Submitted:
16 February 2025
Posted:
17 February 2025
You are already at the latest version
Abstract
Keywords:
Introduction
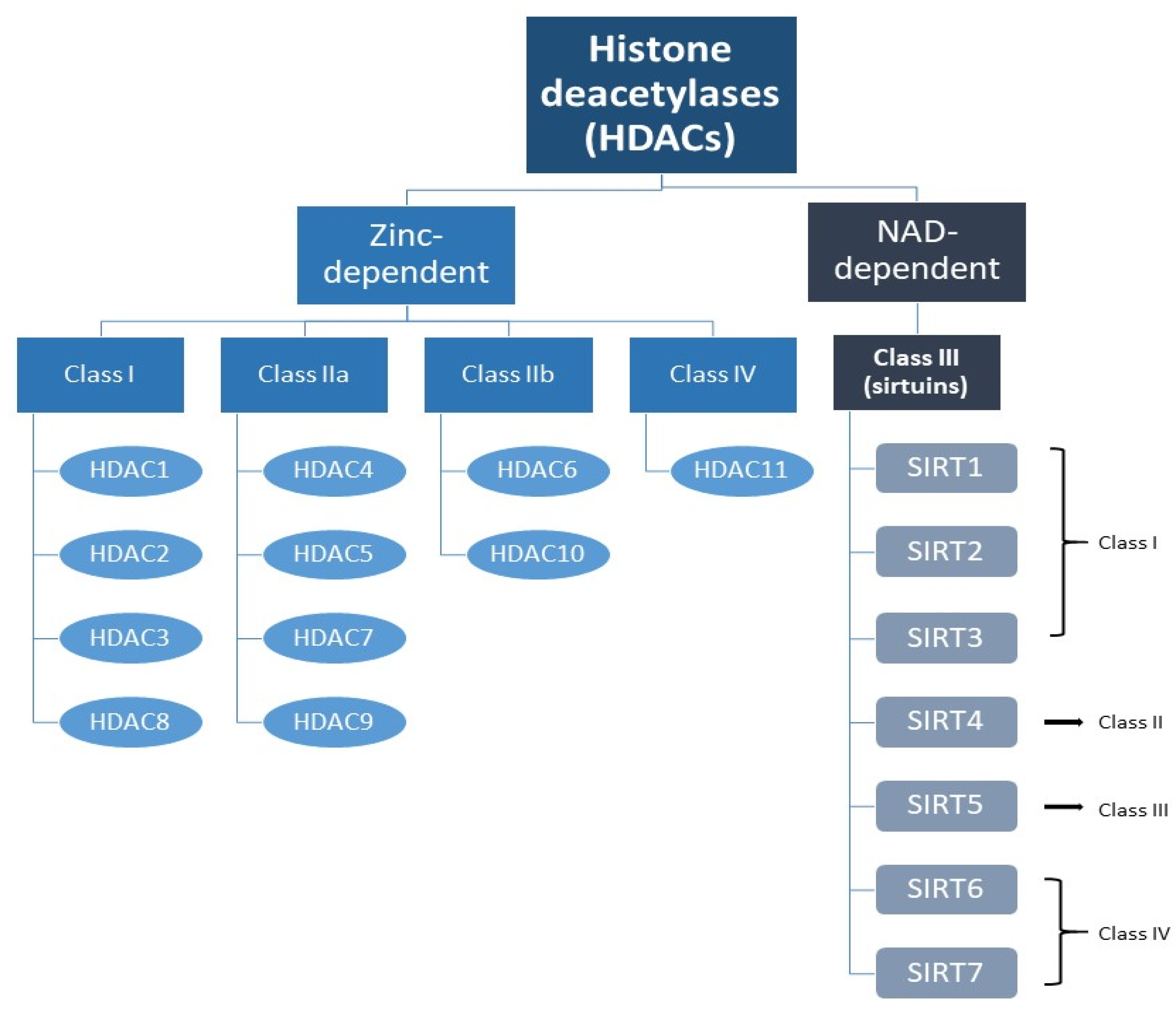
The Sirtuin Family
Classification of Sirtuins
Overview of the Role of the Sirtuin Family in AML
Sirtuin 1
Sirtuin 2
Sirtuin 3
Sirtuin 4
Sirtuin 5
Sirtuin 6
Sirtuin 7
Therapeutic Applications/Implications
Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Velten, L.; Story, B.A.; Hernández-Malmierca, P.; Raffel, S.; Leonce, D.R.; Milbank, J.; Paulsen, M.; Demir, A.; Szu-Tu, C.; Frömel, R.; Lutz, C.; Nowak, D.; Jann, J.C.; Pabst, C.; Boch, T.; Hofmann, W.K.; Müller-Tidow, C.; Trumpp, A.; Haas, S.; Steinmetz, L.M. Identification of leukemic and pre-leukemic stem cells by clonal tracking from single-cell transcriptomics. Nat Commun. 2021, 12, 1366. [Google Scholar] [CrossRef]
- Stelmach, P.; Trumpp, A. Leukemic stem cells and therapy resistance in acute myeloid leukemia. Haematologica. 2023, 108, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.T.; Sun, J.; Zhang, L.; He, X.; Zhu, Y.H.; Dong, H.J.; Wang, H.Y.; Zhu, L.; Zou, J.Y.; Huang, J.W.; Li, L. Role of SIRT1 in hematologic malignancies. J Zhejiang Univ Sci B. 2019, 20, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Batsivari, A.; Grey, W.; Bonnet, D. Understanding of the crosstalk between normal residual hematopoietic stem cells and the leukemic niche in acute myeloid leukemia. Exp Hematol. 2021, 95, 23–30. [Google Scholar] [CrossRef]
- Chen, Y.; Li, J.; Xu, L.; Găman, M.A.; Zou, Z. The genesis and evolution of acute myeloid leukemia stem cells in the microenvironment: From biology to therapeutic targeting. Cell Death Discov. 2022, 8, 397. [Google Scholar] [CrossRef]
- Shlush, L.I.; Mitchell, A.; Heisler, L.; Abelson, S.; Ng, S.W.K.; Trotman-Grant, A.; Medeiros, J.J.F.; Rao-Bhatia, A.; Jaciw-Zurakowsky, I.; Marke, R.; McLeod, J.L.; Doedens, M.; Bader, G.; Voisin, V.; Xu, C.; McPherson, J.D.; Hudson, T.J.; Wang, J.C.Y.; Minden, M.D.; Dick, J.E. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 2017, 547, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Khaldoyanidi, S.K.; Hindoyan, A.; Stein, A.; Subklewe, M. Leukemic stem cells as a target for eliminating acute myeloid leukemia: Gaps in translational research. Crit Rev Oncol Hematol. 2022, 175, 103710. [Google Scholar] [CrossRef]
- Kumar, B.; Garcia, M.; Weng, L.; Jung, X.; Murakami, J.L.; Hu, X.; McDonald, T.; Lin, A.; Kumar, A.R.; DiGiusto, D.L.; Stein, A.S.; Pullarkat, V.A.; Hui, S.K.; Carlesso, N.; Kuo, Y.H.; Bhatia, R.; Marcucci, G.; Chen, C.C. Acute myeloid leukemia transforms the bone marrow niche into a leukemia-permissive microenvironment through exosome secretion. Leukemia. 2018, 32, 575–587. [Google Scholar] [CrossRef]
- Lagadinou, E.D.; Sach, A.; Callahan, K.; Rossi, R.M.; Neering, S.J.; Minhajuddin, M.; Ashton, J.M.; Pei, S.; Grose, V.; O'Dwyer, K.M.; Liesveld, J.L.; Brookes, P.S.; Becker, M.W.; Jordan, C.T. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013, 12, 329–41. [Google Scholar] [CrossRef]
- Carafa, V.; Altucci, L.; Nebbioso, A. Dual Tumor Suppressor and Tumor Promoter Action of Sirtuins in Determining Malignant Phenotype. Front Pharmacol. 2019, 10, 38. [Google Scholar] [CrossRef]
- Lin, Z.; Fang, D. The Roles of SIRT1 in Cancer. Genes Cancer. 2013, 4, 97–104. [Google Scholar] [CrossRef]
- Chalkiadaki, A.; Guarente, L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer. 2015, 15, 608–24. [Google Scholar] [CrossRef] [PubMed]
- Mei, Z.; Zhang, X.; Yi, J.; Huang, J.; He, J.; Tao, Y. Sirtuins in metabolism; DNA repair and cancer. J Exp Clin Cancer Res. 2016, 35, 182. [Google Scholar] [CrossRef]
- Dong, Z.; Cui, H. Function of Sirtuins in Cancer Stem Cells. Int J Stem Cell Res Ther. 2016, 3, 024. [Google Scholar] [CrossRef]
- Wu, Q.J.; Zhang, T.N.; Chen, H.H.; Yu, X.F.; Lv, J.L.; Liu, Y.Y.; Liu, Y.S.; Zheng, G.; Zhao, J.Q.; Wei, Y.F.; Guo, J.Y.; Liu, F.H.; Chang, Q.; Zhang, Y.X.; Liu, C.G.; Zhao, Y.H. The sirtuin family in health and disease. Signal Transduct Target Ther. 2022, 7, 402. [Google Scholar] [CrossRef] [PubMed]
- Haq, M.F.U.; Hussain, M.Z.; Mahjabeen, I.; Akram, Z.; Saeed, N.; Shafique, R.; Abbasi, S.F.; Kayani, M.A. Oncometabolic role of mitochondrial sirtuins in glioma patients. PLoS One. 2023, 18, e0281840. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhan, H.; Ren, Y.; Feng, M.; Wang, Q.; Jiao, Q.; Wang, Y.; Liu, X.; Zhang, S.; Du, L.; Wang, Y.; Wang, C. Sirtuin 4 activates autophagy and inhibits tumorigenesis by upregulating the p53 signaling pathway. Cell Death Differ. 2023, 30, 313–326. [Google Scholar] [CrossRef]
- Shi, Q.; Liu, T.; Zhang, X.; Geng, J.; He, X.; Nu, M.; Pang, D. Decreased sirtuin 4 expression is associated with poor prognosis in patients with invasive breast cancer. Oncol Lett. 2016, 12, 2606–2612. [Google Scholar] [CrossRef]
- Tang, X.; Li, Y.; Liu, L.; Guo, R.; Zhang, P.; Zhang, Y.; Zhang, Y.; Zhao, J.; Su, J.; Sun, L.; Liu, Y. Sirtuin 3 induces apoptosis and necroptosis by regulating mutant p53 expression in small-cell lung cancer. Oncol Rep. 2020, 43, 591–600. [Google Scholar] [CrossRef]
- Wu, X.; Wang, S.; Zhao, X.; Lai, S.; Yuan, Z.; Zhan, Y.; Ni, K.; Liu, Z.; Liu, L.; Xin, R.; Zhou, X.; Yin, X.; Liu, X.; Zhang, X.; Cui, W.; Zhang, C. Clinicopathological and prognostic value of SIRT6 in patients with solid tumors: a meta-analysis and TCGA data review. Cancer Cell Int. 2022, 22, 84. [Google Scholar] [CrossRef]
- Goes, J.V.C.; Carvalho, L.G.; de Oliveira, R.T.G.; Melo, M.M.L.; Novaes, L.A.C.; Moreno, D.A.; Gonçalves, P.G.; Montefusco-Pereira, C.V.; Pinheiro, R.F.; Ribeiro Junior, H.L. Role of Sirtuins in the Pathobiology of Onco-Hematological Diseases: A PROSPERO-Registered Study and In Silico Analysis. Cancers. 2022, 14, 4611. [Google Scholar] [CrossRef] [PubMed]
- Costa-Machado, L.F.; Fernandez-Marcos, P.J. The sirtuin family in cancer. Cell Cycle. 2019, 18, 2164–2196. [Google Scholar] [CrossRef]
- Sauve, A.A. Sirtuin chemical mechanisms. Biochim Biophys Acta. 2010, 1804, 1591–603. [Google Scholar] [CrossRef]
- Teixeira, C.S.S.; Cerqueira, N.M.F.S.A.; Gomes, P.; Sousa, S.F. A Molecular Perspective on Sirtuin Activity. Int J Mol Sci. 2020, 21, 8609. [Google Scholar] [CrossRef]
- Towler, D.A.; Gordon, J.I.; Adams, S.P.; Glaser, L. The biology and enzymology of eukaryotic protein acylation. Annu Rev Biochem. 1988, 57, 69–99. [Google Scholar] [CrossRef] [PubMed]
