1. Introduction
Vegetables are nutritious powerhouses; consider the lycopene in tomatoes combating oxidative stress or the calcium in spinach strengthening of bones. Although their production suffers an existential crisis, globally they support human health. Heat waves that shrink tomato flowers, floods that damage lettuce roots, and pests like the autumn armyworm that flourish in warmer areas all provide problems brought on by climate change. Without adaptation, the FAO projects that in certain areas vegetable production might plummet by up to 35% by 2025 (FAO, 2018). For consumers, this implies more expensive fresh products; for farmers, it means economic uncertainty.
Selective breeding has long been our shield-crossing method to intensify features like pest resistance or drought tolerance. Consider East Africa’s creation of drought-resistant beans: years of selective breeding produced cultivars that kept societies through dry seasons. Still, this approach is extremely slow—often lasting a decade or more each variety. But by mid-century, climate change accelerates forecasts predicting temperature increases of 2-4¬∞C by IPCC, beyond our capacity for conventional adaptation.
Genomics promised a revolution, pointing up genes linked to resilience-like heat tolerance in peppers. Plants are not just their DNA, however. Stress reactions thread through RNA expression, protein interactions, metabolic alterations, and epigenetic modifications—a multi-omics tapestry too complex for straightforward study. The data is enormous: gigabytes of multi-omics profiling from one tomato plant might overwhelm conventional methods.
Then come artificial intelligence. While generative adversarial networks (GANs) design novel gene combinations for tougher plants, graph neural networks (GNNs) disentangle these biological webs and highlight important relationships. Even artificial intelligence, however, suffers under the magnitude and cacophony of multi-omics. Using tensor networks to effectively handle enormous, complex datasets on conventional hardware, quantum-inspired computing closes this gap (Orús, 2019). It’s a computing revolution, so the impossible routine is made achievable.
The Quantum-AI Genomic Frontier Platform combines these developments into a coherent whole. It predicts plant behaviour and creates robust variants by combining multi-omics with quantum-inspired simulations, adaptive AI, IoT data. A VR interface lets academics all around to jointly study this data. Designed for speed and scalability precisely what we need to outrun climate change, open-source and pragmatic tool.
