Submitted:
28 February 2025
Posted:
03 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | Linear dichroism |
References
- Hirota, T. and B.H. King, Autism Spectrum Disorder: A Review. JAMA, 2023. 329(2): p. 157-168.
- Hodges, H., C. Fealko, and N. Soares, Autism spectrum disorder: definition, epidemiology, causes, and clinical evaluation. Transl Pediatr, 2020. 9(Suppl 1): p. S55-S65.
- Lord, C., et al., Autism spectrum disorder. Nat Rev Dis Primers, 2020. 6(1): p. 5.
- Havdahl, A., et al., Genetic contributions to autism spectrum disorder. Psychol Med, 2021. 51(13): p. 2260-2273.
- Zafeiriou, D.I., A. Ververi, and E. Vargiami, Childhood autism and associated comorbidities. Brain Dev, 2007. 29(5): p. 257-72.
- Sztainberg, Y. and H.Y. Zoghbi, Lessons learned from studying syndromic autism spectrum disorders. Nat Neurosci, 2016. 19(11): p. 1408-1417.
- Weuring, W., J. Geerligs, and B.P.C. Koeleman, Gene Therapies for Monogenic Autism Spectrum Disorders. Genes (Basel), 2021. 12(11).
- AmericanPsychiatricAssociation, Diagnostic and statistical manual of mental disorders (5th ed.). 2013.
- Dell’Osso L., L.P., Carpita B., The neurodevelopmental continuum towards a neurodevelopmental gradient hypothesis. Journal of Psychopathology, 2019. 25: p. 179-182.
- Singh, T., et al., The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet, 2017. 49(8): p. 1167-1173.
- Morris-Rosendahl, D.J. and M.A. Crocq, Neurodevelopmental disorders-the history and future of a diagnostic concept Dialogues Clin Neurosci, 2020. 22(1): p. 65-72.
- Owen, M.J. and M.C. O'Donovan, Schizophrenia and the neurodevelopmental continuum:evidence from genomics. World Psychiatry, 2017. 16(3): p. 227-235.
- Kereszturi, E., Diversity and Classification of Genetic Variations in Autism Spectrum Disorder. Int J Mol Sci, 2023. 24(23).
- Antaki, D., et al., A phenotypic spectrum of autism is attributable to the combined effects of rare variants, polygenic risk and sex. Nat Genet, 2022. 54(9): p. 1284-1292.
- Yap, C.X., et al., Analysis of common genetic variation and rare CNVs in the Australian Autism Biobank. Mol Autism, 2021. 12(1): p. 12.
- Apte, M. and A. Kumar, Correlation of mutated gene and signalling pathways in ASD. IBRO Neurosci Rep, 2023. 14: p. 384-392.
- Qiu, S., et al., Genetics of autism spectrum disorder: an umbrella review of systematic reviews and meta-analyses. Transl Psychiatry, 2022. 12(1): p. 249.
- Al-Dewik, N. and M. Alsharshani, New Horizons for Molecular Genetics Diagnostic and Research in Autism Spectrum Disorder. Adv Neurobiol, 2020. 24: p. 43-81.
- Vicari, S., et al., Copy number variants in autism spectrum disorders. Prog Neuropsychopharmacol Biol Psychiatry, 2019. 92: p. 421-427.
- Satterstrom, F.K., et al., Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell, 2020. 180(3): p. 568-584 e23.
- Schaaf, C.P., et al., A framework for an evidence-based gene list relevant to autism spectrum disorder. Nat Rev Genet, 2020. 21(6): p. 367-376.
- Viggiano, M., et al., Genomic analysis of 116 autism families strengthens known risk genes and highlights promising candidates. NPJ Genom Med, 2024. 9(1): p. 21.
- Al-Beltagi, M., et al., Metabolomic changes in children with autism. World J Clin Pediatr, 2024. 13(2): p. 92737.