- Mai, A.; Massa, S.; Rotili, D.; Cerbara, I.; Valente, S.; Pezzi, R.; Simeoni, S.; Ragno, R. Histone deacetylation in epigenetics: an attractive target for anticancer therapy. Med Res Rev. 2005, 25, 261–309. [Google Scholar] [CrossRef]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964, 51, 786–94. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Sprung, R.; Chen, Y.; Xu, Y.; Ball, H.; Pei, J.; Cheng, T.; Kho, Y.; Xiao, H.; Xiao, L.; Grishin, N.V.; White, M.; Yang, X.J.; Zhao, Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006, 23, 607–18. [Google Scholar] [CrossRef]
- Brachmann, C.B.; Sherman, J.M.; Devine, S.E.; Cameron, E.E.; Pillus, L.; Boeke, J.D. The SIR2 gene family; conserved from bacteria to humans; functions in silencing; cell cycle progression; and chromosome stability. Genes Dev. 1995, 9, 2888–902. [Google Scholar] [CrossRef]
- Barneda-Zahonero, B.; Parra, M. Histone deacetylases and cancer. Mol Oncol. 2012, 6, 579–89. [Google Scholar] [CrossRef]
- Rosato, R.R.; Grant, S. Histone deacetylase inhibitors: insights into mechanisms of lethality. Expert Opin Ther Targets. 2005, 9, 809–24. [Google Scholar] [CrossRef] [PubMed]
- Sawas, A.; Radeski, D.; O'Connor, O.A. Belinostat in patients with refractory or relapsed peripheral T-cell lymphoma: a perspective review. Ther Adv Hematol. 2015, 6, 202–8. [Google Scholar] [CrossRef] [PubMed]
- Bose, P.; Dai, Y.; Grant, S. Histone deacetylase inhibitor (HDACI) mechanisms of action: emerging insights. Pharmacol Ther. 2014, 143, 323–36. [Google Scholar] [CrossRef]
- Zhao, E.; Hou, J.; Ke, X.; Abbas, M.N.; Kausar, S.; Zhang, L.; Cui, H. The Roles of Sirtuin Family Proteins in Cancer Progression. Cancers. 2019, 11, 1949. [Google Scholar] [CrossRef]
- Carafa, V.; Altucci, L.; Nebbioso, A. Dual Tumor Suppressor and Tumor Promoter Action of Sirtuins in Determining Malignant Phenotype. Front Pharmacol. 2019, 10, 38. [Google Scholar] [CrossRef]
- Giblin, W.; Lombard, D.B. Sirtuins; Healthspan; and Longevity in Mammals. Handbook of the Biology of Aging; 8th ed.; Kaeberlein, M.R., Martin, M.G.; Academic Press; 2016; Chapter 3; pp. 83-132. [CrossRef]
- Kratz, E.M.; Sołkiewicz, K.; Kubis-Kubiak, A.; Piwowar, A. Sirtuins as Important Factors in Pathological States and the Role of Their Molecular Activity Modulators. Int J Mol Sci. 2021, 22, 630. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, H.; Dessain, S.K.; Ng, E.E.; Imai, S.I.; Frye, R.A.; Pandita, T.K.; Guarente, L.; Weinberg, R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001, 107, 149–59. [Google Scholar] [CrossRef]
- Luo, J.; Nikolaev, A.Y.; Imai, S.; Chen, D.; Su, F.; Shiloh, A.; Guarente, L.; Gu, W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001, 107, 137–48. [Google Scholar] [CrossRef]
- Deng, CX. SIRT1; is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009, 5, 147–52. [Google Scholar] [CrossRef]
- Ng, S.W.; Mitchell, A.; Kennedy, J.A.; Chen, W.C.; McLeod, J.; Ibrahimova, N.; Arruda, A.; Popescu, A.; Gupta, V.; Schimmer, A.D.; Schuh, A.C.; Yee, K.W.; Bullinger, L.; Herold, T.; Görlich, D.; Büchner, T.; Hiddemann, W.; Berdel, W.E.; Wörmann, B.; Cheok, M.; Preudhomme, C.; Dombret, H.; Metzeler, K.; Buske, C.; Löwenberg, B.; Valk, P.J.; Zandstra, P.W.; Minden, M.D.; Dick, J.E.; Wang, J.C. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016, 540, 433–437. [Google Scholar] [CrossRef]
- Li, L.; Osdal, T.; Ho, Y.; Chun, S.; McDonald, T.; Agarwal, P.; Lin, A.; Chu, S.; Qi, J.; Li, L.; Hsieh, Y.T.; Dos Santos, C.; Yuan, H.; Ha, T.Q.; Popa, M.; Hovland, R.; Bruserud, Ø.; Gjertsen, B.T.; Kuo, Y.H.; Chen, W.; Lain, S.; McCormack, E.; Bhatia, R. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell. 2014, 15, 431–446. [Google Scholar] [CrossRef] [PubMed]
- O'Donnell, M.R.; Abboud, C.N.; Altman, J.; Appelbaum, F.R.; Arber, D.A.; Attar, E.; Borate, U.; Coutre, S.E.; Damon, L.E.; Goorha, S.; Lancet, J.; Maness, L.J.; Marcucci, G.; Millenson, M.M.; Moore, J.O.; Ravandi, F.; Shami, P.J.; Smith, B.D.; Stone, R.M.; Strickland, S.A.; Tallman, M.S.; Wang, E.S.; Naganuma, M.; Gregory, K.M. NCCN Clinical Practice Guidelines Acute myeloid leukemia. J Natl Compr Canc Netw. 2012, 10, 984–1021. [Google Scholar] [CrossRef] [PubMed]
- Sasca, D.; Hähnel, P.S.; Szybinski, J.; Khawaja, K.; Kriege, O.; Pante, S.V.; Bullinger, L.; Strand, S.; Strand, D.; Theobald, M.; Kindler, T. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood. 2014, 124, 121–33. [Google Scholar] [CrossRef]
- Daver, N.; Schlenk, R.F.; Russell, N.H.; Levis, M.J. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019, 33, 299–312. [Google Scholar] [CrossRef]
- Patel, J.P.; Gönen, M.; Figueroa, M.E.; Fernandez, H.; Sun, Z.; Racevskis, J.; Van Vlierberghe, P.; Dolgalev, I.; Thomas, S.; Aminova, O.; Huberman, K.; Cheng, J.; Viale, A.; Socci, N.D.; Heguy, A.; Cherry, A.; Vance, G.; Higgins, R.R.; Ketterling, R.P.; Gallagher, R.E.; Litzow, M.; van den Brink, M.R.; Lazarus, H.M.; Rowe, J.M.; Luger, S.; Ferrando, A.; Paietta, E.; Tallman, M.S.; Melnick, A.; Abdel-Wahab, O.; Levine, R.L. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012, 366, 1079–89. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, C.A.; Khanim, F.L.; Hayden, R.; Bunce, C.M.; White, D.A.; Drayson, M.T.; Craddock, C.; Turner, B.M. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005, 19, 1751–9. [Google Scholar] [CrossRef]
- Li, L.C.; Wang, J.D.; Yang, S.S.; Zhou, Z.; Zeng, Q.F.; Zheng, F. [Establishment of Drug-Resistant Cell Lines of Acute Myeloid Leukemia and Correlation of Sirt1 and PGC-1α Expression Levels with Drug Resistance]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2022, 30, 704–710. [Google Scholar] [CrossRef]
- Tian, W.L.; Guo, R.; Wang, F.; Jiang, Z.X.; Tang, P.; Huang, Y.M.; Sun, L. The IRF9-SIRT1-P53 axis is involved in the growth of human acute myeloid leukemia. Exp Cell Res. 2018, 365, 185–193. [Google Scholar] [CrossRef]
- Chen, C.W.; Koche, R.P.; Sinha, A.U.; Deshpande, A.J.; Zhu, N.; Eng, R.; Doench, J.G.; Xu, H.; Chu, S.H.; Qi, J.; Wang, X.; Delaney, C.; Bernt, K.M.; Root, D.E.; Hahn, W.C.; Bradner, J.E.; Armstrong, S.A. DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat Med. 2015, 21, 335–43. [Google Scholar] [CrossRef]
- Levavasseur, F.; Oussous, S.; Zubaidan, T.; Kosmider, O.; Pendino, F.; Rombaut, D.; Bouscary, D.; Fontenay, M.; Lauret, E.; Dusanter-Fourt, I. FOXP1 regulates oxidative stress; SIRT1 expression; and resistance to chemotherapies in acute myeloid leukemia cells. Blood Adv. 2023, 7, 3265–3275. [Google Scholar] [CrossRef]
