Submitted:
12 February 2025
Posted:
13 February 2025
You are already at the latest version
Abstract
Multidrug-resistant (MDR) Acinetobacter baumannii possesses a pressing threat to global healthcare settings, as most antibiotics are ineffective against this nosocomial pathogen. Vaccines, particularly peptide-based vaccines, offer a promising and effective strategy to deal with these infections. This study employed advanced bioinformatic tools to identify potential epitopes for vaccine candidates against A. baumannii infections. Three epitopes (EP1, EP2, and EP3) derived from A. baumannii OmpA were found to effectively bind with specific human leukocyte antigen (HLA) alleles. These epitopes have shown promising potential to elicit both cellular and humoral immune responses. Their physicochemical and immunological properties were thoroughly evaluated, indicating strong antigenic potential, non-toxicity, lack of allergenic properties, good binding affinity, and wide population coverage. The epitopes' two- and three-dimensional structures were predicted, and they were docked with their respective HLA alleles to assess their ability to stimulate innate immune responses. The predicted epitopes and HLA-allelic complexes exhibited excellent binding affinity, optimal Root Mean Square Deviation (RMSD) values, favorable physicochemical properties, and high-quality structural characteristics. Therefore, these in silico screened epitopes hold promise as potential solutions for combating multidrug-resistant A. baumannii, pending validation through wet lab experiments and clinical trials.
