1. Introduction
Avian reovirus remains relevant in the global poultry industry [
1], due to significant economic losses associated with ARV-induced tenosynovitis/arthritis and malabsorption [
2]. The double stranded RNA genome of ARV consists of 10 segments, named for their apparent electrophoretic mobility as large (L1-L3), medium (M1-M3), and small (S1-S4), each encoding various structural and non-structural proteins [
3]. Like many RNA viruses, ARVs expand their genetic diversity through random point mutations and genetic reassortment, with a mean molecular mutation rate of 2.3 × 10
-3 substitutions/site/year as evaluated from 1991 to 2016 [
4] . Such an expansion of genetic diversity gives rise to divergence between field and reference strains. To explore these differences, researchers have utilized both Sanger [
5,
6] and whole genome sequencing (WGS) approaches [
4,
7,
8]. Various distinct genotypic clusters have been identified based on S1 gene sequences encoding the σC protein, with shifts in cluster representation observed over time [
1,
5,
9,
10]. The σC protein remains important for type-specific neutralizing antibodies [
11,
12] and the potential of σC as a promising immunogen with various vaccine technologies has been indicated [
13,
14,
15,
16]. While a correlation between genotype and pathotypes or serotypes has not been clearly established [
17], the sequencing of the σC protein remains important due to its implications in neutralizing antibody production following vaccination [
6,
18,
19].
A significant proportion of clinical ARV isolates are revealed to be coinfections with two or more ARV genotypes [
20]. For experimental purposes, the conventional practice involves two [
21,
22,
23] or three [
24,
25] rounds of plaque purification to obtain a clonal populations of ARV. However, the effectiveness of the approach has not been determined, especially utilizing the advanced sequencing approaches. The objective of the study was to determine purity of a plaque-purified ARV isolate with Sanger and next generation sequencing approaches.
2. Materials and Methods
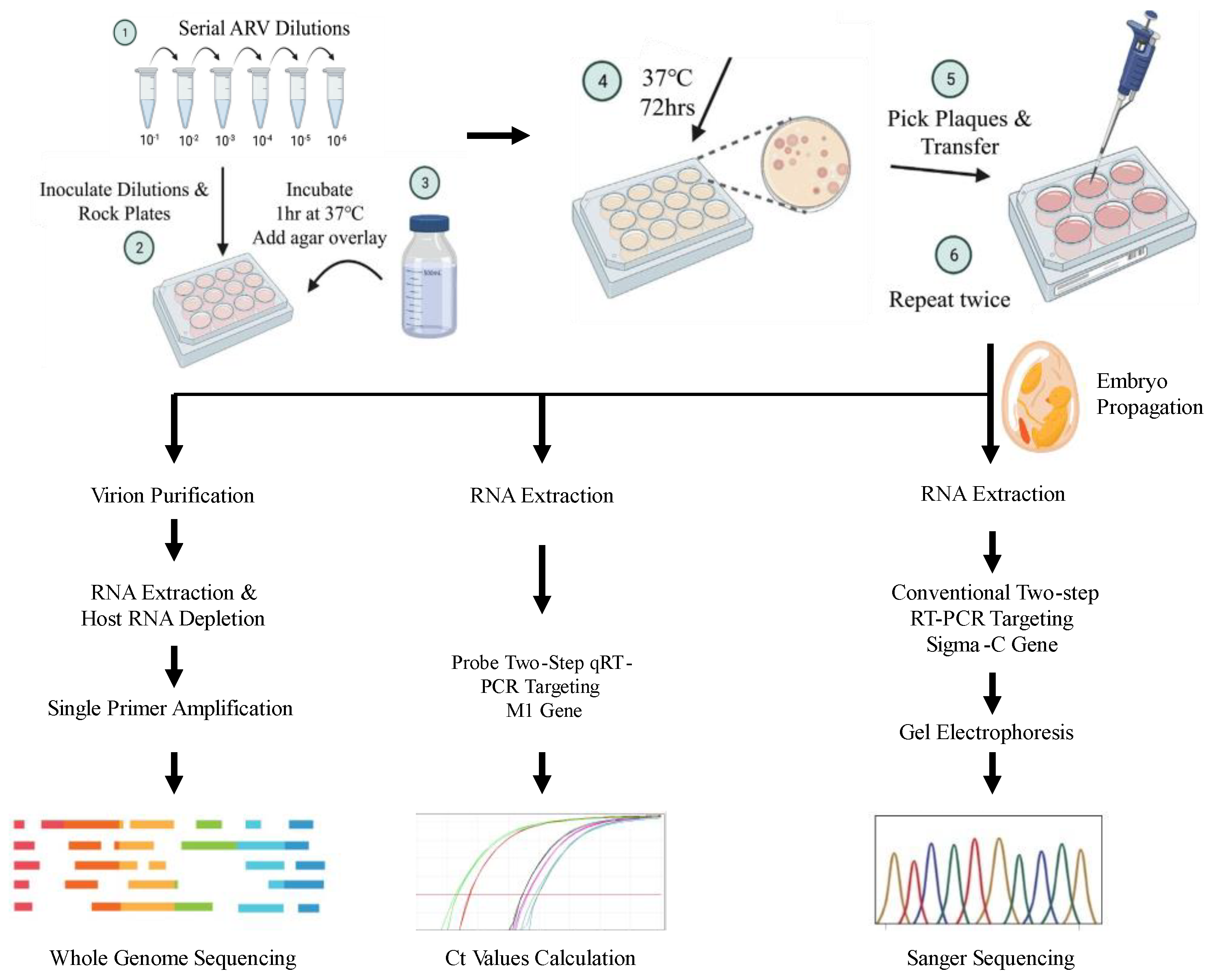
A brief overview of the workflow is shown in
Figure 1.
Clinical History of AL Isolate