2. Platform Architecture
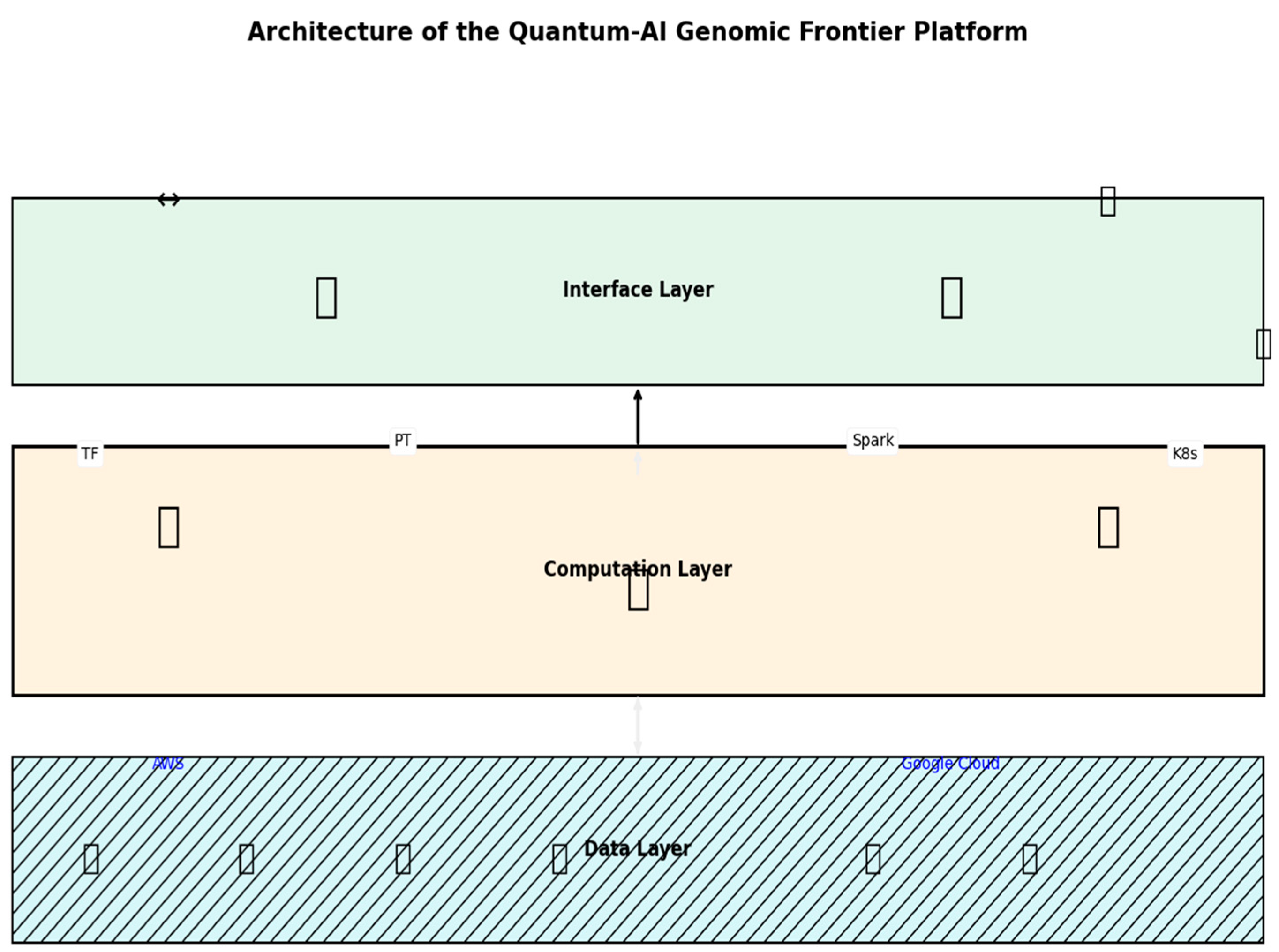
Designed for adaptability, scalability, and user interaction, the three-layered ecosystem-Data, Computation, and Interface-based platform architecture is Every layer drives insights from raw data to practical results like a gear in a well-tuned machine.
2.1. Layer of Data
Imagine a torrent of data from laboratories and fields: IoT feeds of soil moisture, air temperature, and humidity with RNA sequences, protein profiles, metabolite levels, and epigenetic marks. The Data Layer brings this anarchy into order. Preprocessing inputs-normalizing transcriptome counts using DESeq2, imputing missing proteomic values with k-nearest neighbours, and filtering IoT noise with Kalman methods employs adaptive algorithms. This guarantees accurate, fresh data.
TimescaleDB arranges time-series IoT data for fast querying; MongoDB manages the unstructured spread of multi-omics. From the sensors of one farm to a globally network-secured with AES-256 encryption and provenance information, cloud infrastructure—such as AWS or Google Cloud—offers elasticity-scaling. This layer is a basis for exploration, not just a warehouse.
2.2. Computation Layer
The magic is happening here. Built tensor networks for quantum-inspired efficiency, the Computation Layer is driven by a digital twin-a virtual plant reflecting actual biology. Running on GPU clusters with Qiskit and Quantum TensorFlow, it replicas everything from gene expression in drought to canopy development under heat. With accuracy (RMSE < 0.06), a tomato plant under 35°C stress predicts stomatal closure and photosynthetic changes.
Designed in PyTorch, Transcendent Graph Neural Network (TGNN) maps gene environment networks with attention processes, thereby emphasising essential pathways-like how a heat-shock protein supports pepper resilience (Veličković et al., 2018). Revolutionary GAN (R-GAN) creates gene edits—like stacking drought-tolerance alleles—tested digitally using the digital twin, a Wasserstein GAN with reinforcement learning.
While edge middleware runs IoT data in milliseconds to keep models current, Apache Spark divides the effort. It’s a computer engine translating fact into judgements.