- Yuan, H.; Wang, Z.; Li, L.; Zhang, H.; Modi, H.; Horne, D.; Bhatia, R.; Chen, W. Activation of stress response gene SIRT1 by BCR-ABL promotes leukemogenesis. Blood. 2012, 119, 1904–14. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, R.A. Aberrant cytokine signaling in leukemia. Oncogene. 2007, 15, 6738–49. [Google Scholar] [CrossRef]
- Bartel, D.P.; Chen, C.Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef]
- Zhou, L.; Fu, L.; Lv, N.; Chen, X.S.; Liu, J.; Li, Y.; Xu, Q.Y.; Huang, S.; Zhang, X.D.; Dou, L.P.; Wang, L.L.; Li, Y.H.; Yu, L. A minicircuitry comprised of microRNA-9 and SIRT1 contributes to leukemogenesis in t(8;21) acute myeloid leukemia. Eur Rev Med Pharmacol Sci. 2017, 21, 786–794. [Google Scholar] [PubMed]
- Vaquero, A.; Scher, M.B.; Lee, D.H.; Sutton, A.; Cheng, H.L.; Alt, F.W.; Serrano, L.; Sternglanz, R.; Reinberg, D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006, 20, 1256–61. [Google Scholar] [CrossRef]
- Harting, K.; Knöll, B. SIRT2-mediated protein deacetylation: An emerging key regulator in brain physiology and pathology. Eur J Cell Biol. 2010, 89, 262–9. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.A.; O'Callaghan, C.; Chang, E.D.; Jiang, H.; Vassilopoulos, A. Context-Dependent Roles for SIRT2 and SIRT3 in Tumor Development Upon Calorie Restriction or High Fat Diet. Front Oncol. 2020, 9, 1462. [Google Scholar] [CrossRef]
- Chen, G.; Huang, P.; Hu, C. The role of SIRT2 in cancer: A novel therapeutic target. Int J Cancer. 2020, 147, 3297–3304. [Google Scholar] [CrossRef]
- Zhang, K.; Sowers, M.L.; Cherryhomes, E.I.; Singh, V.K.; Mishra, A.; Restrepo, B.I.; Khan, A.; Jagannath, C. Sirtuin-dependent metabolic and epigenetic regulation of macrophages during tuberculosis. Front Immunol. 2023, 14, 1121495. [Google Scholar] [CrossRef]
- Zhao, D.; Zou, S.W.; Liu, Y.; Zhou, X.; Mo, Y.; Wang, P.; Xu, Y.H.; Dong, B.; Xiong, Y.; Lei, Q.Y.; Guan, K.L. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell. 2013, 23, 464–76. [Google Scholar] [CrossRef]
- Kozako, T.; Mellini, P.; Ohsugi, T.; Aikawa, A.; Uchida, Y.I.; Honda, S.I.; Suzuki, T. Novel small molecule SIRT2 inhibitors induce cell death in leukemic cell lines. BMC Cancer. 2018, 18, 791. [Google Scholar] [CrossRef] [PubMed]
- Dan, L.; Klimenkova, O.; Klimiankou, M.; Klusman, J.H.; van den Heuvel-Eibrink, M.M.; Reinhardt, D.; Welte, K.; Skokowa, J. The role of sirtuin 2 activation by nicotinamide phosphoribosyltransferase in the aberrant proliferation and survival of myeloid leukemia cells. Haematologica. 2012, 97, 551–9. [Google Scholar] [CrossRef] [PubMed]
- Deng, A.; Ning, Q.; Zhou, L.; Liang, Y. SIRT2 is an unfavorable prognostic biomarker in patients with acute myeloid leukemia. Sci Rep. 2016, 6, 27694. [Google Scholar] [CrossRef] [PubMed]
- McMahon, K.A.; Hiew, S.Y.; Hadjur, S.; Veiga-Fernandes, H.; Menzel, U.; Price, A.J.; Kioussis, D.; Williams, O.; Brady, H.J. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007, 1, 338–45. [Google Scholar] [CrossRef] [PubMed]
- Jude, C.D.; Climer, L.; Xu, D.; Artinger, E.; Fisher, J.K.; Ernst, P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007, 1, 324–37. [Google Scholar] [CrossRef]
- Hao, C.; Shao, X.; Song, J.; Peng, M.; Lao, Y.; Mack, R.; Zhang, L.; Wei, W.; Liu, N.; Wang, T.; Wu, Y.; Feng, L.; Yin, L.; Wang, S.; Sun, X.; Chen, S.; Zhang, J.; Li, B. SIRT2 regulates proliferation and chemotherapy response of MLL-ENL-driven acute myeloid leukemia. Biochem Biophys Res Commun. 2022, 596, 36–42. [Google Scholar] [CrossRef]
- Xu, H.; Li, Y.; Chen, L.; Wang, C.; Wang, Q.; Zhang, H.; Lin, Y.; Li, Q.; Pang, T. SIRT2 mediates multidrug resistance in acute myelogenous leukemia cells via ERK1/2 signaling pathway. Int J Oncol. 2016, 48, 613–23. [Google Scholar] [CrossRef]
- Yip, P.Y. Phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin (PI3K-Akt-mTOR) signaling pathway in non-small cell lung cancer. Transl Lung Cancer Res. 2015, 4, 165–76. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhao, H.; Zhu, J.; Zhang, L.; Zha, J.; Zhang, L.; Ding, Y.; Jian, X.; Xia, J.; Xu, B.; Qi, Z. SIRT2 inhibitor SirReal2 enhances anti-tumor effects of PI3K/mTOR inhibitor VS-5584 on acute myeloid leukemia cells. Cancer Med. 2023, 12, 18901–18917. [Google Scholar] [CrossRef]
- Dan, L.; Gigina, A.; Welte, K.; Skokowa, J. Nampt/SIRT2 Hyperactivation Lead to Activation of ß-Catenin: a Possible Mechanism of Leukomogenic Transformation In CN Patients. Blood. 2010, 116, 1486. [Google Scholar] [CrossRef]
- Xu, S.N.; Wang, T.S.; Li, X.; Wang, Y.P. SIRT2 activates G6PD to enhance NADPH production and promote leukaemia cell proliferation. Sci Rep. 2016, 6, 32734. [Google Scholar] [CrossRef]
- Russo, C.; Maugeri, A.; De Luca, L.; Gitto, R.; Lombardo, G.E.; Musumeci, L.; De Sarro, G.; Cirmi, S.; Navarra, M. The SIRT2 Pathway Is Involved in the Antiproliferative Effect of Flavanones in Human Leukemia Monocytic THP-1 Cells. Biomedicines. 2022, 10, 2383. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Qi, Y.; Xu, L.; Tao, X.; Han, X.; Yin, L.; Peng, J. MicroRNA-140-5p aggravates doxorubicin-induced cardiotoxicity by promoting myocardial oxidative stress via targeting Nrf2 and Sirt2. Redox Biol. 2018, 15, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Havelange, V.; Stauffer, N.; Heaphy, C.C.; Volinia, S.; Andreeff, M.; Marcucci, G.; Croce, C.M.; Garzon, R. Functional implications of microRNAs in acute myeloid leukemia by integrating microRNA and messenger RNA expression profiling. Cancer. 2011, 117, 4696–706. [Google Scholar] [CrossRef]
- Zhang, J.; Xiang, H.; Liu, J.; Chen, Y.; He, R.R.; Liu, B. Mitochondrial Sirtuin 3: New emerging biological function and therapeutic target. Theranostics. 2020, 10, 8315–8342. [Google Scholar] [CrossRef]
- Chen, Y.; Fu, L.L.; Wen, X.; Wang, X.Y.; Liu, J.; Cheng, Y.; Huang, J. Sirtuin-3 (SIRT3); a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis. 2014, 5, e1047. [Google Scholar] [CrossRef]
- Zhao, K.; Zhou, Y.; Qiao, C.; Ni, T.; Li, Z.; Wang, X.; Guo, Q.; Lu, N.; Wei, L. Oroxylin A promotes PTEN-mediated negative regulation of MDM2 transcription via SIRT3-mediated deacetylation to stabilize p53 and inhibit glycolysis in wt-p53 cancer cells. J Hematol Oncol. 2015, 8, 41. [Google Scholar] [CrossRef]