Keywords:
1. Introduction
2. Methods
2.1. Protein sequence retrieval
2.2. Assessment and selection of helper T-cell epitopes and linear B-cell epitopes
2.3. Final selection of helper T-cell epitopes
2.4. Prediction of cytokine secretion
2.5. Assessment of Allergenicity, Toxicity, Population coverage, and Epitope Identity for Peptide Evaluation
2.6. Tertiary Structure Prediction
2.7. Prediction of physicochemical characteristics, solubility prediction, and structure analyses
2.8. Molecular Docking
3. Result
3.1. Selection of T-cell epitopes
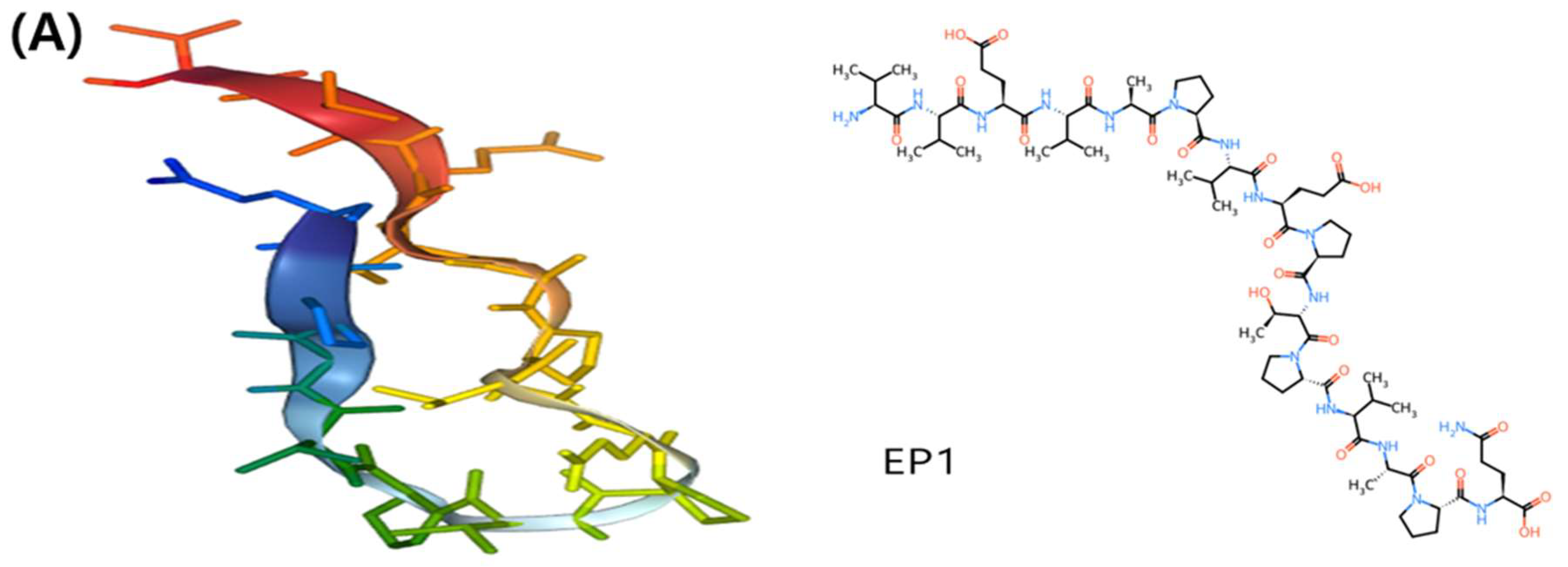
3.2. Assessment of the tertiary structures of the protein and epitopes
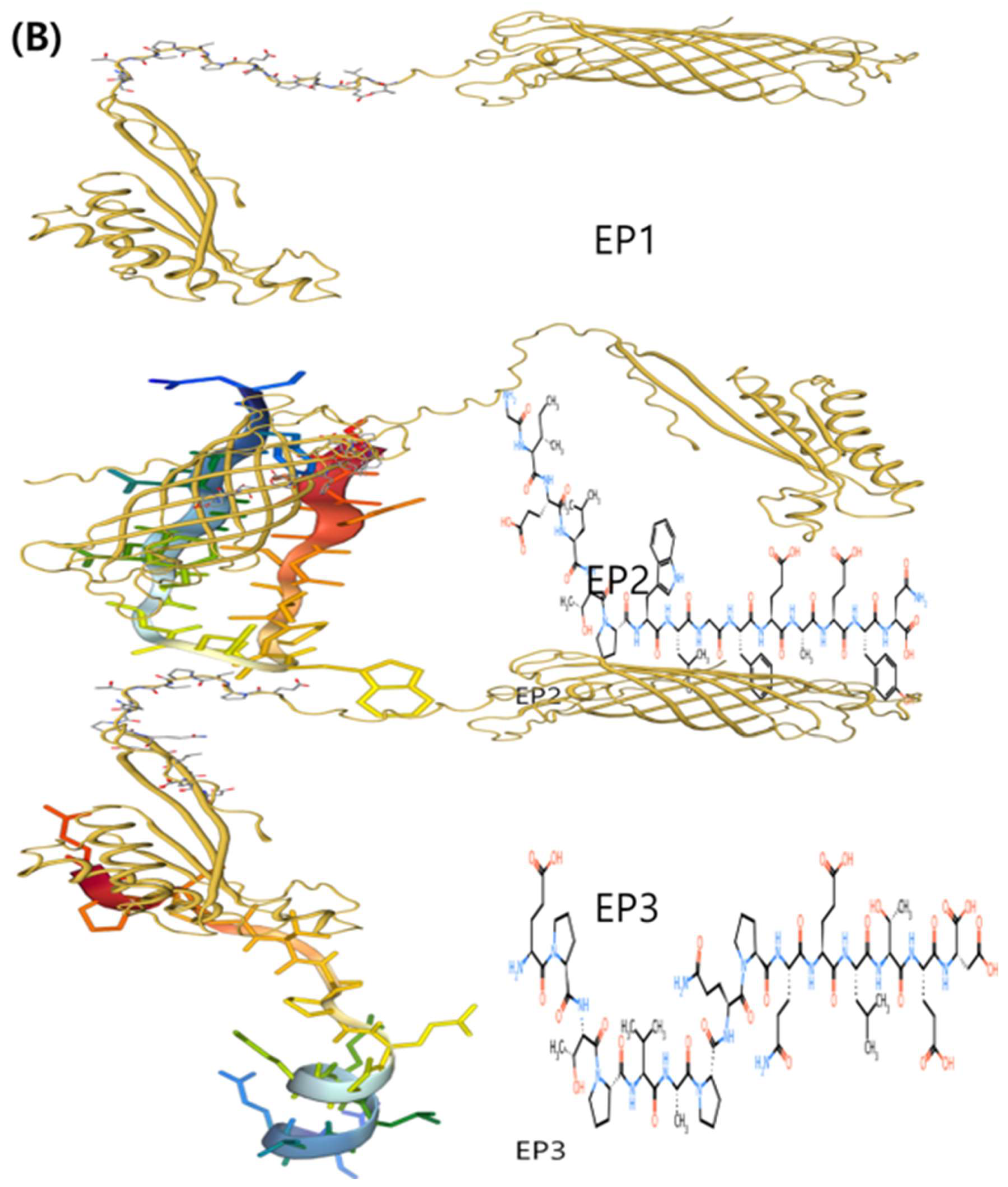
3.3. The structure, antigenic potential, and physicochemical characteristics of the final peptides
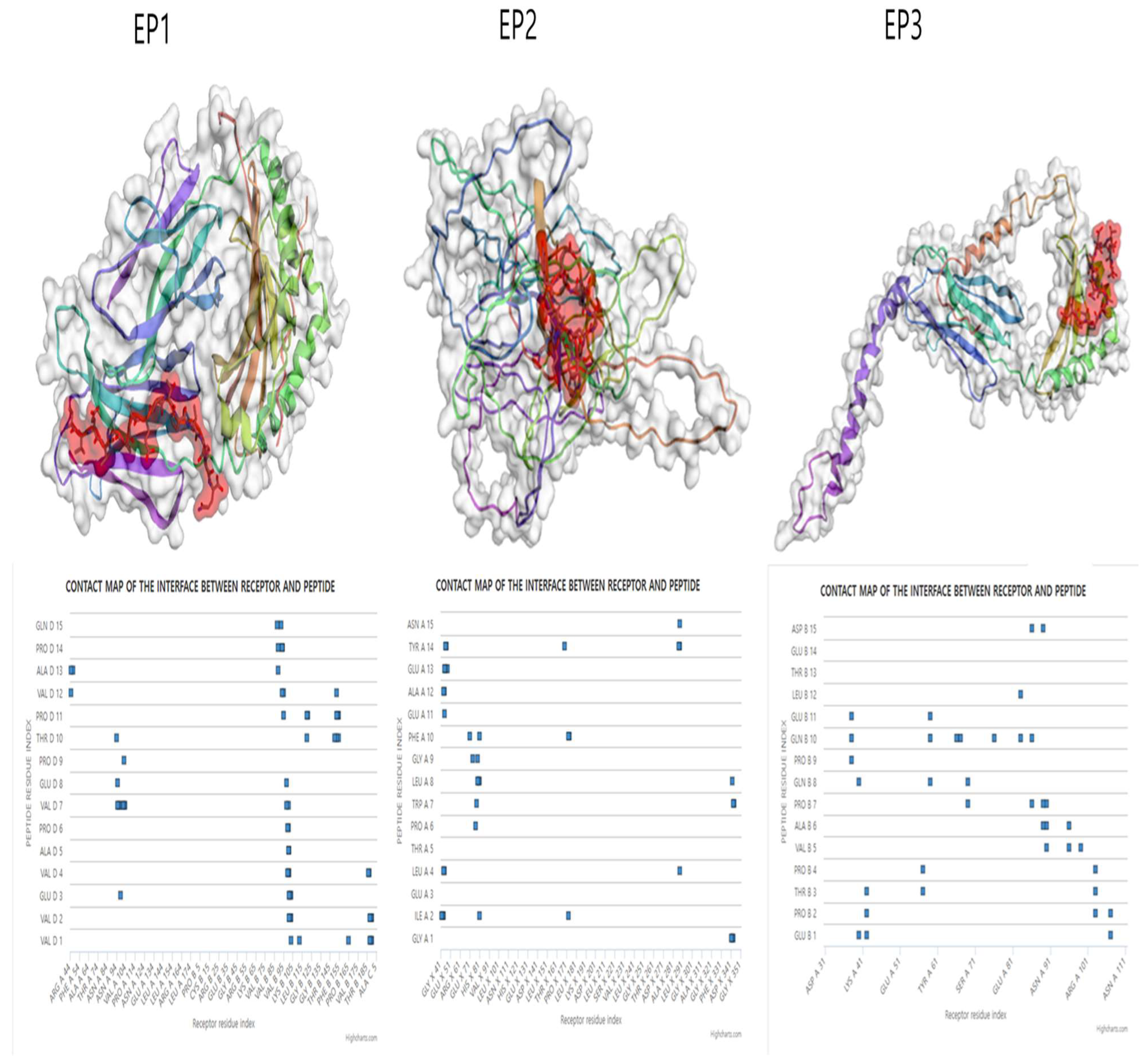
3.4. Molecular Docking Simulation of the Epitopes
4. Discussion
5. Conclusion
Fundings
Data Availability Statement
Acknowledgment
Conflicts of Interest
References
- Islam, M.M.; Mahbub, N.U.; Shin, W.S.; Oh, M.H. Phage-encoded depolymerases as a strategy for combating multidrug-resistant Acinetobacter baumannii. 2024, 14. [Google Scholar] [CrossRef]
- Savin, M.; et al. Tracing clinically-relevant antimicrobial resistances in Acinetobacter baumannii-calcoaceticus complex across diverse environments: A study spanning clinical, livestock, and wastewater treatment settings. Environment International 2024, 186, 108603. [CrossRef]
- Fahy, S., O'Connor, J. A., Lucey, B. & Sleator, R. D. Hospital Reservoirs of Multidrug Resistant Acinetobacter Species-The Elephant in the Room! British journal of biomedical science 2023, 80, 11098. [CrossRef]