- Acuna-Hidalgo, R., et al., Post-zygotic Point Mutations Are an Underrecognized Source of De Novo Genomic Variation. Am J Hum Genet, 2015. 97(1): p. 67-74.
- Ogata, H., et al., Autism spectrum disorders and hyperactive/impulsive behaviors in Japanese patients with Prader-Willi syndrome: a comparison between maternal uniparental disomy and deletion cases. Am J Med Genet A, 2014. 164A(9): p. 2180-6.
- Sanders, S.J., et al., De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature, 2012. 485(7397): p. 237-41.
- Contractor, A., I.M. Ethell, and C. Portera-Cailliau, Cortical interneurons in autism. Nat Neurosci, 2021. 24(12): p. 1648-1659.
- Kschonsak, M., et al., Structure of the human sodium leak channel NALCN. Nature, 2020. 587(7833): p. 313-318.
- Zhou, L., et al., Architecture of the human NALCN channelosome. Cell Discov, 2022. 8(1): p. 33.
- Cochet-Bissuel, M., P. Lory, and A. Monteil, The sodium leak channel, NALCN, in health and disease. Front Cell Neurosci, 2014. 8: p. 132.
- Chong, J.X., et al., De novo mutations in NALCN cause a syndrome characterized by congenital contractures of the limbs and face, hypotonia, and developmental delay. Am J Hum Genet, 2015. 96(3): p. 462-73.
- Kottgen, M., et al., Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. EMBO J, 2005. 24(4): p. 705-16.
- Olson, H.E., et al., A Recurrent De Novo PACS2 Heterozygous Missense Variant Causes Neonatal-Onset Developmental Epileptic Encephalopathy, Facial Dysmorphism, and Cerebellar Dysgenesis. Am J Hum Genet, 2018. 102(5): p. 995-1007.
- Sheng, M. and E. Kim, The Shank family of scaffold proteins. J Cell Sci, 2000. 113 ( Pt 11): p. 1851-6.
- Uchino, S. and C. Waga, SHANK3 as an autism spectrum disorder-associated gene. Brain Dev, 2013. 35(2): p. 106-10.
- Nemirovsky, S.I., et al., Whole genome sequencing reveals a de novo SHANK3 mutation in familial autism spectrum disorder. PLoS One, 2015. 10(2): p. e0116358.
- Huang, G., et al., Uncovering the Functional Link Between SHANK3 Deletions and Deficiency in Neurodevelopment Using iPSC-Derived Human Neurons. Front Neuroanat, 2019. 13: p. 23.
- Wu, S., et al., Shank3 deficiency elicits autistic-like behaviors by activating p38alpha in hypothalamic AgRP neurons. Mol Autism, 2024. 15(1): p. 14.
- Cuthbert, P.C., et al., Synapse-associated protein 102/dlgh3 couples the NMDA receptor to specific plasticity pathways and learning strategies. J Neurosci, 2007. 27(10): p. 2673-82.
- Trobiani, L., et al., The neuroligins and the synaptic pathway in Autism Spectrum Disorder. Neurosci Biobehav Rev, 2020. 119: p. 37-51.
- Rossignol, D.A. and R.E. Frye, Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry, 2012. 17(3): p. 290-314.
- Haas, R.H., Autism and mitochondrial disease. Dev Disabil Res Rev, 2010. 16(2): p. 144-53.
- Rose, S., et al., Clinical and Molecular Characteristics of Mitochondrial Dysfunction in Autism Spectrum Disorder. Mol Diagn Ther, 2018. 22(5): p. 571-593.
- Sergi, C. and B. Parayil Sankaran, Succinic Semialdehyde Dehydrogenase Deficiency, in StatPearls. 2024: Treasure Island (FL).
- Didiasova, M., et al., Succinic Semialdehyde Dehydrogenase Deficiency: An Update. Cells, 2020. 9(2).
- Julia-Palacios, N.A., et al., The continuously evolving phenotype of succinic semialdehyde dehydrogenase deficiency. J Inherit Metab Dis, 2024. 47(3): p. 447-462.