- Wei, L.; Zhou, Y.; Dai, Q.; Qiao, C.; Zhao, L.; Hui, H.; Lu, N.; Guo, Q.L. Oroxylin A induces dissociation of hexokinase II from the mitochondria and inhibits glycolysis by SIRT3-mediated deacetylation of cyclophilin D in breast carcinoma. Cell Death Dis. 2013, 4, e601. [Google Scholar] [CrossRef] [PubMed]
- Bergaggio, E.; Riganti, C.; Garaffo, G.; Vitale, N.; Mereu, E.; Bandini, C.; Pellegrino, E.; Pullano, V.; Omedè, P.; Todoerti, K.; Cascione, L.; Audrito, V.; Riccio, A.; Rossi, A.; Bertoni, F.; Deaglio, S.; Neri, A.; Palumbo, A.; Piva, R. IDH2 inhibition enhances proteasome inhibitor responsiveness in hematological malignancies. Blood. 2019, 133, 156–167. [Google Scholar] [CrossRef]
- O'Brien, C.; Berman, J.; Culp-Hill, R.; Reisz, J.; Ling, T.; Rondeau, V.; Li, M.; Yang, M.; Hong, J.Y.; Lin, H.; Melnick, A.; Arruda, A.; Minden, M.D.; D'Alessandro, A.; Jones, C.L. Sirtuin 3 Inhibition Targets AML Stem Cells through Perturbation of Fatty Acid Oxidation. Blood. 2021, 138 (Suppl. S1). [Google Scholar] [CrossRef]
- O'Brien, C.; Ling, T.; Berman, J.M.; Culp-Hill, R.; Reisz, J.A.; Rondeau, V.; Jahangiri, S.; St-Germain, J.; Macwan, V.; Astori, A.; Zeng, A.; Hong, J.Y.; Li, M.; Yang, M.; Jana, S.; Gamboni, F.; Tsao, E.; Liu, W.; Dick, J.E.; Lin, H.; Melnick, A.; Tikhonova, A.; Arruda, A.; Minden, M.D.; Raught, B.; D'Alessandro, A.; Jones, C.L. Simultaneous inhibition of Sirtuin 3 and cholesterol homeostasis targets acute myeloid leukemia stem cells by perturbing fatty acid β-oxidation and inducing lipotoxicity. Haematologica. 2023, 108, 2343–2357. [Google Scholar] [CrossRef]
- Ma, J.; Liu, B.; Yu, D.; Zuo, Y.; Cai, R.; Yang, J.; Cheng, J. SIRT3 deacetylase activity confers chemoresistance in AML via regulation of mitochondrial oxidative phosphorylation. Br J Haematol. 2019, 187, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jiang, Z.; Yang, Y.; Zhang, C.; Liu, H.; Wan, J. The functions and mechanisms of post-translational modification in protein regulators of RNA methylation: Current status and future perspectives. Int J Biol Macromol. 2023, 253, 126773. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shen, Y.; Wei, W.; Wang, W.; Jiang, D.; Ren, Y.; Peng, Z.; Fan, Q.; Cheng, J.; Ma, J. Dysregulation of SIRT3 SUMOylation Confers AML Chemoresistance via Controlling HES1-Dependent Fatty Acid Oxidation. Int J Mol Sci. 2022, 23, 8282. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, B.; Yu, D.; Tam, W.; Yang, J.; Cheng, J. SIRT3 Sumoylation Contributes to Chemoresistance in AML. Blood. 2018, 132 (Suppl. S1). [Google Scholar] [CrossRef]
- Strzałka, P.; Krawiec, K.; Jarych, D.; Wiśnik, A.; Soin, M.; Góralska, M.; Mikulski, D.; Czemerska, M.; Zawlik, I.; Pluta, A.; Wierzbowska, A. Assessment of SIRT1-SIRT7 and TP53 Genes Expression in Patients with Acute Myeloid Leukemia. Blood. 2023, 142 (Suppl. S1). [Google Scholar] [CrossRef]
- Goes, J.V.C.; Carvalho, L.G.; de Oliveira, R.T.G.; Melo, M.M.L.; Novaes, L.A.C.; Moreno, D.A.; Gonçalves, P.G.; Montefusco-Pereira, C.V.; Pinheiro, R.F.; Ribeiro, Junior H. L. Role of Sirtuins in the Pathobiology of Onco-Hematological Diseases: A PROSPERO-Registered Study and In Silico Analysis. Cancers. 2022, 14, 4611. [Google Scholar] [CrossRef]
- Haigis, M.C.; Mostoslavsky, R.; Haigis, K.M.; Fahie, K.; Christodoulou, D.C.; Murphy, A.J.; Valenzuela, D.M.; Yancopoulos, G.D.; Karow, M.; Blander, G.; Wolberger, C.; Prolla, T.A.; Weindruch, R.; Alt, F.W.; Guarente, L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006, 126, 941–54. [Google Scholar] [CrossRef]
- Huang, G.; Zhu, G. Sirtuin-4 (SIRT4), a therapeutic target with oncogenic and tumor-suppressive activity in cancer. Onco Targets Ther. 2018, 11, 3395–3400. [Google Scholar] [CrossRef]
- Frye, RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999, 260, 273–9. [Google Scholar] [CrossRef]
- Jeong, S.M.; Xiao, C.; Finley, L.W.; Lahusen, T.; Souza, A.L.; Pierce, K.; Li, Y.H.; Wang, X.; Laurent, G.; German, N.J.; Xu, X.; Li, C.; Wang, R.H. , Lee, J.; Csibi, A.; Cerione, R.; Blenis, J.; Clish, C.B.; Kimmelman, A.; Deng, C.X.; Haigis, M.C. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013, 23, 450–63. [Google Scholar] [CrossRef]
- Jeong, S.M.; Lee, A.; Lee, J.; Haigis, M.C. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J Biol Chem. 2014, 289, 4135–44. [Google Scholar] [CrossRef] [PubMed]
- Csibi, A.; Fendt, S.M.; Li, C.; Poulogiannis, G.; Choo, A.Y.; Chapski, D.J.; Jeong, S.M.; Dempsey, J.M.; Parkhitko, A.; Morrison, T.; Henske, E.P.; Haigis, M.C.; Cantley, L.C.; Stephanopoulos, G.; Yu, J.; Blenis, J. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013, 153, 840–54. [Google Scholar] [CrossRef]
- Shi, Q.; Liu, T.; Zhang, X.; Geng, J.; He, X.; Nu, M.; Pang, D. Decreased sirtuin 4 expression is associated with poor prognosis in patients with invasive breast cancer. Oncol Lett. 2016, 12, 2606–2612. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Yu,, Z. ; Chen X.; Huang, G. SIRT4 is upregulated in Chinese patients with esophageal cancer. Int J Clin Exp Pathol. 2016, 9, 10543–10549. [Google Scholar]
- Bradbury, C.A.; Khanim, F.L.; Hayden, R.; Bunce, C.M.; White, D.A.; Drayson, M.T.; Craddock, C.; Turner, B.M. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005, 19, 1751–9. [Google Scholar] [CrossRef]
- Strzałka, P.; Krawiec, K.; Jarych, D.; Wiśnik, A.; Góralska, M.; Soin, M.; Mikulski, D.; Czemerska, M.; Zawlik, I.; Pluta, A.; Wierzbowska, A. Evaluation of Sirtuins and TP53 Gene Expression and Their Impact on Prognosis in Acute Myeloid Leukemia. Clin Lymphoma Myeloma Leuk. 2023, 23 (Suppl. S1), S314–S315. [Google Scholar] [CrossRef]
- Vaquero, A.; Reinberg, D. Sirtuins in Biology and Disease. Epigenetics in Biology and Medicine; 1st ed.; Esteller; M.; CRC Press; 2009; Chapter 6; pp. 73-104. [CrossRef]
- Yue, X.; Shi, Y.; Luo, Q. Advances of SIRT4 in cancer metabolism and therapy. Pediatr Discov. 2023, 1, e17. [Google Scholar] [CrossRef]
- Jeong, S.M.; Hwang, S.; Seong, R.H. SIRT4 regulates cancer cell survival and growth after stress. Biochem Biophys Res Commun. 2016, 470, 251–256. [Google Scholar] [CrossRef]
- Du, J.; Zhou, Y.; Su, X.; Yu, J.J.; Khan, S.; Jiang, H.; Kim, J.; Woo, J.; Kim, J.H.; Choi, B.H.; He, B.; Chen, W.; Zhang, S.; Cerione, R.A.; Auwerx, J.; Hao, Q.; Lin, H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011, 334, 806–9. [Google Scholar] [CrossRef]