- Benaissa, E.; Belouad, E.; Maleb, A.; Elouennass, M. Risk factors for acquiring Acinetobacter baumannii infection in the intensive care unit: experience from a Moroccan hospital. Access microbiology 2023, 5. [Google Scholar] [CrossRef]
- Corcione, S.; Longo, B.M.; Scabini, S.; Pivetta, E.; Curtoni, A.; Shbaklo, N.; Costa, C.; De Rosa, F.G. Risk factors for mortality in Acinetobacter baumannii bloodstream infections and development of a predictive mortality model. Journal of Global Antimicrobial Resistance 2024, 38, 317–326. [Google Scholar] [CrossRef]
- Wei, Z.; Zhou, S.; Zhang, Y.; Zheng, L.; Zhao, L.; Cui, Y.; Xie, K. Microbiological characteristics and risk factors on prognosis associated with Acinetobacter baumannii bacteremia in general hospital: A single-center retrospective study. Frontiers in microbiology 2022, 13, 1051364. [Google Scholar] [CrossRef]
- Russo, A.; Bassetti, M.; Ceccarelli, G.; Carannante, N.; Losito, A.R.; Bartoletti, M.; Corcione, S.; Granata, G.; Santoro, A.; Giacobbe, D.R.; et al. Bloodstream infections caused by carbapenem-resistant Acinetobacter baumannii: Clinical features, therapy and outcome from a multicenter study. The Journal of infection 2019, 79, 130–138. [Google Scholar] [CrossRef]
- Lee, H.; Lee, H. Clinical and Economic Evaluation of Multidrug-Resistant Acinetobacter baumannii Colonization in the Intensive Care Unit. Infection & chemotherapy 2016, 48, 174–180. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Lucidi, M.; Visaggio, D.; Migliaccio, A.; Capecchi, G.; Visca, P.; Imperi, F.; Zarrilli, R. Pathogenicity and virulence of Acinetobacter baumannii: Factors contributing to the fitness in healthcare settings and the infected host. Virulence 2024, 15, 2289769. [Google Scholar] [CrossRef]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nature reviews. Microbiology 2018, 16, 91–102. [Google Scholar] [CrossRef]
- Singh, S.; Singh, S.; Trivedi, M.; Dwivedi, M. An insight into MDR Acinetobacter baumannii infection and its pathogenesis: Potential therapeutic targets and challenges. Microbial Pathogenesis 2024, 192, 106674. [Google Scholar] [CrossRef] [PubMed]
- Erol, H.B.; Kaskatepe, B.; Yildiz, S.; Altanlar, N. The effect of phage-antibiotic combination strategy on multidrug-resistant Acinetobacter baumannii biofilms. Journal of Microbiological Methods 2023, 210, 106752. [Google Scholar] [CrossRef] [PubMed]
- Shadan, A.; Pathak, A.; Ma, Y.; Pathania, R.; Singh, R.P. Deciphering the virulence factors, regulation, and immune response to Acinetobacter baumannii infection. 2023, 13. [Google Scholar] [CrossRef]
- McConnell, M.J.; Actis, L.; Pachón, J. Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiology Reviews 2013, 37, 130–155. [Google Scholar] [CrossRef] [PubMed]
- Cook-Libin, S.; Sykes, E.M.E.; Kornelsen, V.; Kumar, A. Iron Acquisition Mechanisms and Their Role in the Virulence of Acinetobacter baumannii. 2022; 90, e00223-00222. [Google Scholar] [CrossRef]
- WHO Bacterial Priority Pathogens List, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. Geneva: World Health Organization; 2024. Licence: CC BY-NC-SA 3.0 IGO.