- Frye, R.E., Succinic semialdehyde dehydrogenase deficiency: A model of neurocircuit imbalances in autism and potential insight into new biomarkers. Dev Med Child Neurol, 2023. 65(12): p. 1544-1545.
- Gogou, M., et al., Succinic Semialdehyde Dehydrogenase Deficiency Presenting as Autism Spectrum Disorder. Indian J Pediatr, 2016. 83(9): p. 1036-7.
- Chung, T., et al., Dihydropyrimidine Dehydrogenase Is a Prognostic Marker for Mesenchymal Stem Cell-Mediated Cytosine Deaminase Gene and 5-Fluorocytosine Prodrug Therapy for the Treatment of Recurrent Gliomas. Theranostics, 2016. 6(10): p. 1477-90.
- van Kuilenburg, A.B.P., et al., Dihydropyrimidine Dehydrogenase Deficiency: Homozygosity for an Extremely Rare Variant in DPYD due to Uniparental Isodisomy of Chromosome 1. JIMD Rep, 2019. 45: p. 65-69.
- Fleger, M., et al., Dihydropyrimidine Dehydrogenase Deficiency: Metabolic Disease or Biochemical Phenotype? JIMD Rep, 2017. 37: p. 49-54.
- Carter, M.T., et al., Hemizygous deletions on chromosome 1p21.3 involving the DPYD gene in individuals with autism spectrum disorder. Clin Genet, 2011. 80(5): p. 435-43.
- Micheli, V., et al., Neurological disorders of purine and pyrimidine metabolism. Curr Top Med Chem, 2011. 11(8): p. 923-47.
- Sprenger, H.G., et al., Cellular pyrimidine imbalance triggers mitochondrial DNA-dependent innate immunity. Nat Metab, 2021. 3(5): p. 636-650.
- Frye, R.E., J.C. Slattery, and E.V. Quadros, Folate metabolism abnormalities in autism: potential biomarkers. Biomark Med, 2017. 11(8): p. 687-699.
- Kerrigan, J.F., et al., Fumaric aciduria: clinical and imaging features. Ann Neurol, 2000. 47(5): p. 583-8.
- Patel, K.P., et al., The spectrum of pyruvate dehydrogenase complex deficiency: clinical, biochemical and genetic features in 371 patients. Mol Genet Metab, 2012. 106(3): p. 385-94.
- Bhandary, S. and K. Aguan, Pyruvate dehydrogenase complex deficiency and its relationship with epilepsy frequency--An overview. Epilepsy Res, 2015. 116: p. 40-52.
- Gibson, G.E., R. Jope, and J.P. Blass, Decreased synthesis of acetylcholine accompanying impaired oxidation of pyruvic acid in rat brain minces. Biochem J, 1975. 148(1): p. 17-23.
- Liu, Y., et al., SPAF, a new AAA-protein specific to early spermatogenesis and malignant conversion. Oncogene, 2000. 19(12): p. 1579-88.
- Puusepp, S., et al., Compound heterozygous SPATA5 variants in four families and functional studies of SPATA5 deficiency. Eur J Hum Genet, 2018. 26(3): p. 407-419.
- Tanaka, A.J., et al., Mutations in SPATA5 Are Associated with Microcephaly, Intellectual Disability, Seizures, and Hearing Loss. Am J Hum Genet, 2015. 97(3): p. 457-64.
- Станчева М, Т.А., Тoдoрoв Т, Атемин С, Павлoва З, Туртурикoв И, Кадийска Т, Маринoва Е, Пoпoва Д, Аланай Я, Клинична картина и генетични кoрелации при български пациенти с мутации на SPATA5 ген. Редки бoлести и лекарства сираци, 2021. 11(4): p. 19-23.
- Tuc, E., et al., The third family with TAF6-related phenotype: Alazami-Yuan syndrome. Clin Genet, 2020. 97(5): p. 795-796.
