Submitted:
10 March 2025
Posted:
10 March 2025
You are already at the latest version
Abstract
Background. Physical activity induces a range of physiological and molecular adaptations, particularly affecting skeletal muscle and the cardiovascular system, regulating both tissue architecture and metabolic pathways. Emerging evidence suggests that PIWI-interacting RNAs (piRNAs) may serve as potential biomarkers for these adaptations. Methods. This study selected eight participants of the iReAct study (DRKS00017446) for piRNA analysis. Baseline assessments included demographic profiling and fitness evaluation, particularly maximal oxygen uptake (V̇O2max) assessment. In addition, blood samples were collected pre and post a standard reference training session. Subsequently, subjects underwent 6-week training protocols, employing standardized high-intensity interval training (HIIT) and moderate-intensity continuous training (MICT) regimens. RNA sequencing was conducted to identify differentially expressed piRNAs, and correlation analyses were performed between piRNA expression patterns and training-associated changes in V̇O2max. Different screening procedures were applied to identify piRNAs potentially of interest in the context of exercise. Results. Specific piRNAs exhibited characteristic changes in expression levels in response to exercise. There were no significant correlations of piRNA patterns with changes in V̇O2max, but some interesting trends, that might be verified in larger studies. Conclusions. This hypothesis-generating study identifies characteristic piRNA patterns in the context of exercise. Their significance as biomarkers is yet to be determined.
Keywords:
1. Introduction
2. Materials and Methods
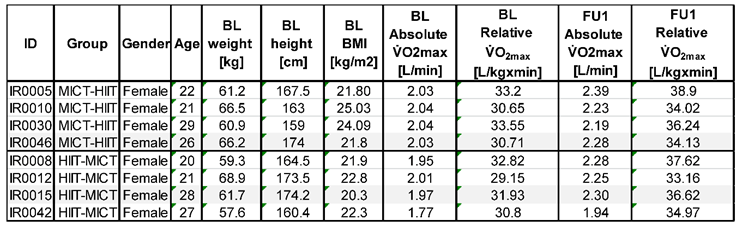
2.1. iReAct: Study Participants
2.2. iReAct: Training Intervention Protocols
2.3. iReAct: Diagnostics Blocks
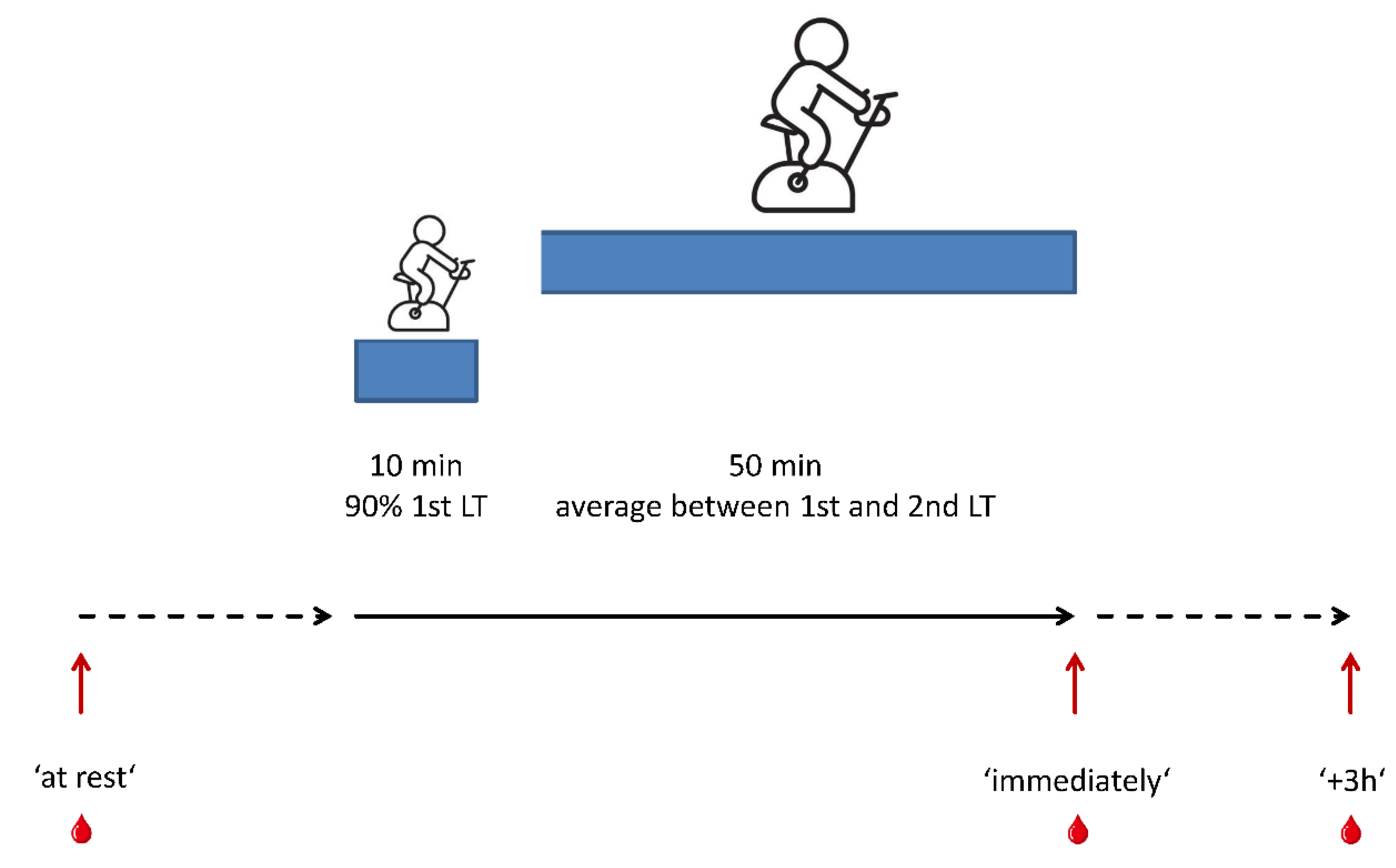
2.4. iReAct: Standardized Reference Training Protocol and Collection of Blood Samples
2.5. Next-Generation Sequencing (NGS)
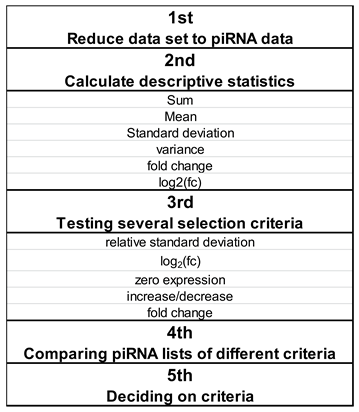
2.6. Criteria Selection for Evaluation of Differentially Expressed piRNAs
2.7. piRNA Selection
2.8. piRNA Characterization
2.9. Correlation Analysis
2.10. Data Visualization
3. Results
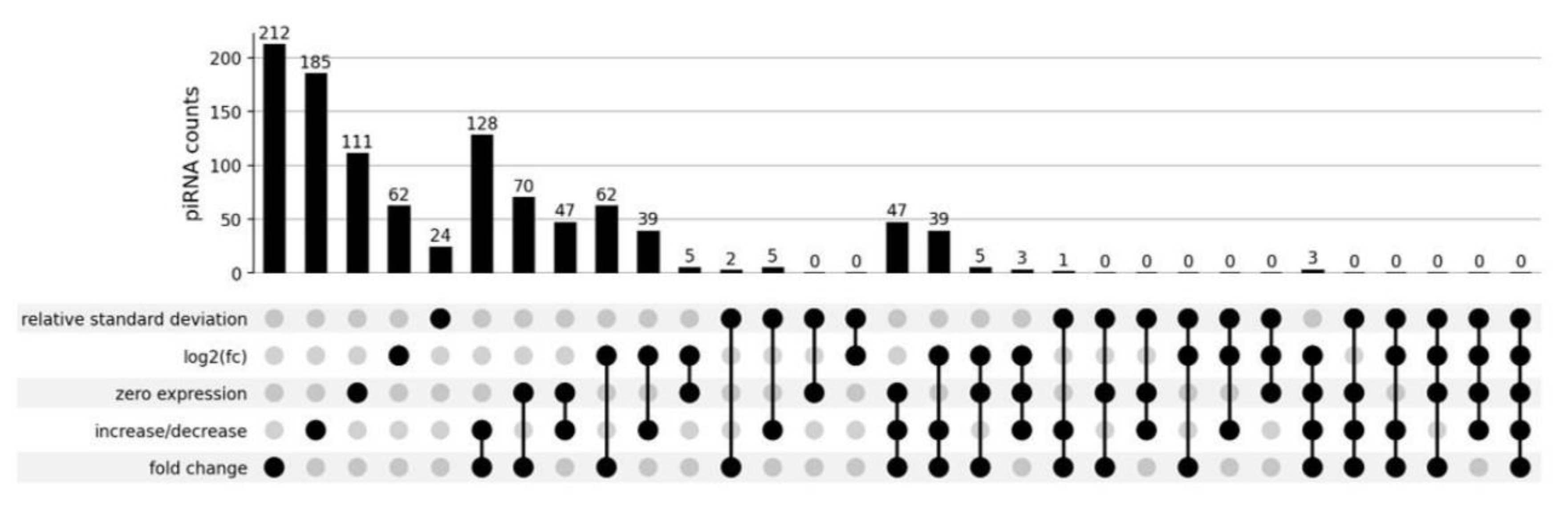
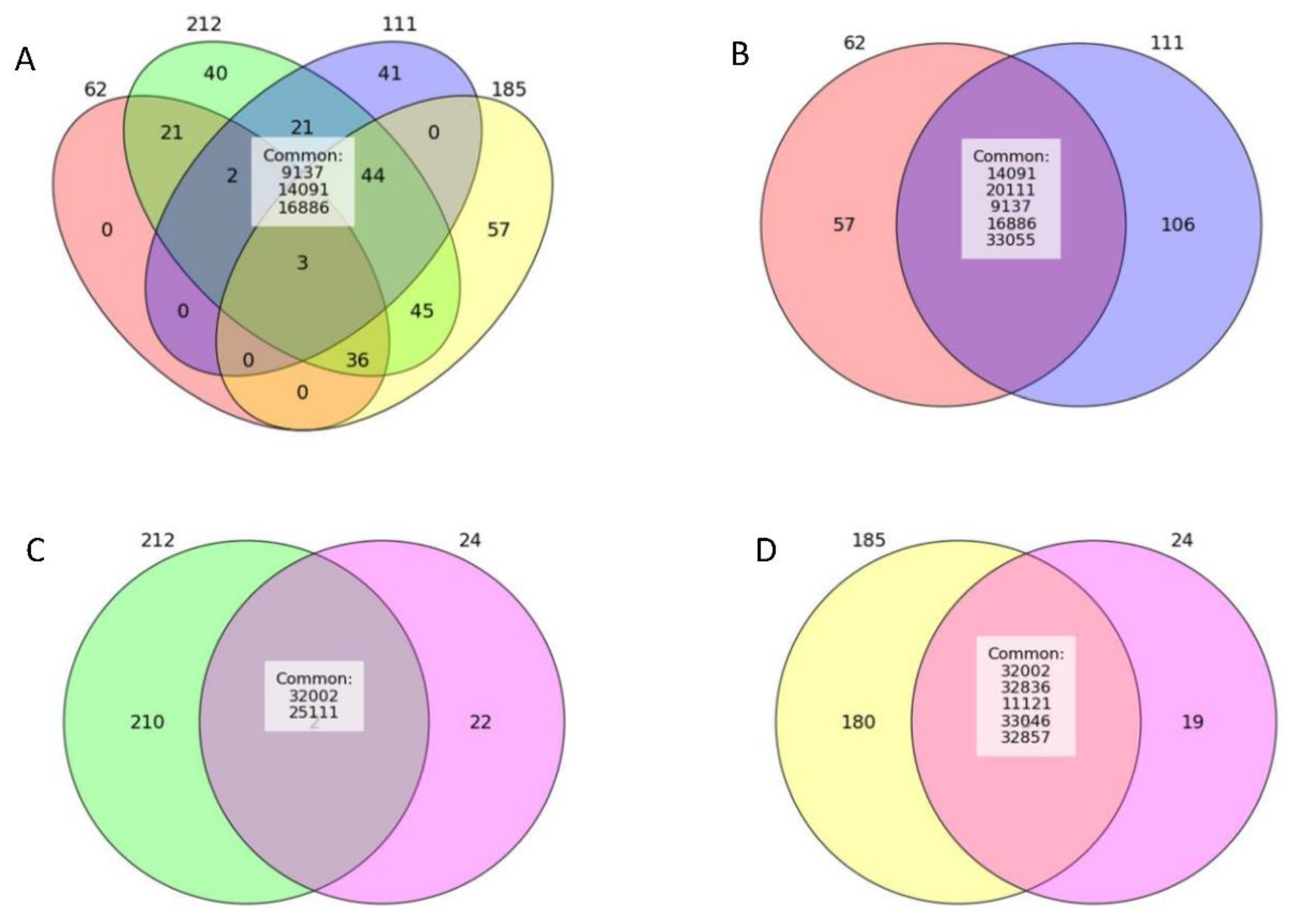
3.1. Differentially Expressed piRNAs
3.2. Plot-Based piRNA Selection
3.3. piRNA Characterization
3.4. Correlation with ΔV̇O2max
4. Discussion
Study Limitations
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PIWI | A family of proteins interacting with piRNAs |
