, C., Mao, S., Huang, H., Zhong, M., Cui, Y., Zhou, X., Yin, F., Theodoridis, S., Zhang, Z.: From english to pcsel: Llm helps design and optimize photonic crystal surface emitting lasers (2023).
- Li, Z., Song, D., Yang, Z., Wang, D., Li, F., Zhang, X., Kinahan, P.E., Qiao, Y.: Visionunite: A vision-language foundation model for ophthalmology enhanced with clinical knowledge. arXiv preprint arXiv:2408.02865 (2024).
- Liang, X., Li, X., Li, F., Jiang, J., Dong, Q., Wang, W., Wang, K., Dong, S., Luo, G., Li, S.: Medfilip: Medical fine-grained language-image pre-training. IEEE Journal of Biomedical and Health Informatics (2025). [CrossRef]
- Lin, J., Xia, Y., Zhang, J., Yan, K., Lu, L., Luo, J., Zhang, L.: Ct-glip: 3d grounded language-image pretraining with ct scans and radiology reports for full-body scenarios. arXiv preprint arXiv:2404.15272 (2024).
- Liu, B., Zhan, L.M., Xu, L., Ma, L., Yang, Y., Wu, X.M.: Slake: A semantically-labeled knowledge-enhanced dataset for medical visual question answering. In: 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI). pp. 1650–1654. IEEE (2021).
- Liu, H., Li, C., Wu, Q., Lee, Y.J.: Visual instruction tuning. Advances in neural information processing systems 36 (2024).
- Low, C.H., Wang, Z., Zhang, T., Zeng, Z., Zhuo, Z., Mazomenos, E.B., Jin, Y.: Surgraw: Multi-agent workflow with chain-of-thought reasoning for surgical intelligence. arXiv preprint arXiv:2503.10265 (2025).
- M. Bran, A., Cox, S., Schilter, O., Baldassari, C., White, A.D., Schwaller, P.: Augmenting large language models with chemistry tools. Nature Machine Intelligence pp. 1–11 (2024).
- Ma, J., He, Y., Li, F., Han, L., You, C., Wang, B.: Segment anything in medical images. Nature Communications 15(1), 654 (2024).
- Ma, Z., Mei, Y., Su, Z.: Understanding the benefits and challenges of using large language model-based conversational agents for mental well-being support. In: AMIA Annual Symposium Proceedings. vol. 2023, p. 1105 (2024).
- McPhee, S.J., Papadakis, M.A., Rabow, M.W., et al.: Current medical diagnosis & treatment 2010. McGraw-Hill Medical New York: (2010).
- of Medicine (US). Friends, N.L.: MedlinePlus, vol. 5. National Institutes of Health and the Friends of the National Library of … (2006).
- Mehta, N., Teruel, M., Sanz, P.F., Deng, X., Awadallah, A.H., Kiseleva, J.: Improving grounded language understanding in a collaborative environment by interacting with agents through help feedback. arXiv preprint arXiv:2304.10750 (2023).
- Miller, N., Lacroix, E.M., Backus, J.E.: Medlineplus: building and maintaining the national library of medicine’s consumer health web service. Bulletin of the Medical Library Association 88(1), 11 (2000).
- Moor, M., Huang, Q., Wu, S., Yasunaga, M., Dalmia, Y., Leskovec, J., Zakka, C., Reis, E.P., Rajpurkar, P.: Med-flamingo: a multimodal medical few-shot learner. In: Machine Learning for Health (ML4H). pp. 353–367. PMLR (2023).
- Moor, M., Huang, Q., Wu, S., Yasunaga, M., Dalmia, Y., Leskovec, J., Zakka, C., Reis, E.P., Rajpurkar, P.: Med-flamingo: a multimodal medical few-shot learner. In: Machine Learning for Health (ML4H). pp. 353–367. PMLR (2023).
- Qin, G., Hu, R., Liu, Y., Zheng, X., Liu, H., Li, X., Zhang, Y.: Data-efficient image quality assessment with attention-panel decoder. In: Proceedings of the AAAI Conference on Artificial Intelligence. vol. 37, pp. 2091–2100 (2023). [CrossRef]
- Ranella, N., Eger, M.: Towards automated video game commentary using generative ai. In: EXAG@ AIIDE (2023).
- Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany, October 5-9, 2015, proceedings, part III 18. pp. 234–241. Springer (2015).
- Shinn, N., Cassano, F., Gopinath, A., Narasimhan, K., Yao, S.: Reflexion: Language agents with verbal reinforcement learning. Advances in Neural Information Processing Systems 36 (2024).
- Steinberg, E., Greenfield, S., Wolman, D.M., Mancher, M., Graham, R.: Clinical practice guidelines we can trust. national academies press (2011).
- Szolovits, P., Patil, R.S., Schwartz, W.B.: Artificial intelligence in medical diagnosis. Annals of internal medicine 108(1), 80–87 (1988).
- Tang, X., Zou, A., Zhang, Z., Li, Z., Zhao, Y., Zhang, X., Cohan, A., Gerstein, M.: Medagents: Large language models as collaborators for zero-shot medical reasoning. arXiv preprint arXiv:2311.10537 (2023).