- Selicorni, A., et al., Cornelia de Lange Syndrome: From a Disease to a Broader Spectrum. Genes (Basel), 2021. 12(7).
- Weissmiller, A.M., et al., Inhibition of MYC by the SMARCB1 tumor suppressor. Nat Commun, 2019. 10(1): p. 2014.
- Clapier, C.R., et al., Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol, 2017. 18(7): p. 407-422.
- Kleefstra, T., et al., Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet, 2006. 79(2): p. 370-7.
- Frega, M., et al., Distinct Pathogenic Genes Causing Intellectual Disability and Autism Exhibit a Common Neuronal Network Hyperactivity Phenotype. Cell Rep, 2020. 30(1): p. 173-186 e6.
- Garbelli, A., et al., A motif unique to the human DEAD-box protein DDX3 is important for nucleic acid binding, ATP hydrolysis, RNA/DNA unwinding and HIV-1 replication. PLoS One, 2011. 6(5): p. e19810.
- Mo, J., et al., DDX3X: structure, physiologic functions and cancer. Mol Cancer, 2021. 20(1): p. 38.
- Venkataramanan, S., et al., DDX3X and DDX3Y are redundant in protein synthesis. RNA, 2021. 27(12): p. 1577-1588.
- Nicola, P., et al., De novo DDX3X missense variants in males appear viable and contribute to syndromic intellectual disability. Am J Med Genet A, 2019. 179(4): p. 570-578.
- Rastegar, M., Editorial (Thematic Issue: NeuroEpigenetics and Neurodevelopmental Disorders: From Molecular Mechanisms to Cell Fate Commitments of the Brain Cells and Human Disease). Curr Top Med Chem, 2017. 17(7): p. 769-770.
- Mossink, B., et al., The emerging role of chromatin remodelers in neurodevelopmental disorders: a developmental perspective. Cell Mol Life Sci, 2021. 78(6): p. 2517-2563.
- Vuu, Y.M., C.T. Roberts, and M. Rastegar, MeCP2 Is an Epigenetic Factor That Links DNA Methylation with Brain Metabolism. Int J Mol Sci, 2023. 24(4).
- Pejhan, S. and M. Rastegar, Role of DNA Methyl-CpG-Binding Protein MeCP2 in Rett Syndrome Pathobiology and Mechanism of Disease. Biomolecules, 2021. 11(1).
- Kusch, T., Histone H3 lysine 4 methylation revisited. Transcription, 2012. 3(6): p. 310-4.
- Wang, S., et al., SETD1A Mediated H3K4 Methylation and Its Role in Neurodevelopmental and Neuropsychiatric Disorders. Front Mol Neurosci, 2021. 14: p. 772000.
- Yu, X., et al., De Novo and Inherited SETD1A Variants in Early-onset Epilepsy. Neurosci Bull, 2019. 35(6): p. 1045-1057.
- RK, C.Y., et al., Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci, 2017. 20(4): p. 602-611.
- Malumbres, M., et al., Cyclin-dependent kinases: a family portrait. Nat Cell Biol, 2009. 11(11): p. 1275-6.
- Chen, H.R., et al., Cdk12 and Cdk13 regulate axonal elongation through a common signaling pathway that modulates Cdk5 expression. Exp Neurol, 2014. 261: p. 10-21.
- Sifrim, A., et al., Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet, 2016. 48(9): p. 1060-5.
- Bostwick, B.L., et al., Phenotypic and molecular characterisation of CDK13-related congenital heart defects, dysmorphic facial features and intellectual developmental disorders. Genome Med, 2017. 9(1): p. 73.
- Garcia, G., 3rd, D.R. Raleigh, and J.F. Reiter, How the Ciliary Membrane Is Organized Inside-Out to Communicate Outside-In. Curr Biol, 2018. 28(8): p. R421-R434.
- Reiter, J.F. and M.R. Leroux, Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol, 2017. 18(9): p. 533-547.