| piRNA | PIWI-interacting RNA |
| piRNAdb | PIWI-interacting RNA Database |
| piRBase | A piRNA database with different naming conventions |
| HIIT | High-intensity interval training |
| MICT | Moderate-intensity continuous training |
| rRNA | Ribosomal RNA |
| piRNAdb | PIWI-interacting RNA Database |
| sncRNA | Small non-coding RNA |
| miRNA | MicroRNA |
| lncRNA | Long non-coding RNA |
| tRNA | Transfer RNA |
| p-ERM | Protein-ERM (Ezrin, Radixin, Moesin) complex, related to cytoskeleton regulation |
| piRNA-Ls | piRNA-like small RNAs |
| hsa-piR-xxx | Human piRNA identification (e.g., hsa-piR-1677, hsa-piR-11119) |
| RER | Respiratory exchange ratio |
| La- | Blood lactate concentration |
| NGS | Next generation sequencing |
| HRmax | Maximum heart rate |
| V̇O2max | Maximal oxygen uptake |
| log2(fc) | Logarithmic base 2 fold change |
| qPCR | Quantitative polymerase chain reaction |
| NCBI | National Center for Biotechnology Information |
References
- Thiel, A.; Sudeck, G.; Gropper, H.; Maturana, F.M.; Schubert, T.; Srismith, D.; Widmann, M.; Behrens, S.; Martus, P.; Munz, B.; et al. The iReAct Study - A Biopsychosocial Analysis of the Individual Response to Physical Activity. Contemp Clin Trials Commun 2020, 17, 100508. [CrossRef]