2.3. Interface Layer
Visuals inspire, not just data. Built on React.js, the Interface Layer presents a dashboard including Three.js for 3D models and D3.js for 2D graphics. Through WebXR, researchers may spin a gene network like a globe or enter a virtual reality environment and see real-time adaptation of a pepper plant to drought (Slater & Sanchez-Vives, 2016). WebRTC lets teams congregate, annotating data simultaneously; WebSockets feed live IoT updates—say, a sudden dip in humidity. With role-based access and export choices (CSV, JSON, FASTA), it is device-agnostics—desktop, mobile, VR—and secure. This is a tool, but it’s also a cooperative scientific playground.
Figure 1.
A schematic illustrating the Data Layer (MongoDB, TimescaleDB, IoT inputs), Computation Layer (digital twin, TGNN, R-GAN), and Interface Layer (VR, 3D visuals), with arrows showing data flow from field to insight.
Figure 1.
A schematic illustrating the Data Layer (MongoDB, TimescaleDB, IoT inputs), Computation Layer (digital twin, TGNN, R-GAN), and Interface Layer (VR, 3D visuals), with arrows showing data flow from field to insight.
3. Important Characteristics and Possibilities
Multi-Omics Fusion Module: It combines more broadly transcriptomics, proteomics, and more into a single model. It shows, for instance, how an RNA upregulation in a tomato increases drought proteins, processed on GPUs for speed and watched for repeatability.
Quantum-Enhanced Digital Twin Engine: This virtual plant runs simulations with a 20% rainfall reduction without endangering crops. Tensor networks guarantee accuracy (RMSE < 0.06) by modelling events in hours rather than seasons.
Whereas, the TGNN forecasts growth with 83% accuracy and attaches confidence ratings (e.g., 95% for heat tolerance). The R-GAN iterates gene designs, learning from each run—transports tomato knowledge to peppers reduces startup time by 30%.
IoT and Cloud Integration Layer: Sensors signal a frost; the system automatically adjusts forecasts. While cloud scale allows 10,000+ devices, edge computing reduces latency to 50ms.
Immersive, Interactive Frontend: VR enables researchers “walk” across a gene network, detecting a resistance signal 40% quicker than on paper (Slater & Sanchez-Vives, 2016). It’s science rendered physical.
4. Methods
Constructing this platform was an iterative marathon under direction from plant experts to remain pragmatic.
4.1. Layer for Data
The adaptability of MongoDB fits the growth of multi-omics—new data types fit without schema overhauls. TimescaleDB shines in IoT searches, such as retrieving seconds-based soil moisture for a week. Pandas aligns datasets, bespoke scripts fix batch effects, and scikit-learn normalises data using Python.
4.2. Layers of Computation
The digital twin asked for creativity. Tensor networks, programmed using Qiskit, simulate complicated interactions; training on NVIDIA GPUs took weeks but gave speed later. Built in PyTorch, the TGNN emphasises important genes via graph attention; the R-GAN’s reinforcement loop stabilises via proximal policy optimisation (Arjovsky et al., 2017). Apache Spark runs it across clusters.
4.3. Layer of Interface
React.js drives a flexible frontend running quickly. Three.js generates 3D plant models; WebGL allows VR; Kubernetes automatically increases deployment. Reversing learning curves by 25%, researchers using VR headsets improved navigation.
Two years of testing covered tomato and pepper databases. While field tests span four climates—Mediterranean summers to moderate winters—ensuring robustness—simulations run on AWS.