- Nishida, Y.; Rardin, M.J.; Carrico, C.; He, W.; Sahu, A.K.; Gut, P.; Najjar, R.; Fitch, M.; Hellerstein, M.; Gibson, B.W.; Verdin, E. SIRT5 Regulates both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Mol Cell. 2015, 59, 321–32. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Peng, C.; Anderson, K.A.; Chhoy, P.; Xie, Z.; Dai, L.; Park, J.; Chen, Y.; Huang, H.; Zhang, Y.; Ro, J.; Wagner, G.R.; Green, M.F.; Madsen, A.S.; Schmiesing, J.; Peterson, B.S.; Xu, G.; Ilkayeva, O.R.; Muehlbauer, M.J.; Braulke, T.; Mühlhausen, C.; Backos, D.S.; Olsen, C.A.; McGuire, P.J.; Pletcher, S.D.; Lombard, D.B.; Hirschey, M.D.; Zhao, Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014, 19, 605–17. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Chen, Y.; Tishkoff, D.X.; Peng, C.; Tan, M.; Dai, L.; Xie, Z.; Zhang, Y.; Zwaans, B.M.; Skinner, M.E.; Lombard, D.B.; Zhao, Y. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013, 50, 919–30. [Google Scholar] [CrossRef]
- Wątroba, M.; Szukiewicz, D. Sirtuins in the biology of aging. Sirtuin Biology in Medicine; Maiese, K.; Academic Press, 2021; Chapter 5, pp. 79-90. [CrossRef]
- Zhao, L.; Cao, J.; Hu, K.; He, X.; Yun, D.; Tong, T.; Han, L. Sirtuins and their Biological Relevance in Aging and Age-Related Diseases. Aging Dis. 2020, 11, 927–945. [Google Scholar] [CrossRef]
- Liu, B.; Che, W.; Zheng, C.; Liu, W.; Wen, J.; Fu, H.; Tang, K.; Zhang, J.; Xu, Y. SIRT5: a safeguard against oxidative stress-induced apoptosis in cardiomyocytes. Cell Physiol Biochem. 2013, 32, 1050–9. [Google Scholar] [CrossRef]
- Liang, F.; Wang, X.; Ow, S.H.; Chen, W.; Ong, W.C. Sirtuin 5 is Anti-apoptotic and Anti-oxidative in Cultured SH-EP Neuroblastoma Cells. Neurotox Res. 2017, 31, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, Y.; Li, Y.; Zhao, Y.; Jiang, H. Sirt5 Attenuates Cisplatin-Induced Acute Kidney Injury through Regulation of Nrf2/HO-1 and Bcl-2. Biomed Res Int. 2019, 2019, 4745132. [Google Scholar] [CrossRef]
- Krawiec, K.; Strzałka, P.; Czemerska, M.; Wiśnik, A.; Zawlik, I.; Wierzbowska, A.; Pluta, A. Targeting Apoptosis in AML: Where Do We Stand? Cancers. 2022, 14, 4995. [Google Scholar] [CrossRef]
- Li, M.; Melnick, A.M. Non-oncogene Addiction to SIRT5 in Acute Myeloid Leukemia. Blood Cancer Discov. 2021, 2, 198–200. [Google Scholar] [CrossRef]
- Yan, D.; Franzini, A.; Pomicter, A.D.; Halverson, B.J.; Antelope, O.; Mason, C.C.; Ahmann, J.M.; Senina, A.V.; Vellore, N.A.; Jones, C.L.; Zabriskie, M.S.; Than, H.; Xiao, M.J.; van Scoyk, A.; Patel, A.B.; Clair, P.M.; Heaton, W.L.; Owen, S.C.; Andersen, J.L.; Egbert, C.M.; Reisz, J.A.; D'Alessandro, A.; Cox, J.E.; Gantz, K.C.; Redwine, H.M.; Iyer, S.M.; Khorashad, J.S.; Rajabi, N.; Olsen, C.A.; O'Hare, T.; Deininger, M.W. SIRT5 is a druggable metabolic vulnerability in acute myeloid leukemia. Blood Cancer Discov. 2021, 2, 266–287. [Google Scholar] [CrossRef]
- Yan, D.; Pomicter, A.D.; Heaton, W.L.; Mason, C.C.; Ahmann, J.; Senina, A.; Franzini, A.; Halverson, B.; Than, H.; Clair, P.M.; Khorashad, J.S.; O'Hare, T.; Deininger, M.W. SIRT5 As a Therapeutic Target in Acute Myeloid Leukemia. Blood. 2018, 132 (Suppl. S1). [Google Scholar] [CrossRef]
- Rajabi, N.; Hansen, T.N.; Nielsen, A.L.; Nguyen, H.T.; Bæk, M.; Bolding, J.E.; Bahlke, O.Ø.; Petersen, S.E.G.; Bartling, C.R.O.; Strømgaard, K.; Olsen, C.A. Angew. Chem. Int. Ed. 2022, 61, e202115805. [CrossRef]
- Arévalo, C.M.; Cruz-Rodriguez, N.; Quijano, S.; Fiorentino, S. Plant-derived extracts and metabolic modulation in leukemia: a promising approach to overcome treatment resistance. Front Mol Biosci. 2023, 10, 1229760. [Google Scholar] [CrossRef]
- Wang, M.; Shi, W.; Zhu, R.; Small, D.; Li, L.; Ma, S.; Hu, Y. Targeting SIRT5 By Succinylating Hadha Synergizes with Venetoclax in Acute Myeloid Leukemia. Blood. 2023, 142 (Suppl. S1). [Google Scholar] [CrossRef]
- Salehi, B.; Mishra, A.P.; Nigam, M.; Sener, B.; Kilic, M.; Sharifi-Rad, M.; Fokou, P.V.T.; Martins, N.; Sharifi-Rad, J. Resveratrol: A Double-Edged Sword in Health Benefits. Biomedicines. 2018, 6, 91. [Google Scholar] [CrossRef]
- Ozkan, T.; Koc, A.; Karabay, A.Z.; Hekmatshoar, Y.; Sunguroglu, A. An investigation on the effects of SIRT5 modulators on SIRT5 and Cytochrome C protein expressions in K562 chronic myeloid leukemia cell line. J. Fac. Pharm. 2022, 46, 805–816. [Google Scholar] [CrossRef]
- Bateman, B.; Bates, B.; Deininger, M.; Zhao, H.G.G. Chronic Mitochondrial Dysfunction Renders Acute Myeloid Leukemia Resistant to Multipronged Inhibition of Mitochondrial Metabolism. Blood. 2023, 142 (Suppl. S1). [Google Scholar] [CrossRef]
- Li, F.; He, X.; Ye, D.; Lin, Y.; Yu, H.; Yao, C.; Huang, L.; Zhang, J.; Wang, F.; Xu, S.; Wu, X.; Liu, L.; Yang, C.; Shi, J.; He, X.; Liu, J.; Qu, Y.; Guo, F.; Zhao, J.; Xu, W.; Zhao, S. NADP(+)-IDH Mutations Promote Hypersuccinylation that Impairs Mitochondria Respiration and Induces Apoptosis Resistance. Mol Cell. 2015, 60, 661–75. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.H.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.T.; Liu, L.X.; Jiang, W.Q.; Liu, J.; Zhang, J.Y.; Wang, B.; Frye, S.; Zhang, Y.; Xu, Y.H.; Lei, Q.Y.; Guan, K.L.; Zhao, S.M.; Xiong, Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011, 19, 17–30. [Google Scholar] [CrossRef]
- Fabbrizi, E.; Fiorentino, F.; Carafa, V.; Altucci, L.; Mai, A.; Rotili, D. Emerging Roles of SIRT5 in Metabolism, Cancer, and SARS-CoV-2 Infection. Cells. 2023, 12, 852. [Google Scholar] [CrossRef]
- Frye, R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000, 273, 793–8. [Google Scholar] [CrossRef] [PubMed]
- Beauharnois, J.M.; Bolívar, B.E.; Welch, J.T. Sirtuin 6: a review of biological effects and potential therapeutic properties. Mol Biosyst. 2013, 9, 1789–806. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.A.; Denu, J.M. Biological and catalytic functions of sirtuin 6 as targets for small-molecule modulators. J Biol Chem. 2020, 295, 11021–11041. [Google Scholar] [CrossRef]
- Banerjee, A.; Pavane, M.S.; Banu, L.H.; Gopikar, A.S.R.; Elizabeth, K.R.; Pathak, S. Traditional medicine for aging-related disorders: Implications for drug discovery. Stem Cells and Aging; Pathak, S.; Banerjee, A. Academic Press, 2021; Chapter 20, pp. 281-297. [CrossRef]
- Zhang, J.; Yin, X.J.; Xu, C.J.; Ning, Y.X.; Chen, M.; Zhang, H.; Chen, S.F.; Yao, L.Q. The histone deacetylase SIRT6 inhibits ovarian cancer cell proliferation via down-regulation of Notch 3 expression. Eur Rev Med Pharmacol Sci. 2015, 19, 818–24. [Google Scholar] [PubMed]