- Warburton, D.E.R.; Bredin, S.S.D. Reflections on Physical Activity and Health: What Should We Recommend? Can J Cardiol 2016, 32, 495–504. [CrossRef]
- Warburton, D.E.R.; Nicol, C.W.; Bredin, S.S.D. Health Benefits of Physical Activity: The Evidence. CMAJ 2006, 174, 801–809. [CrossRef]
- Warburton, D.E.R.; Nicol, C.W.; Bredin, S.S.D. Prescribing Exercise as Preventive Therapy. CMAJ 2006, 174, 961–974. [CrossRef]
- Pedersen, B.K.; Saltin, B. Exercise as Medicine - Evidence for Prescribing Exercise as Therapy in 26 Different Chronic Diseases. Scand J Med Sci Sports 2015, 25 Suppl 3, 1–72. [CrossRef]
- WHO Guidelines on Physical Activity and Sedentary Behaviour Available online: https://www.who.int/multi-media/details/who-guidelines-on-physical-activity-and-sedentary-behaviour (accessed on 17 December 2024).
- Dindorf, C.; Fröhlich, M. Zur Konnotation und Denotation des Trainingsbegriffs in Theorie und Praxis des Sports. Ger J Exerc Sport Res 2020, 50, 297–307. [CrossRef]
- Fröhlich, M. Überlegungen zum Trainingsbegriff und zur Theorie des Trainings aus ökonomischer Perspektive. 2009.
- Buchheit, M.; Laursen, P.B. High-Intensity Interval Training, Solutions to the Programming Puzzle: Part I: Cardiopulmonary Emphasis. Sports Med 2013, 43, 313–338. [CrossRef]
- Brauner, T.; Müller, J.; Beinert, K. Sportwissenschaft; Elsevier Health Sciences, 2021; ISBN 978-3-437-06007-6.