- Yang, N.; Jin, X.; Zhu, C.; Gao, F.; Weng, Z.; Du, X.; Feng, G. Subunit vaccines for Acinetobacter baumannii. 2023, 13. [Google Scholar] [CrossRef]
- Olawade, D.B.; Teke, J.; Fapohunda, O.; Weerasinghe, K.; Usman, S.O.; Ige, A.O.; Clement David-Olawade, A. Leveraging artificial intelligence in vaccine development: A narrative review. Journal of Microbiological Methods 2024, 224, 106998. [Google Scholar] [CrossRef]
- Shawan, M.; Sharma, A.R.; Halder, S.K.; Arian, T.A.; Shuvo, M.N.; Sarker, S.R.; Hasan, M.A. Advances in Computational and Bioinformatics Tools and Databases for Designing and Developing a Multi-Epitope-Based Peptide Vaccine. International journal of peptide research and therapeutics 2023, 29, 60. [Google Scholar] [CrossRef]
- Khalid, K.; Poh, C.L. The Promising Potential of Reverse Vaccinology-Based Next-Generation Vaccine Development over Conventional Vaccines against Antibiotic-Resistant Bacteria. 2023, 11, 1264. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, F.; Zhou, Z.; Ma, S.; Zhang, P.; Tan, C.; Luo, Y.; Qin, R.; Chen, J.; Pan, P. A novel mRNA multi-epitope vaccine of Acinetobacter baumannii based on multi-target protein design in immunoinformatic approach. BMC Genomics 2024, 25, 791. [Google Scholar] [CrossRef]
- Mukhopadhyay, H.; Bairagi, A.; Mukherjee, A.; Prasad, A.K.; Roy, A.D.; Nayak, A. Multidrug resistant Acinetobacter baumannii: A study on its pathogenesis and therapeutics. Current Research in Microbial Sciences 2025, 8, 100331. [Google Scholar] [CrossRef]
- Ahmad, T.A.; Tawfik, D.M.; Sheweita, S.A.; Haroun, M.; El-Sayed, L.H. Development of immunization trials against Acinetobacter baumannii. Trials in Vaccinology 2016, 5, 53–60. [Google Scholar] [CrossRef]
- Nie, D.; Hu, Y.; Chen, Z.; Li, M.; Hou, Z.; Luo, X.; Mao, X.; Xue, X. Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. Journal of Biomedical Science 2020, 27, 26. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Lin, L.; Ibrahim, A.S.; Baquir, B.; Pantapalangkoor, P.; Bonomo, R.A.; Doi, Y.; Adams, M.D.; Russo, T.A.; Spellberg, B. Active and passive immunization protects against lethal, extreme drug resistant-Acinetobacter baumannii infection. PloS one 2012, 7, e29446. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Capalash, N.; Sharma, P. Vaccine development to control the rising scourge of antibiotic-resistant Acinetobacter baumannii: a systematic review. 3 Biotech 2022, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Bravi, B. Development and use of machine learning algorithms in vaccine target selection. npj Vaccines 2024, 9, 15. [Google Scholar] [CrossRef]