- Choe, M.; Brusgard, J.L.; Chumsri, S.; Bhandary, L.; Zhao, X.F.; Lu, S.; Goloubeva, O.G.; Polster, B.M.; Fiskum, G.M.; Girnun, G.D.; Kim, M.S.; Passaniti, A. The RUNX2 Transcription Factor Negatively Regulates SIRT6 Expression to Alter Glucose Metabolism in Breast Cancer Cells. J Cell Biochem. 2015, 116, 2210–26. [Google Scholar] [CrossRef]
- Qi, J.; Cui, C.; Deng, Q.; Wang, L.; Chen, R.; Zhai, D.; Xie, L.; Yu, J. Downregulated SIRT6 and upregulated NMNAT2 are associated with the presence; depth and stage of colorectal cancer. Oncol Lett. 2018, 16, 5829–5837. [Google Scholar] [CrossRef]
- Min, L.; Ji, Y.; Bakiri, L.; Qiu, Z.; Cen, J.; Chen, X.; Chen, L.; Scheuch, H.; Zheng, H.; Qin, L.; Zatloukal, K.; Hui, L.; Wagner, E.F. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat Cell Biol. 2012, 14, 1203–11. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; Reddy, A.; Liu, M.; Murray, L.; Berger, M.F.; Monahan, J.E.; Morais, P.; Meltzer, J.; Korejwa, A.; Jané-Valbuena, J.; Mapa, F.A.; Thibault, J.; Bric-Furlong, E.; Raman, P.; Shipway, A.; Engels, I.H.; Cheng, J.; Yu, G.K.; Yu, J.; Aspesi, P. Jr.; de Silva, M.; Jagtap, K.; Jones, M.D.; Wang, L.; Hatton, C.; Palescandolo, E.; Gupta, S.; Mahan, S.; Sougnez, C.; Onofrio, R.C.; Liefeld, T.; MacConaill, L.; Winckler, W.; Reich, M.; Li, N.; Mesirov, J.P.; Gabriel, S.B.; Getz, G.; Ardlie, K.; Chan, V.; Myer, V.E.; Weber, B.L.; Porter, J.; Warmuth, M.; Finan, P.; Harris, J.L.; Meyerson, M.; Golub, T.R.; Morrissey, M.P.; Sellers, W.R.; Schlegel, R.; Garraway, L.A. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012, 483, 603–7. [Google Scholar] [CrossRef]
- Lai, CC.; Lin, P.M.; Lin, S.F.; Hsu, C.H.; Lin, H.C.; Hu, M.L.; Hsu, C.M.; Yang, M.Y. Altered expression of SIRT gene family in head and neck squamous cell carcinoma. Tumour Biol. 2013, 34, 1847–54. [Google Scholar] [CrossRef]
- Lefort, K.; Brooks, Y.; Ostano, P.; Cario-André, M.; Calpini, V.; Guinea-Viniegra, J.; Albinger-Hegyi, A.; Hoetzenecker, W.; Kolfschoten, I.; Wagner, E.F.; Werner, S.; Dotto, G.P. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 2013, 32, 2248–63. [Google Scholar] [CrossRef]
- Cea, M.; Cagnetta, A.; Adamia, S.; Acharya, C.; Tai, Y.T.; Fulciniti, M.; Ohguchi, H.; Munshi, A.; Acharya, P.; Bhasin, M.K.; Zhong, L.; Carrasco, R.; Monacelli, F.; Ballestrero, A.; Richardson, P.; Gobbi, M.; Lemoli, R.M.; Munshi, N.; Hideshima, T.; Nencioni, A.; Chauhan, D.; Anderson, K.C. Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood. 2016, 127, 1138–50. [Google Scholar] [CrossRef]
- Wang, J.C.; Kafeel, M.I.; Avezbakiyev, B.; Chen, C.; Sun, Y.; Rathnasabapathy, C.; Kalavar, M.; He, Z.; Burton, J.; Lichter, S. Histone deacetylase in chronic lymphocytic leukemia. Oncology. 2011, 81, 325–9. [Google Scholar] [CrossRef] [PubMed]
- Cagnetta, A.; Soncini, D.; Orecchioni, S.; Talarico, G.; Minetto, P.; Guolo, F.; Retali, V.; Colombo, N.; Carminati, E.; Clavio, M.; Miglino, M.; Bergamaschi, M.; Nahimana, A.; Duchosal, M.; Todoerti, K.; Neri, A.; Passalacqua, M.; Bruzzone, S.; Nencioni, A.; Bertolini, F.; Gobbi, M.; Lemoli, R.M.; Cea, M. Depletion of SIRT6 enzymatic activity increases acute myeloid leukemia cells' vulnerability to DNA-damaging agents. Haematologica. 2018, 103, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Cea, M.; Cagnetta, A.; Soncini, D.; Minetto, P.; Lovera, D.; Guolo, F.; Colombo, N.; Ballerini, F.; Clavio, M.; Miglino, M.; Gobbi, M.; Lemoli, R.M. SIRT6 Inhibition As a Novel Approach for Treating Acute Myeloid Leukemia. Blood. 2016, 128, 5222. [Google Scholar] [CrossRef]
- Cea, M.; Cagnetta, A.; Lovera, D.; Grasso, R.; Colombo, N.; Bergamaschi, M.; Aquino, S.; Guolo, F.; Minetto, P.; Ballerini, F.; Canepa, L.; Adamia, S.; Nencioni, A.; Pierri, I.; Clavio, M.; Miglino, M.; Gobbi, M.; Lemoli, R.M. A Novel Synthetic Lethal Approach Targeting SIRT6 in Acute Myeloid Leukemia. Blood. 2015, 126, 1375. [Google Scholar] [CrossRef]
- Hubner, S.E.; de Camargo Magalhães, E.S.; Hoff, F.W.; Brown, B.D.; Qiu, Y.; Horton, T.M.; Kornblau, S.M. DNA Damage Response-Related Proteins Are Prognostic for Outcome in Both Adult and Pediatric Acute Myelogenous Leukemia Patients: Samples from Adults and from Children Enrolled in a Children's Oncology Group Study. Int J Mol Sci. 2023, 24, 5898. [Google Scholar] [CrossRef]
- Carraway, H.E.; Malkaram, S.A.; Cen, Y.; Shatnawi, A.; Fan, J.; Ali, H.E.A.; Abd Elmageed, Z.Y.; Buttolph, T.; Denvir, J.; Primerano, D.A.; Fandy, T.E. Activation of SIRT6 by DNA hypomethylating agents and clinical consequences on combination therapy in leukemia. Sci Rep. 2020, 10, 10325. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, Z.; Sheng, F.; Yin, Z. MYC upregulated LINC00319 promotes human acute myeloid leukemia (AML) cells growth through stabilizing SIRT6. Biochem Biophys Res Commun. 2019, 509, 314–321. [Google Scholar] [CrossRef]
- Zhang, P.; Brinton, L.T.; Williams, K.; Sher, S.; Orwick, S.; Tzung-Huei, L.; Mims, A.S.; Coss, C.C.; Kulp, S.K.; Youssef, Y.; Chan, W.K.; Mitchell, S.; Mustonen, A.; Cannon, M.; Phillips, H.; Lehman, A.M.; Kauffman, T.; Beaver, L.; Canfield, D.; Grieselhuber, N.R.; Alinari, L.; Sampath, D.; Yan, P.; Byrd, J.C.; Blachly, J.S.; Lapalombella, R. Targeting DNA Damage Repair Functions of Two Histone Deacetylases; HDAC8 and SIRT6; Sensitizes Acute Myeloid Leukemia to NAMPT Inhibition. Clin Cancer Res. 2021, 27, 2352–2366. [Google Scholar] [CrossRef]
- Shi, X.; Jiang, Y.; Kitano, A.; Hu, T.; Murdaugh, R.L.; Li, Y.; Hoegenauer, K.A.; Chen, R.; Takahashi, K.; Nakada, D. Nuclear NAD+ homeostasis governed by NMNAT1 prevents apoptosis of acute myeloid leukemia stem cells. Sci Adv. 2021, 7, eabf3895. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.; Yang, Y.; Li, Z.; Chen, Y.; Shang, S.; Wang, Y. DNMT3A mutation promotes leukemia development through NAM-NAD metabolic reprogramming. J Transl Med. 2023, 21, 481. [Google Scholar] [CrossRef] [PubMed]
- Blank, M.F.; Grummt, I. The seven faces of SIRT7. Transcription. 2017, 8, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, B.N.; Thackray, J.K.; Simonet, N.G.; Kane-Goldsmith, N.; Martinez-Redondo, P.; Nguyen, T.; Bunting, S.; Vaquero, A.; Tischfield, J.A.; Serrano, L. SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J. 2016, 35, 1488–503. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, L.; Yang, S.; Yan, R.; Zhang, D.; Yang, J.; He, L.; Li, W.; Yi, X.; Sun, L.; Liang, J.; Cheng, Z.; Shi, L.; Shang, Y.; Yu, W. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun. 2016, 7, 12235. [Google Scholar] [CrossRef]