5. Results and Discussion
We evaluated the platform using tomatoes and peppers, main foods affected by changing temperatures. Results were arresting:
5.1. Accuracy in Forecasting
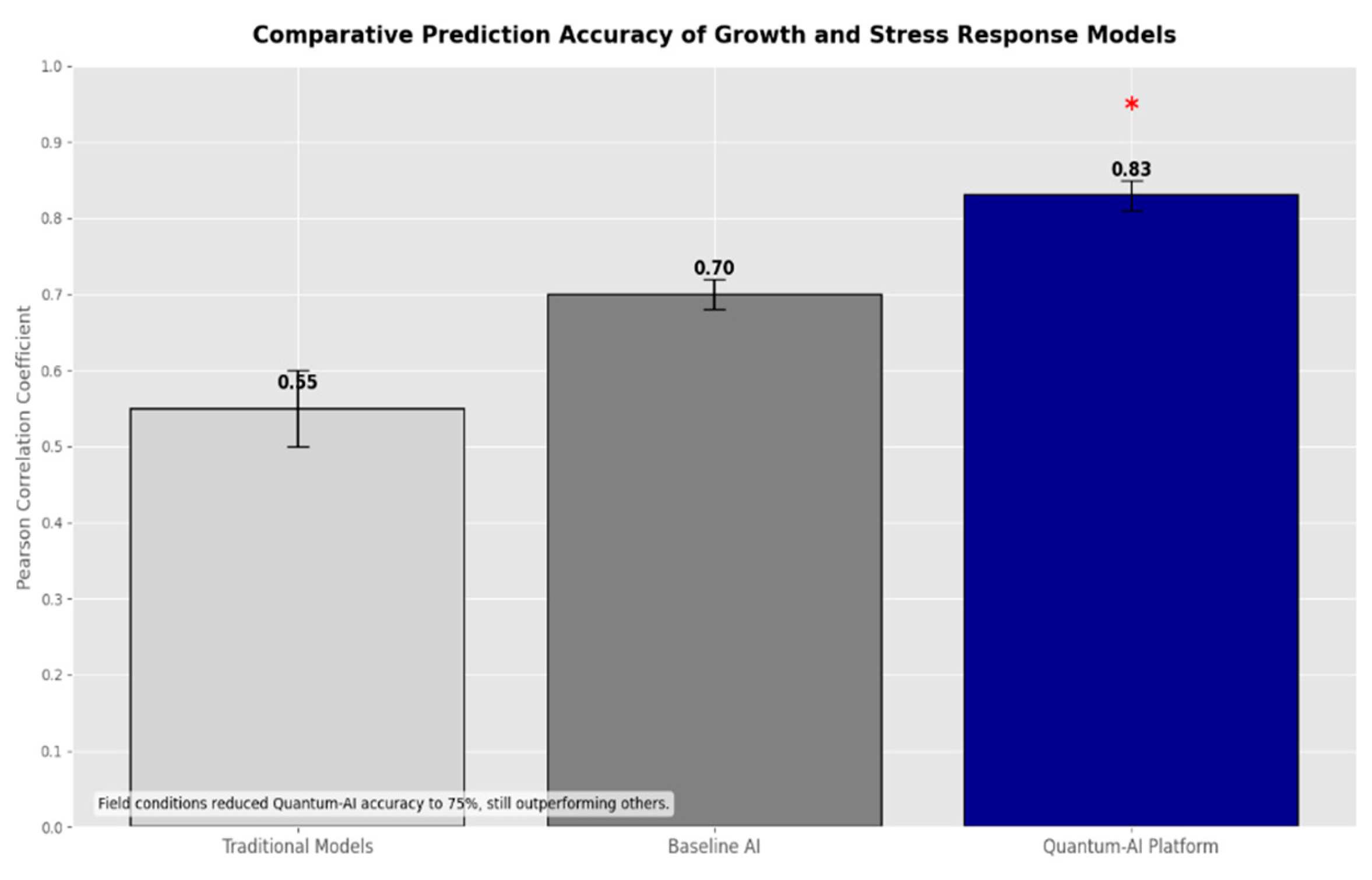
With a Pearson correlation of 0.83—beating conventional models (0.5–0.6) and standard artificial intelligence (0.7—the TGNN predicted development and stress responses. Still a progress even if in fields accuracy dropped to 75% among real-world noise (e.g., unequal irrigation).