- Dey, J.; Mahapatra, S.R.; Singh, P.K.; Prabhuswamimath, S.C.; Misra, N.; Suar, M. Designing of multi-epitope peptide vaccine against Acinetobacter baumannii through combined immunoinformatics and protein interaction–based approaches. Immunologic Research 2023, 71, 639–662. [Google Scholar] [CrossRef]
- Monterrubio-López, G. P. Monterrubio-López, G. P., González, Y. M. J. A. & Ribas-Aparicio, R. M. Identification of Novel Potential Vaccine Candidates against Tuberculosis Based on Reverse Vaccinology. BioMed research international 2015, 483150. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Vir, P.; Raghava, G.P.S. Designing of interferon-gamma inducing MHC class-II binders. Biology Direct 2013, 8, 30. [Google Scholar] [CrossRef]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic acids research 2016, 44, W449–454. [Google Scholar] [CrossRef]
- Wodak, S.J.; Vajda, S.; Lensink, M.F.; Kozakov, D.; Bates, P.A. Critical Assessment of Methods for Predicting the 3D Structure of Proteins and Protein Complexes. Annual review of biophysics 2023, 52, 183–206. [Google Scholar] [CrossRef]
- Colovos, C.; Yeates, T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein science : a publication of the Protein Society 1993, 2, 1511–1519. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Gasteiger, E. et al. in The Proteomics Protocols Handbook (ed John M. Walker) 571-607 (Humana Press, 2005).
- Ciemny, M. P., Kurcinski, M., Kozak, K. J., Kolinski, A. & Kmiecik, S. in Modeling Peptide-Protein Interactions: Methods and Protocols (eds Ora Schueler-Furman & Nir London) 69-94 (Springer New York, 2017).
- Rafiei Delfan, R.; Fekrirad, Z.; Jalali Nadoushan, M.; Rasooli, I. Adherence and cytotoxicity of Acinetobacter baumannii on human cervical carcinoma epithelial cells: Exploring the role of anti-OmpA antibodies. Medicine in Microecology 2024, 22, 100113. [Google Scholar] [CrossRef]