- Ianni, A.; Hoelper, S.; Krueger, M.; Braun, T.; Bober, E. Sirt7 stabilizes rDNA heterochromatin through recruitment of DNMT1 and Sirt1. Biochem Biophys Res Commun. 2017, 492, 434–440. [Google Scholar] [CrossRef]
- Ianni, A.; Kumari, P.; Tarighi, S.; Braun, T.; Vaquero, A. SIRT7: a novel molecular target for personalized cancer treatment? Oncogene 2024, 43, 993–1006. [Google Scholar] [CrossRef]
- Barber, M.F.; Michishita-Kioi, E.; Xi, Y.; Tasselli, L.; Kioi, M.; Moqtaderi, Z.; Tennen, R.I.; Paredes, S.; Young, N.L.; Chen, K.; Struhl, K.; Garcia, B.A.; Gozani, O.; Li, W.; Chua, K.F. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012, 487, 114–8. [Google Scholar] [CrossRef]
- Li, L.; Bhatia, R. The controversial role of Sirtuins in tumorigenesis - SIRT7 joins the debate. Cell Res. 2013, 23, 10–2. [Google Scholar] [CrossRef]
- Ford, E.; Voit, R.; Liszt, G.; Magin, C.; Grummt, I.; Guarente, L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006, 20, 1075–80. [Google Scholar] [CrossRef]
- Malik, S.; Villanova, L.; Tanaka, S.; Aonuma, M.; Roy, N.; Berber, E.; Pollack, J.R.; Michishita-Kioi, E.; Chua, K.F. SIRT7 inactivation reverses metastatic phenotypes in epithelial and mesenchymal tumors. Sci Rep. 2015, 5, 9841. [Google Scholar] [CrossRef]
- Poniewierska-Baran. A.; Warias, P.; Zgutka, K. Sirtuins (SIRTs) As a Novel Target in Gastric Cancer. Int J Mol Sci. 2022, 23, 15119. [Google Scholar] [CrossRef]
- Kim, J.K.; Noh, J.H.; Jung, K.H.; Eun, J.W.; Bae, H.J.; Kim, M.G.; Chang, Y.G.; Shen, Q.; Park, W.S.; Lee, J.Y.; Borlak, J.; Nam, S.W. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013, 57, 1055–67. [Google Scholar] [CrossRef]
- Wang, H.L.; Lu, R.Q.; Xie, S.H.; Zheng, H.; Wen, X.M.; Gao, X.; Guo, L. SIRT7 Exhibits Oncogenic Potential in Human Ovarian Cancer Cells. Asian Pac J Cancer Prev. 2015, 16, 3573–7. [Google Scholar] [CrossRef]
- Aljada, A.; Saleh, A.M.; Alkathiri, M.; Shamsa, H.B.; Al-Bawab, A.; Nasr, A. Altered Sirtuin 7 Expression is Associated with Early Stage Breast Cancer. Breast Cancer (Auckl). 2015, 9, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ji, Y.; Zhang, D.; Liu, Y.; Fang, P. MicroRNA-3666-induced suppression of SIRT7 inhibits the growth of non-small cell lung cancer cells. Oncol Rep. 2016, 36, 3051–3057. [Google Scholar] [CrossRef]
- Li, W.; Zhu, D.; Qin, S. SIRT7 suppresses the epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis by promoting SMAD4 deacetylation. J Exp Clin Cancer Res. 2018, 37, 148. [Google Scholar] [CrossRef] [PubMed]
- Raza, U.; Tang, X.; Liu, Z.; Liu, B. SIRT7: the seventh key to unlocking the mystery of aging. Physiol Rev. 2024, 104, 253–280. [Google Scholar] [CrossRef]
- Wang, Y.; Maruichi, A.; Song, Z.; Greenberg, P.; Chen, D. SIRT7 improves hematopoiesis in myelodysplastic syndrome through regulating mitochondrial stress. Experimental Hematology. 2023, 124, S157. [Google Scholar]
- Nowicki, M.; Wierzbowska, A.; Stec-Martyna, E.; Kulczycka-Wojdala, D.; Nowicki, G.; Szmigielska-Kapłon, A. SIRT1-SIRT7 Expression in Patients with Lymphoproliferative Disorders Undergoing Hematopoietic Stem Cell Mobilization. Cancers. 2022, 14, 1213. [Google Scholar] [CrossRef]
- Kaiser, A.; Schmidt, M.; Huber, O.; Frietsch, J.J.; Scholl, S.; Heidel, F.H.; Hochhaus, A.; Müller, J.P.; Ernst, T. SIRT7: an influence factor in healthy aging and the development of age-dependent myeloid stem-cell disorders. Leukemia. 2020, 34, 2206–2216. [Google Scholar] [CrossRef]
- Metzeler, K.H.; Hummel, M.; Bloomfield, C.D.; Spiekermann, K.; Braess, J.; Sauerland, M.C.; Heinecke, A.; Radmacher, M.; Marcucci, G.; Whitman, S.P.; Maharry, K.; Paschka, P.; Larson, R.A.; Berdel, W.E.; Büchner, T.; Wörmann, B.; Mansmann, U.; Hiddemann, W.; Bohlander, S.K.; Buske, C.; Cancer and Leukemia Group B; German AML Cooperative Group. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008, 112, 4193–201. [Google Scholar] [CrossRef]
- Vakhrusheva, O.; Smolka, C.; Gajawada, P.; Kostin, S.; Boettger, T.; Kubin, T.; Braun, T.; Bober, E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008, 102, 703–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Kim, D.K.; Kim, E.S.; Park, S.J.; Kwon, J.H.; Shin, J.; Park, S.M.; Moon, Y.H.; Wang, H.J.; Gho, Y.S.; Choi, K.Y. Comparative interactomes of SIRT6 and SIRT7: Implication of functional links to aging. Proteomics. 2014, 14, 1610–22. [Google Scholar] [CrossRef] [PubMed]
- Ianni, A.; Kumari, P.; Tarighi, S.; Simonet, N.G.; Popescu, D.; Guenther, S.; Hölper, S.; Schmidt, A.; Smolka, C.; Yue, S.; Krüger, M.; Fiorillo, C.; Vaquero, A.; Bober, E.; Braun, T. SIRT7-dependent deacetylation of NPM promotes p53 stabilization following UV-induced genotoxic stress. Proc Natl Acad Sci U S A. 2021, 118, e2015339118. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Tian, J.; Zheng, G.; Zhao, J. Sirtuin7 is involved in protecting neurons against oxygen-glucose deprivation and reoxygenation-induced injury through regulation of the p53 signaling pathway. J Biochem Mol Toxicol. 2017, 31. [Google Scholar] [CrossRef]
- Sun, M.; Zhai, M.; Zhang, N.; Wang, R.; Liang, H.; Han, Q.; Jia, Y.; Jiao, L. MicroRNA-148b-3p is involved in regulating hypoxia/reoxygenation-induced injury of cardiomyocytes in vitro through modulating SIRT7/p53 signaling. Chem Biol Interact. 2018, 296, 211–219. [Google Scholar] [CrossRef]
- Yu, M.; Shi, X.; Ren, M.; Liu, L.; Qi, H.; Zhang, C.; Zou, J.; Qiu, X.; Zhu, W.G.; Zhang, Y.E.; Wang, W.; Luo, J. SIRT7 Deacetylates STRAP to Regulate p53 Activity and Stability. Int J Mol Sci. 2020, 21, 4122. [Google Scholar] [CrossRef]
- Olsen, E.A.; Kim, Y.H.; Kuzel, T.M.; Pacheco, T.R.; Foss, F.M.; Parker, S.; Frankel, S.R.; Chen, C.; Ricker, J.L.; Arduino, J.M.; Duvic, M. Phase IIb multicenter trial of vorinostat in patients with persistent; progressive; or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007, 25, 3109–15. [Google Scholar] [CrossRef]
- O'Connor, O.A.; Horwitz, S.; Masszi, T.; Van Hoof, A.; Brown, P.; Doorduijn, J.; Hess, G.; Jurczak, W.; Knoblauch, P.; Chawla, S.; Bhat, G.; Choi, M.R.; Walewski, J.; Savage, K.; Foss, F.; Allen, L.F.; Shustov, A. Belinostat in Patients With Relapsed or Refractory Peripheral T-Cell Lymphoma: Results of the Pivotal Phase II BELIEF (CLN-19) Study. J Clin Oncol. 2015, 33, 2492–9. [Google Scholar] [CrossRef]