- Topsakal, O., Akinci, T.C.: Creating large language model applications utilizing langchain: A primer on developing llm apps fast. In: International Conference on Applied Engineering and Natural Sciences. vol. 1, pp. 1050–1056 (2023). [CrossRef]
- Wang, D., Zhang, Y., Zhang, K., Wang, L.: Focalmix: Semi-supervised learning for 3d medical image detection. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 3951–3960 (2020).
- Wang, K., Lu, Y., Santacroce, M., Gong, Y., Zhang, C., Shen, Y.: Adapting llm agents through communication. arXiv preprint arXiv:2310.01444 (2023).
- Wang, M., Lin, T., Lin, A., Yu, K., Peng, Y., Wang, L., Chen, C., Zou, K., Liang, H., Chen, M., et al.: Common and rare fundus diseases identification using vision-language foundation model with knowledge of over 400 diseases. arXiv preprint arXiv:2406.09317 (2024).
- Wang, Z., Zhang, Y., Wang, Y., Cai, L., Zhang, Y.: Dynamic pseudo label optimization in point-supervised nuclei segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 220–230. Springer (2024).
- Wu, J., Antonova, R., Kan, A., Lepert, M., Zeng, A., Song, S., Bohg, J., Rusinkiewicz, S., Funkhouser, T.: Tidybot: Personalized robot assistance with large language models. Autonomous Robots 47(8), 1087–1102 (2023). [CrossRef]
- Wu, J., Ji, W., Liu, Y., Fu, H., Xu, M., Xu, Y., Jin, Y.: Medical sam adapter: Adapting segment anything model for medical image segmentation. arXiv preprint arXiv:2304.12620 (2023). [CrossRef]
- Xia, Y., Shenoy, M., Jazdi, N., Weyrich, M.: Towards autonomous system: flexible modular production system enhanced with large language model agents. In: 2023 IEEE 28th International Conference on Emerging Technologies and Factory Automation (ETFA). pp. 1–8. IEEE (2023).
- Yang, S., Chen, Y., Tian, Z., Wang, C., Li, J., Yu, B., Jia, J.: Visionzip: Longer is better but not necessary in vision language models. arXiv preprint arXiv:2412.04467 (2024).
- Yang, S., Liu, J., Zhang, R., Pan, M., Guo, Z., Li, X., Chen, Z., Gao, P., Guo, Y., Zhang, S.: Lidar-llm: Exploring the potential of large language models for 3d lidar understanding. arXiv preprint arXiv:2312.14074 (2023).
- Yang, S., Qu, T., Lai, X., Tian, Z., Peng, B., Liu, S., Jia, J.: Lisa++: An improved baseline for reasoning segmentation with large language model. arXiv preprint arXiv:2312.17240 (2023).
- Zhan, L.M., Liu, B., Fan, L., Chen, J., Wu, X.M.: Medical visual question answering via conditional reasoning. In: Proceedings of the 28th ACM International Conference on Multimedia. pp. 2345–2354 (2020).
- Zhang, C., Yang, K., Hu, S., Wang, Z., Li, G., Sun, Y., Zhang, C., Zhang, Z., Liu, A., Zhu, S.C., et al.: Proagent: building proactive cooperative agents with large language models. In: Proceedings of the AAAI Conference on Artificial Intelligence. vol. 38, pp. 17591–17599 (2024). [CrossRef]
- Zhang, J., Xie, Y., Wu, Q., Xia, Y.: Medical image classification using synergic deep learning. Medical image analysis 54, 10–19 (2019). [CrossRef]
- Zhang, S., Xu, Y., Usuyama, N., Xu, H., Bagga, J., Tinn, R., Preston, S., Rao, R., Wei, M., Valluri, N., et al.: Biomedclip: a multimodal biomedical foundation model pretrained from fifteen million scientific image-text pairs. arXiv preprint arXiv:2303.00915 (2023).
- Zhang, X., Wu, C., Zhao, Z., Lin, W., Zhang, Y., Wang, Y., Xie, W.: Pmc-vqa: Visual instruction tuning for medical visual question answering. arXiv preprint arXiv:2305.10415 (2023).
- Zhang, Y., Wang, Y., Fang, Z., Bian, H., Cai, L., Wang, Z., Zhang, Y.: Dawn: Domain-adaptive weakly supervised nuclei segmentation via cross-task interactions. IEEE Transactions on Circuits and Systems for Video Technology (2024). [CrossRef]
- Zhao, D., Ferdian, E., Maso Talou, G.D., Quill, G.M., Gilbert, K., Wang, V.Y., Babarenda Gamage, T.P., Pedrosa, J., D’hooge, J., Sutton, T.M., et al.: Mitea: A dataset for machine learning segmentation of the left ventricle in 3d echocardiography using subject-specific labels from cardiac magnetic resonance imaging. Frontiers in Cardiovascular Medicine 9, 1016703 (2023). [CrossRef]
- Zhu, J., Qi, Y., Wu, J.: Medical sam 2: Segment medical images as video via segment anything model 2. arXiv preprint arXiv:2408.00874 (2024).
- Zuo, K., Jiang, Y., Mo, F., Lio, P.: Kg4diagnosis: A hierarchical multi-agent llm framework with knowledge graph enhancement for medical diagnosis. arXiv preprint arXiv:2412.16833 (2024).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).