Submitted:
03 March 2025
Posted:
04 March 2025
You are already at the latest version
Abstract
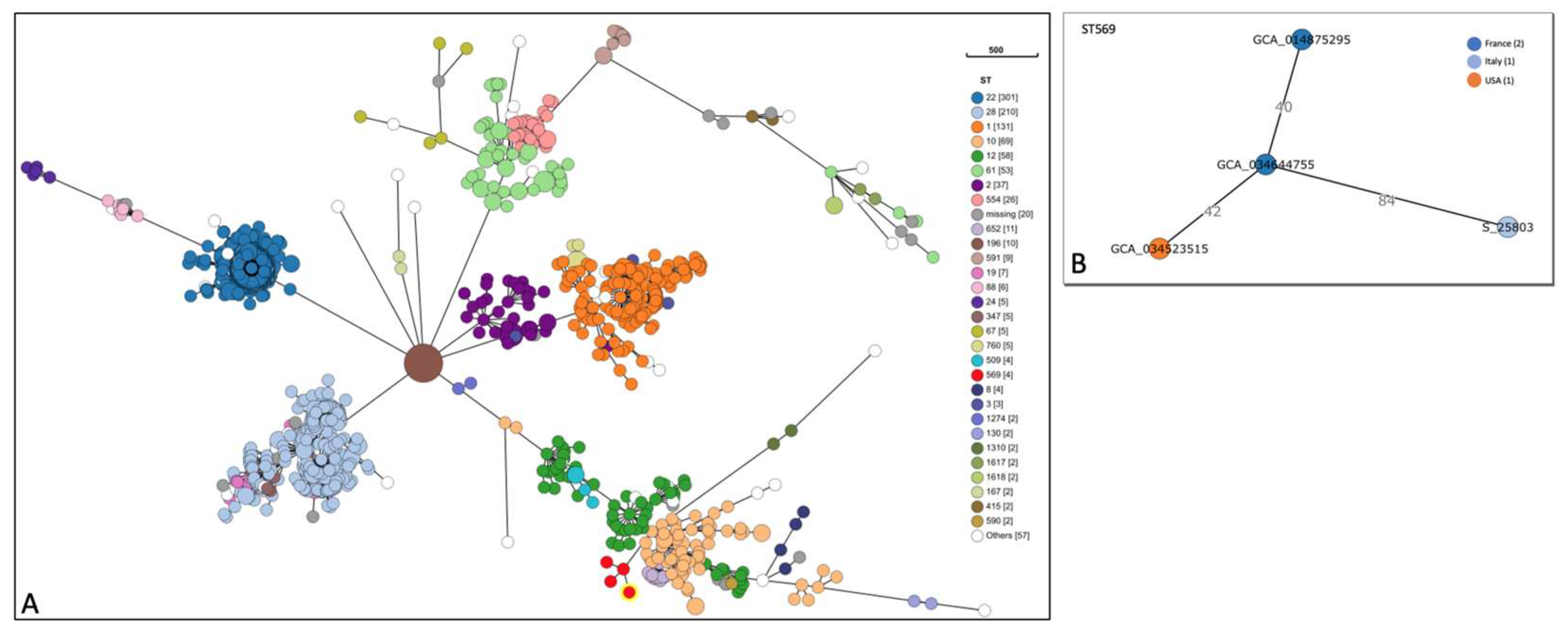
This study, for the first time in Italy, analyses by WGS a Streptococcus agalactiae strain isolated from a non-pregnant adult affected by meningitis and without common risk factors. The S. agalactiae strain was classified as a serotype II (SS2), sequence type ST569. Molecular characterization evidenced the presence of resistance genes to tetracycline and macrolide (tet(M) and mre(A)) and several virulence genes coding for adhesion and immune evasion factors (bca, cps family, neu family, scpB, gbs family, pil family and hylB), toxins (cfa/cfb, cyl family), pro-inflammatory factors (lepA), and two homologous genes that contributed to bacterial escape from the host immune system (lmb, luxS). SNPs analysis showed 18 different alleles, with 9 missense SNP mutations related to genes involved in cellular metabolism (dhaS, ftsE, ligA, nrdD and secA), virulence (bgrR and galE) and antimicrobial resistance (glpK and mutL). SNPs in glpK and mutL genes might reduce susceptibility to drugs. The SNPs analysis highlighted the presence of mutations conferring pathogenicity to the strain. The evidence in this study could explain the development of meningitis in a healthy patient. This case highlights the importance of using molecular methods to characterize the complete genome of a bacterial species that could seriously affect human health.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Case Review
2.2. Bacteriological Examination
2.3. DNA Extraction
2.4. PCR Typing
2.5. Whole Genome Sequencing Analysis
3. Results
3.1. Biochemical Analysis of CSF Samples
3.2. Microbiological Examination and Molecular Analysis of CSF Samples
3.3. Whole Genome Sequencing Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| WGS | Whole Genome Sequencing |
| S. agalactiae | Streptococcus agalactiae |
| MLST | Multi Locus Sequence Type |
| SNP | Single Nucleotide Polymorphism |
| GBS | group B Streptococcus |
| CSF | cerebrospinal fluid |
| cgMLST | core genome multilocus sequence typing |
| CPS | capsular polysaccharide |
References
- Lancefield RC. A serological differentiation of human and other groups of hemolytic streptococci. J Exp Med. 1933 Mar 354 31;57(4):571-95. [CrossRef]
- Burcham LR, Spencer BL, Keeler LR, et al. Determinants of Group B streptococcal virulence potential amongst vaginal clinical 320 isolates from pregnant women. PLoS One. 2019 Dec 18;14(12):e0226699. [CrossRef]
- Furfaro LL, Chang BJ, Kahler CM, et al. Genomic characterisation of perinatal Western Australian Streptococcus agalactiae 344 isolates. PLoS One. 2019 Oct 2;14(10):e0223256. [CrossRef]
- Hanna M, Noor A. Streptococcus Group B. Stat Pearls Publishing. 2023 Jan 16. PMID: 31985936.351.