The AL isolate of ARV was isolated from 4-weeks-old broiler chickens submitted to Thompson-Bishop State Diagnostic Lab, Auburn, Alabama, where initial testing was performed as follows. Upon histopathological examination, multifocal, lymphocytic myocarditis, with formation of lymphoid nodules, was observed in the heart. Using homogenates of cardiac tissue samples, the virus isolation was performed in chicken embryo kidney cell culture. The presence of ARV antigens was confirmed by direct immunofluorescence assay using fluorescent antibody (FA) conjugate (reagent code: 680-ADV) purchased from National Veterinary Services Laboratories (Ames, IA).
Chicken Embryo Liver Cell Culture
Chicken embryo liver (CELi) cell culture was prepared from 14-days-old specific-pathogen-free (SPF) chicken embryos (AVS Bio, Norwich, CT). Briefly, liver tissues were collected from embryos in PBS, minced with scissors, and homogenized in 0.05 % Trypsin-EDTA solution (Gibco, Grand Island, New York). The homogenate was filtered with a 40-micron filter, centrifuged at 100 × g for 10 minutes, and resuspended in 50 mL of growth medium containing Dulbecco’s Modified Eagle Medium (DMEM) (Corning, location) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT) and 2% Penicillin-Streptomycin with L-glutamine (Corning, Corning, NY) and 3% sodium pyruvate (Corning, New York, NY). The cells were maintained at 37°C in a humidified incubator with 5% CO₂. To prepare maintenance medium, the same ingredients were used at the concentrations as described above except for FBS, for which the concentration was reduced to 2% in total DMEM solution.
Plaque Purification of AL Isolate
For plaque purification of AL isolate, a modified version of previously described method was utilized for each round [
24]. CELi cell culture was prepared as described above and 500 μL of 1×10
5 cells/mL in growth medium were added to each well of 24 well plates. Upon monolayer formation, growth medium was aspirated, and the adherent cells were washed twice with 250 μL sterile phosphate-buffered saline (PBS) at pH 7.4. 10-fold dilutions of the AL isolate stock were prepared in DMEM and 150 μL of each dilution was gently dispensed onto the monolayer. The plates were placed back into the incubator at 37°C and 5% CO
2 to allow for the adsorption of viral particles. After one hour, the inoculation medium was aspirated and 500 μL of overlay medium maintained at 37°C, containing 1:5 ratio of 4% Sea Plaque agarose (Lonza, Morristown, NJ) and maintenance medium respectively, was pipetted onto the monolayer.