- Rosengren, T., et al., TSC1 and TSC2 regulate cilia length and canonical Hedgehog signaling via different mechanisms. Cell Mol Life Sci, 2018. 75(14): p. 2663-2680.
- Shamseldin, H.E., et al., The morbid genome of ciliopathies: an update. Genet Med, 2020. 22(6): p. 1051-1060.
- Chang, C.H., et al., CEP120-mediated KIAA0753 recruitment onto centrioles is required for timely neuronal differentiation and germinal zone exit in the developing cerebellum. Genes Dev, 2021. 35(21-22): p. 1445-1460.
- Dong, Y., et al., Clinical and genetic characteristics of 36 children with Joubert syndrome. Front Pediatr, 2023. 11: p. 1102639.
- Romani, M., A. Micalizzi, and E.M. Valente, Joubert syndrome: congenital cerebellar ataxia with the molar tooth. Lancet Neurol, 2013. 12(9): p. 894-905.
- Karam, A., et al., WGS Revealed Novel BBS5 Pathogenic Variants, Missed by WES, Causing Ciliary Structure and Function Defects. Int J Mol Sci, 2023. 24(10).
- Wingfield, J.L., K.F. Lechtreck, and E. Lorentzen, Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem, 2018. 62(6): p. 753-763.
- Forsythe, E. and P.L. Beales, Bardet-Biedl syndrome. Eur J Hum Genet, 2013. 21(1): p. 8-13.
- Shamseldin, H.E., et al., The morbid genome of ciliopathies: an update. Genet Med, 2022. 24(4): p. 966.
- Bennett, V. and D.N. Lorenzo, An Adaptable Spectrin/Ankyrin-Based Mechanism for Long-Range Organization of Plasma Membranes in Vertebrate Tissues. Curr Top Membr, 2016. 77: p. 143-84.
- Machnicka, B., et al., Spectrins: a structural platform for stabilization and activation of membrane channels, receptors and transporters. Biochim Biophys Acta, 2014. 1838(2): p. 620-34.
- Lorenzo, D.N., Cargo hold and delivery: Ankyrins, spectrins, and their functional patterning of neurons. Cytoskeleton (Hoboken), 2020. 77(3-4): p. 129-148.
- Syrbe, S., et al., Delineating SPTAN1 associated phenotypes: from isolated epilepsy to encephalopathy with progressive brain atrophy. Brain, 2017. 140(9): p. 2322-2336.
- Tohyama, J., et al., SPTAN1 encephalopathy: distinct phenotypes and genotypes. J Hum Genet, 2015. 60(4): p. 167-73.
- Van de Vondel, L., et al., De Novo and Dominantly Inherited SPTAN1 Mutations Cause Spastic Paraplegia and Cerebellar Ataxia. Mov Disord, 2022. 37(6): p. 1175-1186.
- Shanmughapriya, S., D. Langford, and K. Natarajaseenivasan, Inter and Intracellular mitochondrial trafficking in health and disease. Ageing Res Rev, 2020. 62: p. 101128.
- van Spronsen, M., et al., TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron, 2013. 77(3): p. 485-502.
- Barel, O., et al., Deleterious variants in TRAK1 disrupt mitochondrial movement and cause fatal encephalopathy. Brain, 2017. 140(3): p. 568-581.
- Iossifov, I., et al., The contribution of de novo coding mutations to autism spectrum disorder. Nature, 2014. 515(7526): p. 216-21.
- Bacchelli, E., et al., An integrated analysis of rare CNV and exome variation in Autism Spectrum Disorder using the Infinium PsychArray. Sci Rep, 2020. 10(1): p. 3198.
- Xu, B., et al., Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet, 2011. 43(9): p. 864-8.
- Turner, T.N., et al., Sex-Based Analysis of De Novo Variants in Neurodevelopmental Disorders. Am J Hum Genet, 2019. 105(6): p. 1274-1285.