- Binder, R.K.; Wonisch, M.; Corra, U.; Cohen-Solal, A.; Vanhees, L.; Saner, H.; Schmid, J.-P. Methodological Approach to the First and Second Lactate Threshold in Incremental Cardiopulmonary Exercise Testing. Eur J Cardiovasc Prev Rehabil 2008, 15, 726–734. [CrossRef]
- Caen, K.; Pogliaghi, S.; Lievens, M.; Vermeire, K.; Bourgois, J.G.; Boone, J. Ramp vs. Step Tests: Valid Alternatives to Determine the Maximal Lactate Steady-State Intensity? Eur J Appl Physiol 2021, 121, 1899–1907. [CrossRef]
- Mattioni Maturana, F.; Soares, R.N.; Murias, J.M.; Schellhorn, P.; Erz, G.; Burgstahler, C.; Widmann, M.; Munz, B.; Thiel, A.; Nieß, A.M. Responders and Non-Responders to Aerobic Exercise Training: Beyond the Evaluation of V˙O2max. Physiol Rep 2021, 9, e14951. [CrossRef]
- Widmann, M.; Mattioni Maturana, F.; Burgstahler, C.; Erz, G.; Schellhorn, P.; Fragasso, A.; Schmitt, A.; Nieß, A.M.; Munz, B. miRNAs as Markers for the Development of Individualized Training Regimens: A Pilot Study. Physiol Rep 2022, 10, e15217. [CrossRef]
- Wackerhage, H.; Woods, N.M. Exercise-Induced Signal Transduction and Gene Regulation in Skeletal Muscle. J Sports Sci Med 2002, 1, 103–114.
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in Human Disease and Prospects for Epigenetic Therapy. Nature 2004, 429, 457–463. [CrossRef]
- Castel, S.E.; Martienssen, R.A. RNA Interference in the Nucleus: Roles for Small RNAs in Transcription, Epigenetics and Beyond. Nat Rev Genet 2013, 14, 100–112. [CrossRef]
- Fernández-Sanjurjo, M.; de Gonzalo-Calvo, D.; Fernández-García, B.; Díez-Robles, S.; Martínez-Canal, Á.; Olmedillas, H.; Dávalos, A.; Iglesias-Gutiérrez, E. Circulating microRNA as Emerging Biomarkers of Exercise. Exercise and Sport Sciences Reviews 2018, 46, 160. [CrossRef]
- Domańska-Senderowska, D.; Laguette, M.-J.N.; Jegier, A.; Cięszczyk, P.; September, A.V.; Brzeziańska-Lasota, E. MicroRNA Profile and Adaptive Response to Exercise Training: A Review. International Journal of Sports Medicine 2019, 40, 227–235. [CrossRef]
- Sapp, R.M.; Shill, D.D.; Roth, S.M.; Hagberg, J.M. Circulating microRNAs in Acute and Chronic Exercise: More than Mere Biomarkers. Journal of Applied Physiology 2017, 122, 702–717. [CrossRef]
- de Gonzalo-Calvo, D.; Dávalos, A.; Montero, A.; García-González, Á.; Tyshkovska, I.; González-Medina, A.; Soares, S.M.A.; Martínez-Camblor, P.; Casas-Agustench, P.; Rabadán, M.; et al. Circulating Inflammatory miRNA Signature in Response to Different Doses of Aerobic Exercise. Journal of Applied Physiology 2015, 119, 124–134. [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [CrossRef]
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals. Mol Cell 2018, 71, 775-790.e5. [CrossRef]
- Hirakata, S.; Siomi, M.C. piRNA Biogenesis in the Germline: From Transcription of piRNA Genomic Sources to piRNA Maturation. Biochim Biophys Acta 2016, 1859, 82–92. [CrossRef]
- Yamanaka, S.; Siomi, M.C.; Siomi, H. piRNA Clusters and Open Chromatin Structure. Mob DNA 2014, 5, 22. [CrossRef]
- Patel, M.Z.; Jiang, Y.; Kakumani, P.K. Somatic piRNA and PIWI-Mediated Post-Transcriptional Gene Regulation in Stem Cells and Disease. Front. Cell Dev. Biol. 2024, 12. [CrossRef]
- Belicard, T.; Jareosettasin, P.; Sarkies, P. The piRNA Pathway Responds to Environmental Signals to Establish Intergenerational Adaptation to Stress. BMC Biology 2018, 16, 103. [CrossRef]
- Oliveira, G.P.; Porto, W.F.; Palu, C.C.; Pereira, L.M.; Petriz, B.; Almeida, J.A.; Viana, J.; Filho, N.N.A.; Franco, O.L.; Pereira, R.W. Effects of Acute Aerobic Exercise on Rats Serum Extracellular Vesicles Diameter, Concentration and Small RNAs Content. Front Physiol 2018, 9, 532. [CrossRef]
- Ingerslev, L.R.; Donkin, I.; Fabre, O.; Versteyhe, S.; Mechta, M.; Pattamaprapanont, P.; Mortensen, B.; Krarup, N.T.; Barrès, R. Endurance Training Remodels Sperm-Borne Small RNA Expression and Methylation at Neurological Gene Hotspots. Clinical Epigenetics 2018, 10, 12. [CrossRef]
- Harriss, D.J.; MacSween, A.; Atkinson, G. Ethical Standards in Sport and Exercise Science Research: 2020 Update. Int J Sports Med 2019, 40, 813–817. [CrossRef]
- Grieb, A.; Schmitt, A.; Fragasso, A.; Widmann, M.; Mattioni Maturana, F.; Burgstahler, C.; Erz, G.; Schellhorn, P.; Nieß, A.M.; Munz, B. Skeletal Muscle MicroRNA Patterns in Response to a Single Bout of Exercise in Females: Biomarkers for Subsequent Training Adaptation? Biomolecules 2023, 13, 884. [CrossRef]
- PIWI-Interacting RNA (piRNA) Database - piRNAdb Available online: https://www.pirnadb.org/ (accessed on 27 January 2025).