- Rainard, P.; Répérant-Ferter, M.; Gitton, C.; Gilbert, F.B.; Germon, P. Cellular and humoral immune response to recombinant Escherichia coli OmpA in cows. PloS one 2017, 12, e0187369. [Google Scholar] [CrossRef]
- Heidarinia, H.; Tajbakhsh, E.; Rostamian, M.; Momtaz, H. Epitope mapping of Acinetobacter baumannii outer membrane protein W (OmpW) and laboratory study of an OmpW-derivative peptide. Heliyon 2023, 9, e18614. [Google Scholar] [CrossRef]
- Goetz, K.B.; Pfleiderer, M.; Schneider, C.K. First-in-human clinical trials with vaccines--what regulators want. Nature biotechnology 2010, 28, 910–916. [Google Scholar] [CrossRef]
- Jeffreys, S.; Chambers, J.P.; Yu, J.J.; Hung, C.Y.; Forsthuber, T.; Arulanandam, B.P. Insights into Acinetobacter baumannii protective immunity. Frontiers in immunology 2022, 13, 1070424. [Google Scholar] [CrossRef]
- Kang, M.J.; Jang, A.R.; Park, J.Y.; Ahn, J.H.; Lee, T.S.; Kim, D.Y.; Lee, M.S.; Hwang, S.; Jeong, Y.J.; Park, J.H. IL-10 Protects Mice From the Lung Infection of Acinetobacter baumannii and Contributes to Bacterial Clearance by Regulating STAT3-Mediated MARCO Expression in Macrophages. Frontiers in immunology 2020, 11, 270. [Google Scholar] [CrossRef]
- ud-din, M.; Albutti, A.; Ullah, A.; Ismail, S.; Ahmad, S.; Naz, A.; Khurram, M.; Haq, M.u.; Afsheen, Z.; Bakri, Y.E.; et al. Vaccinomics to Design a Multi-Epitopes Vaccine for Acinetobacter baumannii. 2022; 19, 5568. [Google Scholar]
- Maiti, B.; Dubey, S.; Munang'andu, H.M.; Karunasagar, I.; Karunasagar, I.; Evensen, Ø. Application of Outer Membrane Protein-Based Vaccines Against Major Bacterial Fish Pathogens in India. Frontiers in immunology 2020, 11, 1362. [Google Scholar] [CrossRef] [PubMed]
- Gorai, S.; Das, N.C.; Gupta, P.S.S.; Panda, S.K.; Rana, M.K.; Mukherjee, S. Designing efficient multi-epitope peptide-based vaccine by targeting the antioxidant thioredoxin of bancroftian filarial parasite. Infection, Genetics and Evolution 2022, 98, 105237. [Google Scholar] [CrossRef] [PubMed]
- Girija, A.S.S.; Gunasekaran, S.; Habib, S.; Aljeldah, M.; Al Shammari, B.R.; Alshehri, A.A.; Alwashmi, A.S.S.; Turkistani, S.A.; Alawfi, A.; Alshengeti, A.; et al. Prediction of Putative Epitope Peptides against BaeR Associated with TCS Adaptation in Acinetobacter baumannii Using an In Silico Approach. Medicina (Kaunas, Lithuania) 2023, 59. [Google Scholar] [CrossRef]