Preparation of Viral Stocks in Chicken Embryos
A previously maintained stock of ARV strain S1133 with unknown titer was obtained from the strain collection of Auburn University’s Department of Poultry Science. 100 μL of both S1133 and AL isolate were inoculated into the yolk sac of each of the ten specific pathogen-free (SPF) embryos (Charles River, Wilmington, MA) at seven days of embryonation (DOE). The embryos were each homogenized at 11 DOE with 2 mL of phosphate-buffered saline (PBS) at pH 7.4, pooled, freeze-thawed two times, and centrifuged for 20 minutes at 15000 × g. The supernatant was pipetted and stored as viral stock which was used for the following steps.
Viral RNA Detection and Quantification
Total RNA was extracted from both AL and S1133 stocks using the RNeasy Mini Kit (Qiagen, Hilden, Germany) as per manufacturer’s instructions. For denaturation of viral dsRNA, 5 µL of total RNA was subjected to denaturation at 95°C for 10 minutes. Complementary DNA (cDNA) was synthesized from 2 µL the denatured RNA using LunaScript RT SuperMix Kit (New England Biolabs, Ipswich, MA) as per manufacturer’s instructions. Thermal cycling was conducted as per manufacturer’s instructions for up to 45 cycles using the qTOWER³ PCR Thermal Cycler (Analytik Jena, Jena, Germany).
For detection of σC gene, two unique primer sets were used. A sequence of around 1089 bp of S1 segment spanning roughly from 533 to 1621 bp positions, was targeted using forward and reverse primers 5’-AGTATTTGTGAGTACGATTG-3’ and 5’-GGCGCCACACCTTAGGT-3’, described as P1 and P4, respectively [
5]. Since this primer set did not amplify the σC gene of AL isolate, forward 5’-AGTATTTGTGAATACGACTG-3’ and reverse 5’-GCGAGATGACGTGACACACT-3’ primers were designed de novo based on WGS results for AL isolate. The amplicon spanned over 959 bp, starting from 535 bp to 1493 bp position of the S1 genomic segment. The amplicon sequences for both S1133 and AL isolate have been attached (Supplementary file S1).
For detection of σC gene of S1133 or AL isolate, master mixes were prepared using AccuStart II PCR SuperMix (Quantabio, Beverly, MA) and thermal cycling was performed as per manufacturer’s instructions. Briefly, denaturation was performed at 94ºC for 15 seconds, followed by annealing at 60ºC for 30 seconds. The extension was carried out at 72ºC for 1 minute, enabling DNA polymerase to synthesize the complementary strand efficiently. To confirm successful amplification of targeted segments and ascertain the length of PCR amplicons, agarose gel electrophoresis was performed. The bands were visualized using Gel Doc XR+ System (Bio-Rad, Hercules, CA) and Image Lab software (Bio-Rad, Hercules, CA) version 5.2.1.
The quantification of ARV M1 gene was performed using Forget-Me-Not™ Universal Probe qPCR Master Mix (Biotium, Fremont, CA). The following oligos were utilized for forward and reverse primers and probes respectively: 5’-ATGGCCTMTCTAGCCACACCTG; CAACGARATRGCATCAATAGTAC-3’; 5’-FAM-TGCTAGGAGTCGGTTCTCGTA-BHQ1-3’ [
26]. Thermal cycling was performed as per manufacturer’s instructions. Briefly, one cycle of enzyme activation at 95°C for 2 minutes and 40 cycles of denaturation (95°C for 15 seconds) and combined annealing and extension (60°C for 60 seconds) was performed.
Sanger Sequencing and Data Analysis
The σC genes of both AL isolate and S1133 were amplified using the respective primers and PCR conditions as described above for viral RNA detection. To retrieve amplicons of required length, agarose with low melting point (Promega, Madison, WI) was used for preparing 1.5% gel in TBE. Fifteen microliter of the PCR products were loaded into the wells and electrophoresis was performed at 80 volts for an hour. The bands were UV-illuminated using Gel Doc XR+ System (Bio-Rad, Hercules, CA) and amplicon lengths were analyzed with Image Lab software (Bio-Rad, Hercules, CA) version 5.2.1. The bands corresponding to expected product lengths as described above were excised using a scalpel and melted at 37°C for 10 mins. The DNA was then extracted from the melted gel excisions using QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) following manufacturer’s protocol. The DNA was analyzed with NanoDrop spectrophotometer (Thermo Fischer Scientific, Waltham, MA) and 260/280 nm absorption ratio was determined for each sample. The DNA was then submitted to Center for Computational and Integrative Biology DNA Core, Massachusetts General Hospital for Sanger sequencing.