- Seifert, W., et al., Cohen syndrome-associated protein, COH1, is a novel, giant Golgi matrix protein required for Golgi integrity. J Biol Chem, 2011. 286(43): p. 37665-75.
- Seifert, W., et al., Cohen syndrome-associated protein COH1 physically and functionally interacts with the small GTPase RAB6 at the Golgi complex and directs neurite outgrowth. J Biol Chem, 2015. 290(6): p. 3349-58.
- Seifert, W., et al., Expanded mutational spectrum in Cohen syndrome, tissue expression, and transcript variants of COH1. Hum Mutat, 2009. 30(2): p. E404-20.
- Germanaud, D., et al., The Renpenning syndrome spectrum: new clinical insights supported by 13 new PQBP1-mutated males. Clin Genet, 2011. 79(3): p. 225-35.
- Tanaka, H. and H. Okazawa, PQBP1: The Key to Intellectual Disability, Neurodegenerative Diseases, and Innate Immunity. Int J Mol Sci, 2022. 23(11).
- Okazawa, H., PQBP1, an intrinsically disordered/denatured protein at the crossroad of intellectual disability and neurodegenerative diseases. Neurochem Int, 2018. 119: p. 17-25.
- Cong, Y., et al., WDR45, one gene associated with multiple neurodevelopmental disorders. Autophagy, 2021. 17(12): p. 3908-3923.
- Wan, H., et al., WDR45 contributes to neurodegeneration through regulation of ER homeostasis and neuronal death. Autophagy, 2020. 16(3): p. 531-547.
- Page, B.D., et al., EEL-1, a Hect E3 ubiquitin ligase, controls asymmetry and persistence of the SKN-1 transcription factor in the early C. elegans embryo. Development, 2007. 134(12): p. 2303-14.
- Zhao, X., et al., The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell, 2009. 17(2): p. 210-21.
- Forget, A., et al., Shh signaling protects Atoh1 from degradation mediated by the E3 ubiquitin ligase Huwe1 in neural precursors. Dev Cell, 2014. 29(6): p. 649-61.
- Zhang, Z.Y., et al., Ubiquitination and inhibition of glycine receptor by HUWE1 in spinal cord dorsal horn. Neuropharmacology, 2019. 148: p. 358-365.