- Emaneini M, Khoramian B, Jabalameli F, et al. Comparison of virulence factors and capsular types of Streptococcus agalactiae 336 isolated from human and bovine infections. Microb Pathog. 2016 Feb;91:1-4. [CrossRef]
- He EM, Chen CW, Guo Y, et al. The genome of serotype VI Streptococcus agalactiae serotype VI and comparative analysis. 352 Gene. 2017 Jan 15;597:59-65. [CrossRef]
- Meroni G, Sora VM, Martino PA, et al. Epidemiology of Antimicrobial Resistance Genes in Streptococcus agalactiae Sequences 359 from a Public Database in a One Health Perspective. Antibiotics (Basel). 2022 Sep 12;11(9):1236. [CrossRef]
- Zhou Y, Zhao XC, Wang LQ, et al. Detecting Genetic Variation of Colonizing Streptococcus agalactiae Genomes in Humans: A 391 Precision Protocol. Front Bioinform. 2022 Jun 3;2:813599. [CrossRef]
- Yao K, Poulsen K, Maione D, et al. Capsular gene typing of Streptococcus agalactiae compared to serotyping by latex aggluti- 389 nation. J Clin Microbiol. 2013 Feb;51(2):503-7. Epub 2012 Nov 28. 390. [CrossRef]
- Pang M, Sun L, He T, et al. Molecular and virulence characterization of highly prevalent Streptococcus agalactiae circulated in 361 bovine dairy herds. Vet Res. 2017 Oct 16;48(1):65. [CrossRef]
- Crestani C, Forde TL, Lycett SJ, et al. The fall and rise of group B Streptococcus in dairy cattle: reintroduction due to human-to- 329 cattle host jumps? Microb Genom. 2021 Sep;7(9):000648. [CrossRef]
- Furfaro LL, Chang BJ, Payne MS. A novel one-step real-time multiplex PCR assay to detect Streptococcus agalactiae presence 346 and serotypes Ia, Ib, and III. Diagn Microbiol Infect Dis. 2017 Sep;89(1):7-12. [CrossRef]
- Slotved HC, Møller JK, Khalil MR, et al. The serotype distribution of Streptococcus agalactiae (GBS) carriage isolates among 375 pregnant women having risk factors for early-onset GBS disease: a comparative study with GBS causing invasive infections 376 during the same period in Denmark. BMC Infect Dis. 2021 Nov 1;21(1):1129. [CrossRef]
- D’Angelo M, Boretti I, Quattrocchi S, et al. Lethal infective endocarditis due to Streptococcus agalactiae in a man with a history 331 of alcohol abuse: A case report. Medicine (Baltimore). 2019 Dec;98(51):e18270. [CrossRef]
- Sendi P, Johansson L, Norrby-Teglund A. Invasive group B Streptococcal disease in non-pregnant adults: a review with em- 367 phasis on skin and soft-tissue infections. Infection. 2008 Mar;36(2):100-11. Epub 2008 Jan 12. 368. [CrossRef]
- Al-Bayati A, Douedi S, Alsaoudi G, et al. Meningitis from invasive Streptococcus agalactiae in a healthy young adult. IDCases. 316 2020 Jul 7;21:e00907. [CrossRef]
- Chaiwarith R, Jullaket W, Bunchoo M, et al. Streptococcus agalactiae in adults at Chiang Mai University Hospital: a retrospec- 324 tive study. BMC Infect Dis. 2011 May 25;11:149. [CrossRef]
- Vasikasin V, Changpradub D. Clinical manifestations and prognostic factors for Streptococcus agalactiae bacteremia among 378 nonpregnant adults in Thailand. J Infect Chemother. 2021 Jul;27(7):967-971. Epub 2021 Feb 18. 379. [CrossRef]
- Vuillemin X, Hays C, Plainvert C, Dmytruk N, Louis M, Touak G, Saint-Pierre B, Adoux L, Letourneur F, Frigo A, Poyart C, 384 Tazi A. Invasive group B Streptococcus infections in non-pregnant adults: a retrospective study, France, 2007-2019. Clin Micro- 385 biol Infect. Jan 2021;27(1):129.e1-129.e4. Epub 2020 Sep 29. 386. [CrossRef]
- Diaz MH, Waller JL, Napoliello RA, et al. Optimization of Multiple Pathogen Detection Using the TaqMan Array Card: Appli- 333 cation for a Population-Based Study of Neonatal Infection. PLoS One. 2013 Jun 21;8(6):e66183. [CrossRef]
- Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014 Mar 387 3;15(3):R46. [CrossRef]
- Brettin T, Davis JJ, Disz T, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom 318 annotation pipelines and annotating batches of genomes. Sci Rep. 2015 Feb 10;5:8365. [CrossRef]
- Sheppard AE, Vaughan A, Jones N, et al. Capsular Typing Method for Streptococcus agalactiae Using Whole-Genome Sequence 373 Data. J Clin Microbiol. 2016 May;54(5):1388-90. Epub 2016 Mar 9. 374. [CrossRef]
- Regan JA, Klebanoff MA, Nugent RP. The epidemiology of group B streptococcal colonization in pregnancy. Vaginal Infections 365 and Prematurity Study Group. Obstet Gynecol. 1991 Apr;77(4):604-10. PMID: 2002986. 366.