- Garcia-Manero, G.; Yang, H.; Bueso-Ramos, C.; Ferrajoli, A.; Cortes, J.; Wierda, W.G.; Faderl, S.; Koller, C.; Morris, G.; Rosner, G.; Loboda, A.; Fantin, V.R.; Randolph, S.S.; Hardwick, J.S.; Reilly, J.F.; Chen, C.; Ricker, J.L.; Secrist, J.P.; Richon, V.M.; Frankel, S.R.; Kantarjian, H.M. Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]) in patients with advanced leukemias and myelodysplastic syndromes. Blood. 2008, 111, 1060–6. [Google Scholar] [CrossRef]
- Gu, S.; Hou, Y.; Dovat, K.; Dovat, S.; Song, C.; Ge, Z. Synergistic effect of HDAC inhibitor Chidamide with Cladribine on cell cycle arrest and apoptosis by targeting HDAC2/c-Myc/RCC1 axis in acute myeloid leukemia. Exp Hematol Oncol. 2023, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Maher, K.R.; Schafer, D.; Schaar, D.; Sabo, R.; Bandyopadhyay, D.; Grant, S. Pevonedistat plus belinostat in relapsed/refractory acute myeloid leukemia or myelodysplastic syndrome: A phase I multicenter study. JCO. 2023, 41, e19015–e19015. [Google Scholar] [CrossRef]
- Maher, K.R.; Shafer, D.; Schaar, D.; Bandyopadhyay, D.; Deng, X.; Wright, J.; Piekarz, R.; Rudek, M.A.; Harvey, R.D.; Grant, S. A phase I study of MLN4924 and belinostat in relapsed/refractory acute myeloid leukemia or myelodysplastic syndrome. Cancer Chemother Pharmacol. 2025, 95, 24. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Osdal, T.; Ho, Y.W.; McDonald, T.; Chun, S.; Chu, S.; Lin, A.; Dos Santos, C.; Popa, M.L.; Gjertsen, B.T.; Chen, W.; Lain, S.; McCormack, E.; Bhatia, R. Pharmacological Inhibition Of The SIRT1 Deacetylase With The Small Molecule Inhibitor Tenovin-6 Enhances Ablation Of FLT3-ITD+ LSC In Combination With TKI Treatment. Blood. 2013, 122, 2685. [Google Scholar] [CrossRef]
| Sirtuin (SIRT) |
Class | Main cell localization | Potential signaling pathways impact and affected molecules | Functions | Level | Probable role in AML | Ref. |
|---|---|---|---|---|---|---|---|
| SIRT1 | I | Nucleus | p53, c-Myc, USP22, FOXO1, FOXP1, STAT5, DOT1L, miR-9 | Deacetylase Deacylase | High | Promoter | 38, 39, 42, 50, 51, 52, 53, 55 |
| SIRT2 | I | Cytosol Nucleus (during mitosis) |
NAMPT, MAPK, VEGF, MRP1, ERK1/2, BCL-2, p53, Caspase-1, AKT/GSK3ß/ß-catenin, PI3K/AKT/mTOR, pentose-phosphate cycle, p21, cyclin E1, miR-140-5p, miR-145 | Deacetylase Deacylase | High | Promoter | 63, 64, 68, 69, 70, 71, 72, 73, 74, 75 |
| SIRT3 | I | Mitochondria | PTEN, MDM2, p53, cyclophilin D, IDH2, tricarboxylic acid cycle, Fatty acid oxidation, sumoylation, HES1, Notch1 | Deacetylase Decrotonylase | Low | Undetermined/ Bi-directional |
78, 79, 80, 81, 84, 85 |
| SIRT4 | II | Mitochondria | Glutamine metabolism | Deacetylase Deacylase ADP-rybosil-transferase, |
Low (high according to some sources) | Undetermined/ Bi-directional |
90, 92, 93 |
| SIRT5 | III | Mitochondria | BAX/BCL-2, oxidative phosphorylation | Deacetylase Demalonylase Deglutarylase Desuccinylase | No data | Promoter | 102,103,104,105, 108, 109, 111, 112 |
| SIRT6 | IV | Nucleus | NAMPT, NMNAT1, cyclin-CDK | Deacetylase Deacylase ADP-rybosil-transferase |
High | Promoter | 137, 143, 144, 145 |
| SIRT7 | IV | Nucleus Nucleolus |
NRF1, NPM1, p53 | Deacetylase Dessuccinylase |
Low | Suppressor | 144, 148, 161, 162, 164, 168 |
| Molecule | Mechanism of action | Targeted sirtuin (SIRT) |
Additional mechanisms | Reference |
|---|---|---|---|---|
| Selisistat (EX-527) | Inhibitor | SIRT1 | Westerberg et al., 2015 | |
| MC3482 | Inhibitor | SIRT5 | Polletta L et al., 2015 | |
| SIRT5 inhibitor 1 | Inhibitor | SIRT5 | Wang et al., 2022 | |
| Tenovin-1 | Inhibitor | SIRT1 SIRT2 |
- p53 activation - Dihydroorotate dehydrogenase inhibitor |
Huang et al., 2019 Wan et al., 2021 Lain et al., 2008 Ladds et al., 2021 |
| BZD9Q1 | Inhibitor | SIRT1 SIRT2 SIRT3 |
- apoptosis and necrosis induction, G2/M cycle arrest | Chen et Yeong, 2024 |
| SIRT6-IN-2 (compound 5) | Inhibitor | SIRT6 | - increase acetylation of H3K9 - increase glucose uptake in cultured cells |
Damonte et al., 2017 |
| UBCS039 | Activator | SIRT6 | - anti-inflammatory response - oxidative stress alleviation |
Chen et Yeong, 2024 Jiao et al., 2022 |
| 4'-Bromo-resveratrol | Inhibitor | SIRT1 SIRT3 |
- mitochondrial metabolic reprogramming | George et al., 2019 |
| JFD00244 | Inhibitor | SIRT2 | - Nsp-16 inhibitor | Shankar et al., 2020 |
| Nicotinamide-d4 Nicotinamide-13C6 Nicotinamide-15N,13C3 |
Inhibitors | SIRT1 | - NAD+ redox homeostasis | Bitterman et al., 2002 Hwang et al., 2017 |
| Z26395438 (compound 1) | Inhibitor | SIRT1 | Gryniukova et al., 2023 | |
| Sirt1/2-IN-3 (compound PS9) | Inhibitor | SIRT1 SIRT2 |
- p53 deacetylation blockade | Cai et al., 2023 |
| Sirt1/2-IN-2 (compound hsa55) | Inhibitor | SIRT1 SIRT2 |
- p53 deacetylation blockade | Cai et al., 2023 |
| Antiproliferative agent-17 | Inhibitor | SIRT1 |
- anticancer activity - Gram+ bacteria inhibition |
Warda et al., 2022 |
| SirReal1, SirReal2 | Inhibitors | SIRT2 | Schiedel et al., 2016 | |
| SIRT1 activator 1(compound 3) | Activator | SIRT1 | Yang et al., 2023 | |
| SIRT5 inhibitor 8 (compound 10) | Inhibitor | SIRT5 | - anticancer potential | Wang et al., 2023 |
| 3-aryl-mercapto-butyrylated peptide derivative | Inhibitor | SIRT2 | Kalbas et al., 2022 | |
| SIRT5 Inhibitor 6 (2,4,5-trisubstituted pyrimidine derivative) | Inhibitor | SIRT5 | - modulating sepsis - AKI |
Mou et al., 2023 |
| SIRT2-IN-12 (compound 3) (xanthone derivative) | Inhibitor | SIRT2 | Mazur et al., 2024 | |
| SIRT5 inhibitor 9 (compound 14) | Inhibitor | SIRT5 | - anticancer potential | Wang et al., 2023 |
| Mz325 | Inhibitor | SIRT2 | - HDAC inhibitor | Sinatra et al., 2023 |
| HSP70/SIRT2-IN-2 (Compounds 1a) | Inhibitor | SIRT2 | - HSP70 inhibitor - anticancer activity |
Abbotto et al., 2023 |
| Sirt1/2-IN-4 (compound PS3) | Inhibitor | SIRT1 SIRT2 SIRT3 |
- potential anticancer activity - blocks p53 deacetylation |
Cai et al., 2023 |
| SIRT4-IN-1 (compound 69) | Inhibitor | SIRT4 | Pannek et al., 2024 | |
| SIRT2/6-IN-1 (Compound 5) | Inhibitor | SIRT2 SIRT6 |
- increases H3K9 acetylation - increases glucose uptake - reduces TNF-alfa secretion in cells |
Parenti et al., 2014 |
| SIRT5 inhibitor 7 (compound 58) (2,4,5-trisubstituted pyrimidine derivative) | Inhibitor | SIRT5 | - anti-inflammatory activity | Mou et al., 2023 |
| SIRT1/2/3-IN-1 (compound 10) | Inhibitor | SIRT1 SIRT2 SIRT3 |
- potential anticancer activity |
Li et al., 2020 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