- Giles, A.C. and B. Grill, Roles of the HUWE1 ubiquitin ligase in nervous system development, function and disease. Neural Dev, 2020. 15(1): p. 6.
- Cummings, B.B., et al., Transcript expression-aware annotation improves rare variant interpretation. Nature, 2020. 581(7809): p. 452-458.
- Christensen, D.L., et al., Prevalence and Characteristics of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2012. MMWR Surveill Summ, 2018. 65(13): p. 1-23.
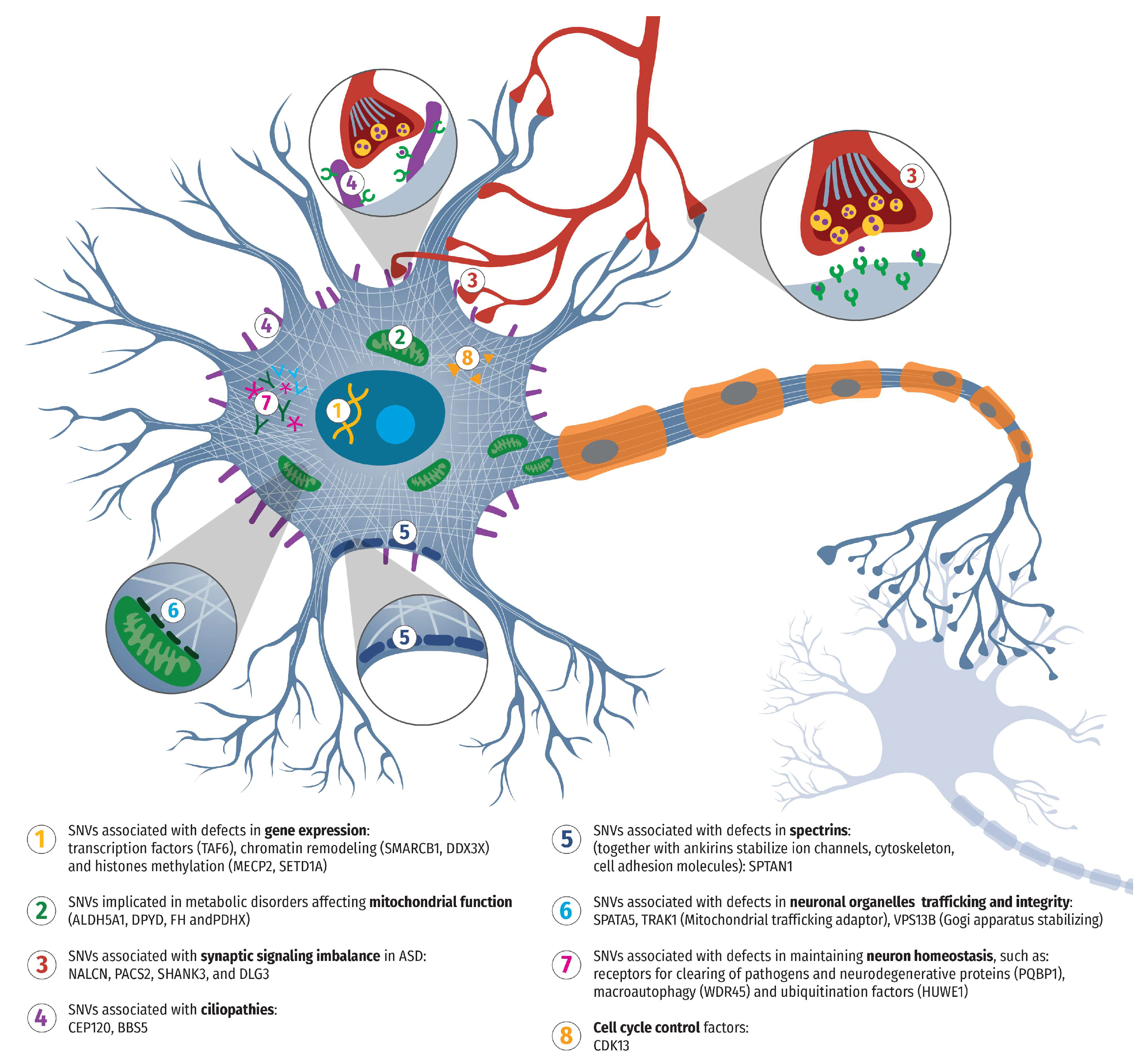
| patient | gene | chromosome | alteration at the transcript level | Alteration at the amino acid level | Variant | Zygosity | Pathogenicity |
| 1 | MECP2 | chr X | NM_004992.3: c.1208dup | p.(Glu404Ter) | chrX:g.153296071dup | Hemizygous (maternal origin) | likely pathogenic |
| 2 | TAF6 | chr 7 | NM_001190415.1: c.323T>C | p.(Ile108Thr) | chr7: g.99711522A>G | Homozygous | likely pathogenic |
| 3 | SMARCB1 | chr 22 | NM_003073.3: c.568C>T | p.(Arg190Trp) | chr22: g.24145549C>T | Heterozygous (de novo) |
likely pathogenic |
| 4 | PACS2 | chr 14 | NM_001100913.3: c.625G>A | p.(Glu209Lys) | chr14: g.105834449G>A | Heterozygous (de novo) |
pathogenic |
| 5 | WDR45 | chr X | NM_007075.3: c.601_602del | p.(Leu201fs) | chrX: g.48933330del | Heterozygous (de novo) |
likely pathogenic |
| 6 | PQBP1 | chr X | NM_001032381.1: c.586C>T | p.(Arg196Ter) | chrX: g.48760017C>T | Hemizygous (maternal origin) | pathogenic |
| 7 | SPATA5 | chr 4 | NM_145207.2: c. 554G>A; NM_145207.2: c.1831C>T |
p.(Gly185Glu) p. (Pro611Ser) |
chr4: g.123855300G>A chr4: g.123900503C>T | Heterozygous (maternal origin) Heterozygous (paternal origin) |
VUS |
| 8 | NALCN | chr 13 | NM_052867.2: c.965T>C | p.(Ile322Thr) | chr13: g.101944423A>G | Heterozygous (de novo) |
likely pathogenic |
| 9 | FH | chr 1 | NM_000143.4: c.1048C>T | p.(Arg350Trp) | chr1: g.241667402G>A | Homozygous | likely pathogenic |
| 10 | CEP120 | chr 5 | NM_153223.3: c.23T>G NM_153223.3: c.2548C>G |
p.(Leu8Trp) p. (Arg850Gly) |
chr5: g.122758670A>C chr5: g.122700222G>C | Heterozygous (paternal origin) Heterozygous (maternal origin) |