- Seale AC, Bianchi-Jassir F, Russell NJ, et al. Estimates of the Burden of Group B Streptococcal Disease Worldwide for Pregnant 369 Women, Stillbirths, and Children. Clin Infect Dis. 2017 Nov 6;65(suppl_2):S200-S219. [CrossRef]
- Schwartz B, Schuchat A, Oxtoby MJ, et al. Invasive group B streptococcal disease in adults. A population-based study in met- 371 ropolitan Atlanta. JAMA. 1991 Aug 28;266(8):1112-4. PMID: 1865545. 372.
- Farley MM, Harvey RC, Stull T, et al. A population-based assessment of invasive disease due to group B Streptococcus in 339 nonpregnant adults. N Engl J Med. 1993 Jun 24;328(25):1807-11. [CrossRef]
- Farley MM. Group B streptococcal disease in nonpregnant adults. Clin Infect Dis. 2001 Aug 15;33(4):556-61. [CrossRef]
- Crespo-Ortiz Mdel P, Castañeda-Ramirez CR, Recalde-Bolaños M, Vélez-Londoño JD. Emerging trends in invasive and noninvasive isolates of Streptococcus agalactiae in a Latin American hospital: a 17-year study. BMC Infect Dis. 2014 Aug 3;14:428. [CrossRef]
- Camuset G, Picot S, Jaubert J, et al. Invasive Group B Streptococcal Disease in Non-pregnant Adults, Réunion Island, 2011. Int 322 J Infect Dis. 2015 Jun;35:46-50. [CrossRef]
- Francois Watkins LK, McGee L, Schrag SJ, et al. Epidemiology of Invasive Group B Streptococcal Infections Among Nonpreg- 341 nant Adults in the United States, 2008-2016. JAMA Intern Med. 2019 Apr 1;179(4):479-488. [CrossRef]
- Coelho T, Pacheco M, Mendes T, Valente J, Gil P. Invasive Streptococcus agalactiae Disease With Meningitis and Septic Arthritis in a Non-pregnant Patient. Cureus. 2022 Nov 4;14(11):e31077. [CrossRef]
- Peechakara B, Demkowicz R, Gupta M. Meningitis by Streptococcus agalactiae secondary to otitis media. QJM. 2018 Dec 363 1;111(12):891-892. [CrossRef]
- Villareal K, Goslin A, Bajracharya H. Group B Streptococcus Meningitis Associated with Acute Otitis Media in an Adult Patient. 382 Am J Case Rep. 2021 Oct 3;22:e933093. [CrossRef]
- Ma J, Zhang Z, Pan Z, et al. Streptococcus suis Uptakes Carbohydrate Source from Host Glycoproteins by N-glycans Degrada- 356 tion System for Optimal Survival and Full Virulence during Infection. Pathogens. 2020 May 18;9(5):387. [CrossRef]
- Vasilyeva A, Santos Sanches I, Florindo C, et al. Natural Mutations in Streptococcus agalactiae Resulting in Abrogation of β 380 Antigen Production. PLoS One. 2015 Jun 5;10(6):e0128426. [CrossRef]
- Chen CL, Cheng MH, Kuo CF, et al. Dextromethorphan Attenuates NADPH Oxidase-Regulated Glycogen Synthase Kinase 3β 326 and NF-κB Activation and Reduces Nitric Oxide Production in Group A Streptococcal Infection. Antimicrob Agents Chemother. 327 2018 May 25;62(6):e02045-17. [CrossRef]
- Gizachew M, Tiruneh M, Moges F, et al. Molecular characterization of Streptococcus agalactiae isolated from pregnant women 348 and newborns at the University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia. BMC Infect Dis. 2020 Jan 349 13;20(1):35. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