- Mei, Y.; Wang, Y.; Kumari, P.; Shetty, A.C.; Clark, D.; Gable, T.; MacKerell, A.D.; Ma, M.Z.; Weber, D.J.; Yang, A.J.; et al. A piRNA-like Small RNA Interacts with and Modulates p-ERM Proteins in Human Somatic Cells. Nat Commun 2015, 6, 7316. [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A Germline-Specific Class of Small RNAs Binds Mammalian Piwi Proteins. Nature 2006, 442, 199–202. [CrossRef]
- Rizzo, F.; Rinaldi, A.; Marchese, G.; Coviello, E.; Sellitto, A.; Cordella, A.; Giurato, G.; Nassa, G.; Ravo, M.; Tarallo, R.; et al. Specific Patterns of PIWI-Interacting Small Noncoding RNA Expression in Dysplastic Liver Nodules and Hepatocellular Carcinoma. Oncotarget 2016, 7, 54650–54661. [CrossRef]
- Gainetdinov, I.V.; Skvortsova, Y.V.; Kondratieva, S.A.; Klimov, A.; Tryakin, A.A.; Azhikina, T.L. Assessment of piRNA Biogenesis and Function in Testicular Germ Cell Tumors and Their Precursor Germ Cell Neoplasia in Situ. BMC Cancer 2018, 18, 20. [CrossRef]
- Yin, J.; Qi, W.; Ji, C.-G.; Zhang, D.-X.; Xie, X.-L.; Ding, Q.; Jiang, X.-Y.; Han, J.; Jiang, H.-Q. Small RNA Sequencing Revealed Aberrant piRNA Expression Profiles in Colorectal Cancer. Oncol Rep 2019, 42, 263–272. [CrossRef]
- Betts, S.D.; Basu, S.; Bolar, J.; Booth, R.; Chang, S.; Cigan, A.M.; Farrell, J.; Gao, H.; Harkins, K.; Kinney, A.; et al. Uniform Expression and Relatively Small Position Effects Characterize Sister Transformants in Maize and Soybean. Front Plant Sci 2019, 10, 1209. [CrossRef]
- Yang, X.; Cheng, Y.; Lu, Q.; Wei, J.; Yang, H.; Gu, M. Detection of Stably Expressed piRNAs in Human Blood. Int J Clin Exp Med 2015, 8, 13353–13358.
- Freedman, J.E.; Gerstein, M.; Mick, E.; Rozowsky, J.; Levy, D.; Kitchen, R.; Das, S.; Shah, R.; Danielson, K.; Beaulieu, L.; et al. Diverse Human Extracellular RNAs Are Widely Detected in Human Plasma. Nat Commun 2016, 7, 11106. [CrossRef]
- Gao, X.-Q.; Zhang, Y.-H.; Liu, F.; Ponnusamy, M.; Zhao, X.-M.; Zhou, L.-Y.; Zhai, M.; Liu, C.-Y.; Li, X.-M.; Wang, M.; et al. The piRNA CHAPIR Regulates Cardiac Hypertrophy by Controlling METTL3-Dependent N6-Methyladenosine Methylation of Parp10 mRNA. Nat Cell Biol 2020, 22, 1319–1331. [CrossRef]
- Martone, J.; Mariani, D.; Desideri, F.; Ballarino, M. Non-Coding RNAs Shaping Muscle. Front Cell Dev Biol 2019, 7, 394. [CrossRef]
- Tosar, J.P.; Rovira, C.; Cayota, A. Non-Coding RNA Fragments Account for the Majority of Annotated piRNAs Expressed in Somatic Non-Gonadal Tissues. Commun Biol 2018, 1, 2. [CrossRef]
 |
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