- Ma, S.; Zhu, F.; Zhang, P.; Xu, Y.; Zhou, Z.; Yang, H.; Tan, C.; Chen, J.; Pan, P. Development of a novel multi-epitope subunit mRNA vaccine candidate to combat Acinetobacter baumannii. Scientific Reports 2025, 15, 1410. [Google Scholar] [CrossRef] [PubMed]
- Mirali, M.; Jahangiri, A.; Jalali Nadoushan, M.; Rasooli, I. A two-protein cocktail elicits a protective immune response against Acinetobacter baumannii in a murine infection model. Microbial Pathogenesis 2023, 182, 106262. [Google Scholar] [CrossRef]
- Raoufi, Z.; Abdollahi, S.; Armand, R. DcaP porin and its epitope-based subunit promise effective vaccines against Acinetobacter baumannii; in-silico and in-vivo approaches. Microbial Pathogenesis 2022, 162, 105346. [Google Scholar] [CrossRef]
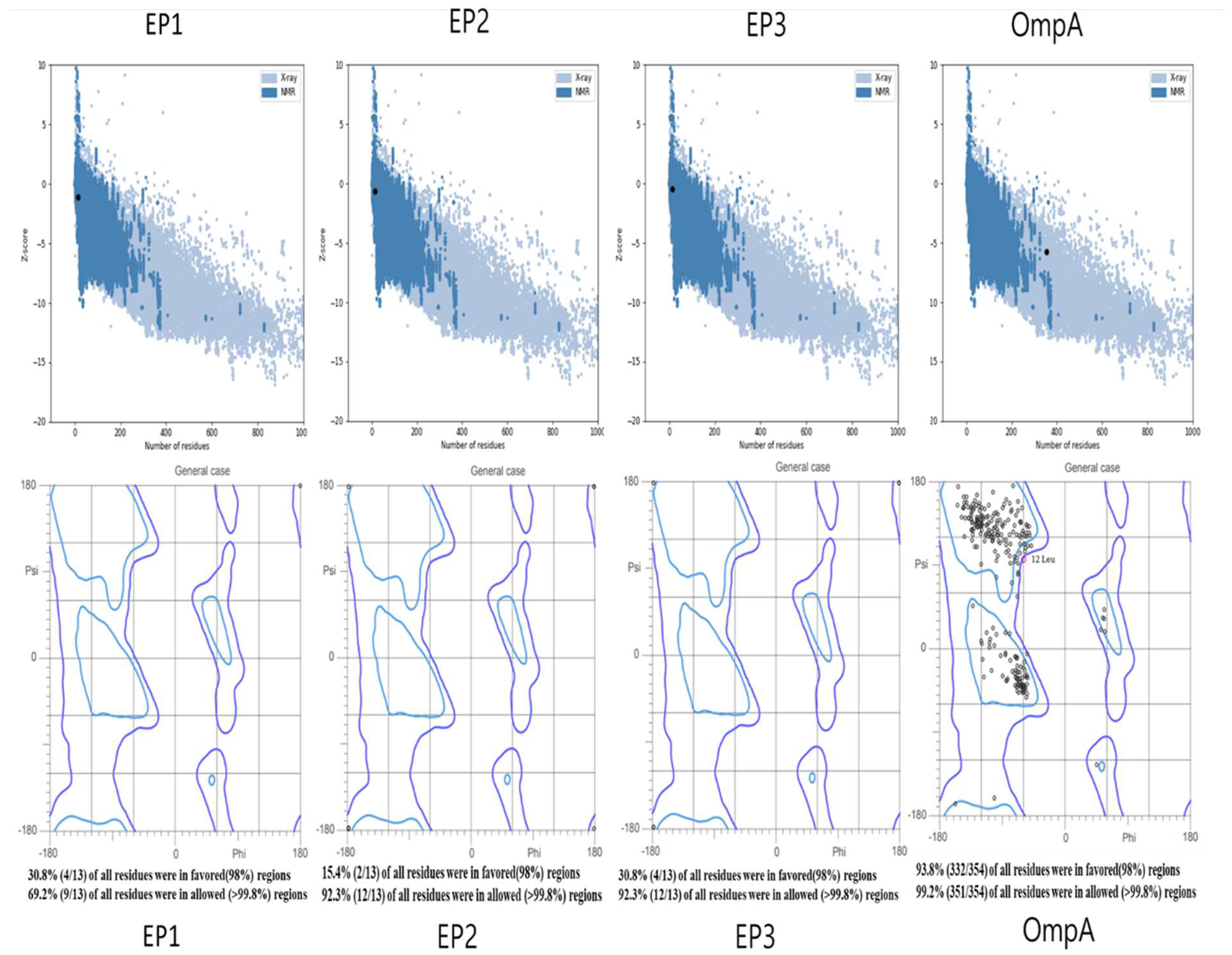
| Epitope | Core peptide | HLA allele | IEDB percentile score | SYFPEITHI | IFN-γ | IL-4 | IL-10 | Toxicity | Allergenicity |
| VVEVAPVEPTPVAPQ | VAPVEPTPV | DRB1*07:01 | 0.96 | 16 | Yes | NO | No | No | No |
| GIELTPWLGFEAEYN | LTPWLGFEA | DRB1*15:01 | 0.59 | 34 | Yes | Yes | No | No | No |
| EPTPVAPQPQELTED | PVAPQPQEL | DRB4*01:01 | 0.85 | N/A | Yes | No | NO | No | No |
| Peptide name | Vaxijen score | Antigen | Molecular weight (g/mol) | Half-life | Hydrophobicity | Net charge at pH 7 | Water solubility | PI | GRAVY | Instability index |
| EP1 | 0.7548 | Yes | 1531.77 | 100 hours (mammalian reticulocytes, in vitro) >20 hours (yeast, in vivo) >10 hours (Escherichia coli, in vivo) | 73.33% | -2 | poor | 3.79 | 0.467 | 86.37 (Unstable) |
| EP2 | 0.9127 | Yes | 1738.91 | 30 hours (mammalian reticulocytes, in vitro) >20 hours (yeast, in vivo) >10 hours (Escherichia coli, in vivo) | 46.67% | -3 | poor | 3.67 | -173 | 39.27 (Stable) |
| EP3 | 0.5131 | Yes | 1650.76 | 1 hour (mammalian reticulocytes, in vitro) 30 min (yeast, in vivo) >10 hours (Escherichia coli, in vivo) | 46.67% | -4 | good | 3.5 | -1.267 | 124.89 (Unstable) |
| Epitope | Cluster density | Average RMSD | Max. RMSD | Number of elements |
| EP1 | 38.9179 | 2.69799 | 13.097 | 105 |
| EP2 | 39.1403 | 2.55491 | 11.7363 | 100 |
| EP3 | 48.824 | 2.04817 | 6.8723 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