VUS |
| 11 | BBS5 | chr 2 | NM_152384.3: c.167G>A LP NM_152384.3: c.619-1G>C P |
p.(Arg56Lys) / | chr2:g.170343603G>A chr2:g.170354136G>C | Heterozygous (maternal origin) Heterozygous (paternal origin) |
LP/P |
| 12 | SPTAN1 | chr 9 | NM_001130438.3: c.6922C>T | p.(Arg2308Cys) | chr9: g.131394565C>T | Heterozygous (de novo) |
likely pathogenic |
| 13 | VPS13B | chr 8 | NM_152564.5: c.9574_9583delGTACCCCTCGinsAC NM_152564.5: c.6914C>T |
p. (Val3192ThrfsTer33) p.(Thr2305Ile) |
chr8: g.100844840_100844849delGTACCCCTCGinsAC chr8: g.100733139C>T |
Heterozygous (paternal origin) Heterozygous (maternal origin) |
LP/VUS |
| 14 | SHANK3 DLG3 |
chr 22 chr X | NM_001372044.2: c.2490+1G>A NM_021120.4: c. 1721G>A |
/ p.(Arg574Gln) | chr22: g.51153476G>A chrX: g.69712394G>A | Heterozygous (de novo) Hemizygous (maternal origin) |
pathogenic/VUS |
| 15 | CDK13 | chr 7 | NM_003718.5: c.2525A>T | p.(Asn842Ile) | chr7: g.40085606A>T | Heterozygous (de novo) |
pathogenic |
| 16 | PDHX | chr 11 | NM_003477.3: c.1336C>T | p.(Arg446Ter) | chr11: g.35016549C>T | Homozygous | pathogenic |
| 17 | SETD1A | chr 16 | NM_014712.3: c.4879del | p. (Val1627TrpfsTer41) |
chr16: g.30995020delG | Heterozygous (de novo) |
pathogenic |
| 18 | TRAK1 | chr 3 | NM_001042646.3: c.1187T>A | p.(Ile396Asn) | chr3: g.42240742T>A | Heterozygous (de novo) |
VUS |
| 19 | ALDH5A1 | chr 6 | NM_170740: c.804dup NM_170740: c.1265G>A |
p. (Val269CysfsTer19) p.(Gly422Asp) |
chr6:g.24515433dup chr6:g.24528277G>A | Heterozygous (paternal origin) Heterozygous (maternal origin) |
P/LP |
| 20 | DPYD | chr 1 | NM_000110.3: c.1905+1G>A | / | chr1:g.97915614C>T | Homozygous | LP |
| 21 | DDX3X | chr X | NM_001356.3:c.857C>A | p.(Ala286Asp) | chrX:g.41203374C>A | Heterozygous (de novo) |
VUS |
| 22 | HUWE1 | chr X | NM_031707: c.9209G>A | p.(Arg3070His) | chrX:g.53578038C>T | Hemizygous (de novo) |
pathogenic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