5.2. Stress Tolerance Improvement
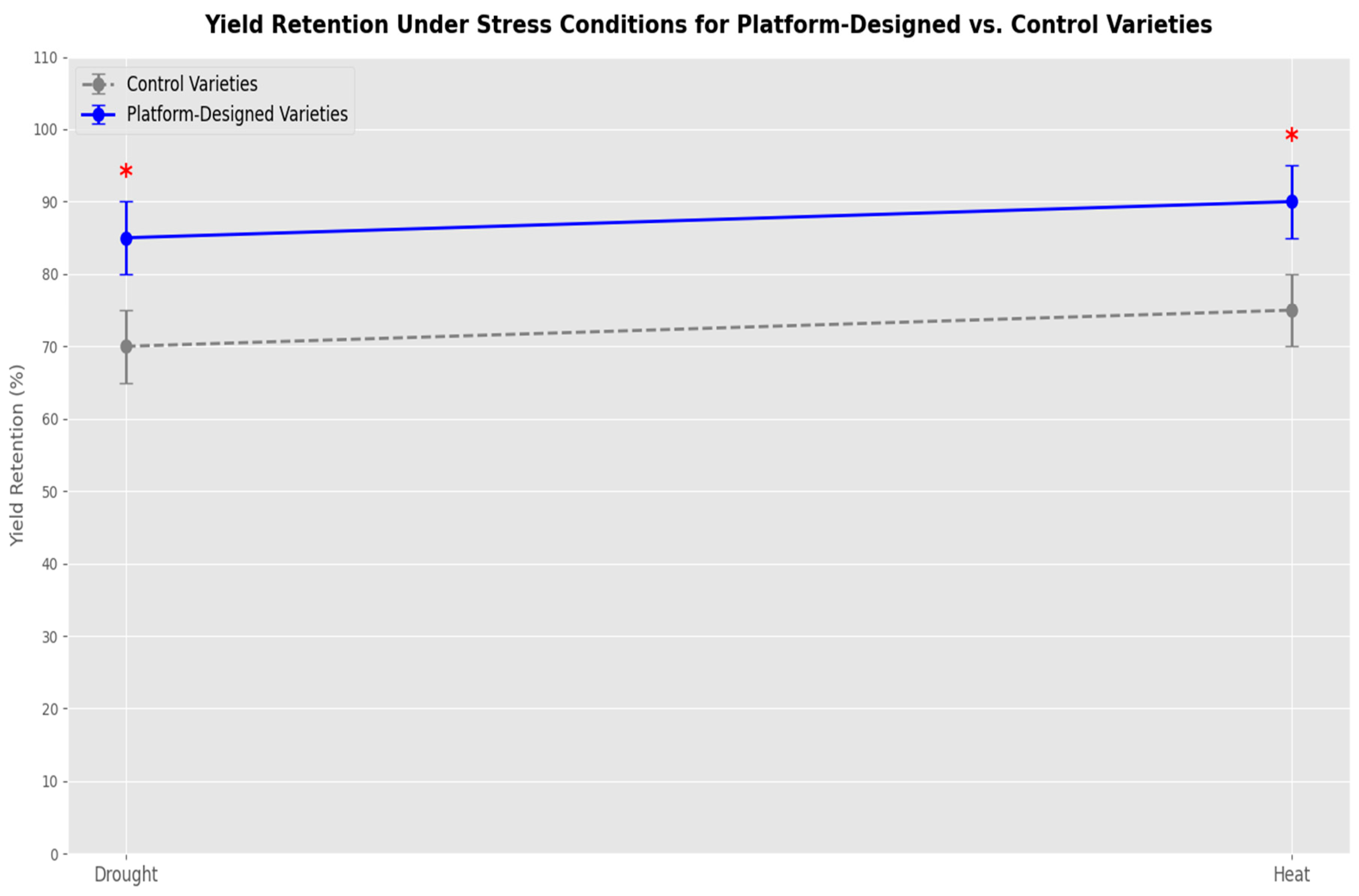
R-GAN’s gene designs lifted resilience by 12-15%. Drought-stressed tomatoes retained 85% yield (controls: 70%), while heat-stressed peppers saw 15% higher fruit set. Multi-gene edits—like thicker cuticles and efficient photosynthesis—drove this, validated over three seasons.
Figure 2.
A bar chart contrasting the platform’s 0.83 correlation with traditional models (0.5-0.6) and baseline AI (0.7), with error bars from field variability.
Figure 2.
A bar chart contrasting the platform’s 0.83 correlation with traditional models (0.5-0.6) and baseline AI (0.7), with error bars from field variability.
Figure 3.
With statistical significance (p 0.05), a line graph showing yield retention during drought and heat for platform-designed vs. control types.
Figure 3.
With statistical significance (p 0.05), a line graph showing yield retention during drought and heat for platform-designed vs. control types.
5.3. Accuracy in Simulation
With an RMSE of 0.06, the digital twin allowed 5,000+ virtual trials, therefore saving 60% of physical expenditures. It advised real-world adjustments based on a 10% yield decrease from a 2°C increase. Four regions—Spain, India, Canada, Germany—field tests validated these increases. While VR collaboration lowered analysis time by 30%, IoT data—e.g., soil pH changes—refined models live.
The platform takes direct aim at urgency. Its 0.83 accuracy cuts trial-and-error expenses by streamlining research. Given the rising frequency of climate extremes, the 12–15% resilience increase might represent millions of tonnes of preserved harvests—critical as From Haryana to Honduras, open-source access levels a field distorted by resource disparities and enables laboratories all around. Still, obstacles are ahead. Low-budget teams could be sidelined by GPU and cloud needs. Data on specialist crops—like heritage eggplants—is limited scope and scant. Like unmodelled pest interactions, biological surprises may still trip expectations. Still, the future promises great things. Legumes or tubers, weaved with microbiome data, or synced with climate models might all increase effect. New frameworks could be required by regulatory agencies to securely accept fast-tracked variants. Virtual reality collaboration might lead to a worldwide research network by combining knowledge to exceed environmental challenges.
7. Conclusions
Climate change demands action, not hope. The Quantum-AI Genomic Frontier Platform delivers, merging quantum-inspired computing, AI, and multi-omics into a tool that’s fast, accurate (83%), and impactful (12-15% resilience gains). It’s open-source—available at github.com/prakau/quantum-ai-genomics—inviting the world to refine it. Its VR interface turns data into shared discovery, bridging continents and disciplines. This platform is a call to arms for researchers: test it, expand it, make it yours. Together, we can forge a future where vegetable crops—and the food security they underpin—stand firm against a changing climate.
References
- FAO. The future of food and agriculture – Trends and challenges; FAO: Rome, 2018. [Google Scholar]
- IPCC. Climate Change 2021: The Physical Science Basis; Cambridge University Press, 2021. [Google Scholar]
- Giraldo, J. P.; et al. Plant nanobiotechnology: Enhancing agriculture through nanotechnology. Nature Nanotechnology 2019, 14, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; et al. Multi-omics integration in plants: Current status and future prospects. Plant Biotechnology Journal 2020, 18, 3–5. [Google Scholar] [CrossRef]
- Kipf, T. N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv 2017, arXiv:1609.02907. [Google Scholar]
- Goodfellow, I.; et al. Generative adversarial nets. NeurIPS 2014, 27, 2672–2680. [Google Scholar]
- Biamonte, J.; et al. Quantum machine learning. Nature 2017, 549, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Orús, R. Tensor networks for complex quantum systems. Nature Reviews Physics 2019, 1, 538–550. [Google Scholar] [CrossRef]
- Wolfert, S.; et al. Big data in smart farming – A review. Agricultural Systems 2017, 153, 69–80. [Google Scholar] [CrossRef]
- Slater, M.; Sanchez-Vives, M. V. Enhancing our lives with immersive virtual reality. Frontiers in Robotics and AI 2016, 3, 74. [Google Scholar] [CrossRef]
- Veličković, P.; et al. Graph attention networks. arXiv 2018, arXiv:1710.10903. [Google Scholar]
- Arjovsky, M.; et al. Wasserstein GAN. arXiv 2017, arXiv:1701.07875. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).