The raw forward and reverse reads were imported into Geneious Prime version 2025.0.2 (
https://www.geneious.com). The chromatograms were visualized, and the terminal low-quality bases were trimmed to obtain unambiguous overlapping assembly. These sequences were then mapped to the S1 segment of reference S1133 strain (GenBank accession: KF741762.1) using MUSCLE alignment algorithm [
27] version 5.1. Consensus sequences for each of the ARVs S1133 and AL isolate were generated from the aligned forward and reverse sequences. The results were compared with WGS alignment (see below). To explore the phenotypic differences, the σC protein sequence for the AL isolate was predicted based on nucleotide sequences and mapped against the reference (UniProt accession: C0M031_9REOV).
For the phylogenetic analysis, σC sequences representing various genetic clusters (GC) were retrieved from GenBank based on previous classification [
9], aligned using MUSCLE [
27] version 5.1. The phylogenetic tree was constructed using the Neighbor-Joining method [
28] and Tamura-Nei model [
29] in Geneious Prime software version 2025.0.2.
A brief overview of the workflow for the methods tested is shown in
Figure 1.
Whole Genome Sequencing: Library Preparation
ARV genome enrichment, genomic library preparation and Illumina whole-genome sequencing were performed at the U.S. National Poultry Research Center, Athens, Georgia, US, as previously described [
7,
30]. First, virions were purified from cell lysates with Capto Core 700 resin (Cytiva, Buckinghamshire, United Kingdom), and RNA was extracted using MagMAX™ Viral RNA Isolation Kit (Applied Biosystems, Foster City, California). Leftover chicken RNA was depleted from samples using custom ssDNA probes [
31], RNase H (New England Biolabs, Ipswich, MA) and DNase I (New England Biolabs, Ipswich, MA). Chicken-depleted ARV RNA was purified using RNA Clean XP beads (Beckman Coulter, Brea, CA), before being converted to cDNA and PCR-amplified using the ARV single primer amplification (R-SPA) approach. Genomic libraries were generated with the Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA) and IDT for Illumina DNA/RNA UD Indexes (Illumina, San Diego, CA). Short-read sequencing was performed on an Illumina MiSeq instrument (Illumina, San Diego, CA) using a MiSeq Reagent Nano Kit v2 500 cycles cartridge (Illumina San Diego, CA).
Whole Genome Sequencing: Data Analysis
The quality of paired-end raw FASTQ files was evaluated using FASTQC [
32] version 0.12.0. Before downstream processing, adapters and low-quality bases were trimmed using Trimmomatic [
33] version 0.39. Specifically, reads having less than a minimum Phred score threshold of 33 and those shorter than 36 bases were removed as part of the trimming procedure. Trimmed, high-quality paired-end reads were assembled de novo using SPAdes program [
34] version 3.15.5. Assemblies were generated using default parameters for read error correction, graph construction, and scaffolding. To evaluate the de novo assembly quality, the trimmed reads were mapped back to the assembled contigs using the BWA-MEM software [
35] version 0.7.17. Summary statistics were obtained with SAMtools [
36] version 1.19.2 to quantify assembly completeness and contiguity, guiding decisions for downstream consensus generation. Resulting alignments in SAM format were converted, sorted, and indexed in the more compact BAM format using SAMtools to visualize alignments in the Integrative Genome Viewer [
37]. The visualize and verify the de novo assembly graphs, Bandage [
38] was utilized. The contigs for each isolate were then imported into Geneious Prime software version 2025.0.2. The assembled contigs were aligned against the reference genome S1133 (GenBank accession: KF741762.1).
3. Results
qPCR Targeting ARV M1 Gene Quantified RNA of Both AL and S1133 Isolates
The qPCR of the viral stocks grown in chicken embryos targeting a conserved ARV M1 gene yielded amplicons for both AL and S1133 isolates with threshold cycle (Ct) values of 23.47 and 20.42, respectively.
PCR Using Kant Primer Amplified S1133 and Sanger Sequencing Confirmed Identity with S1133
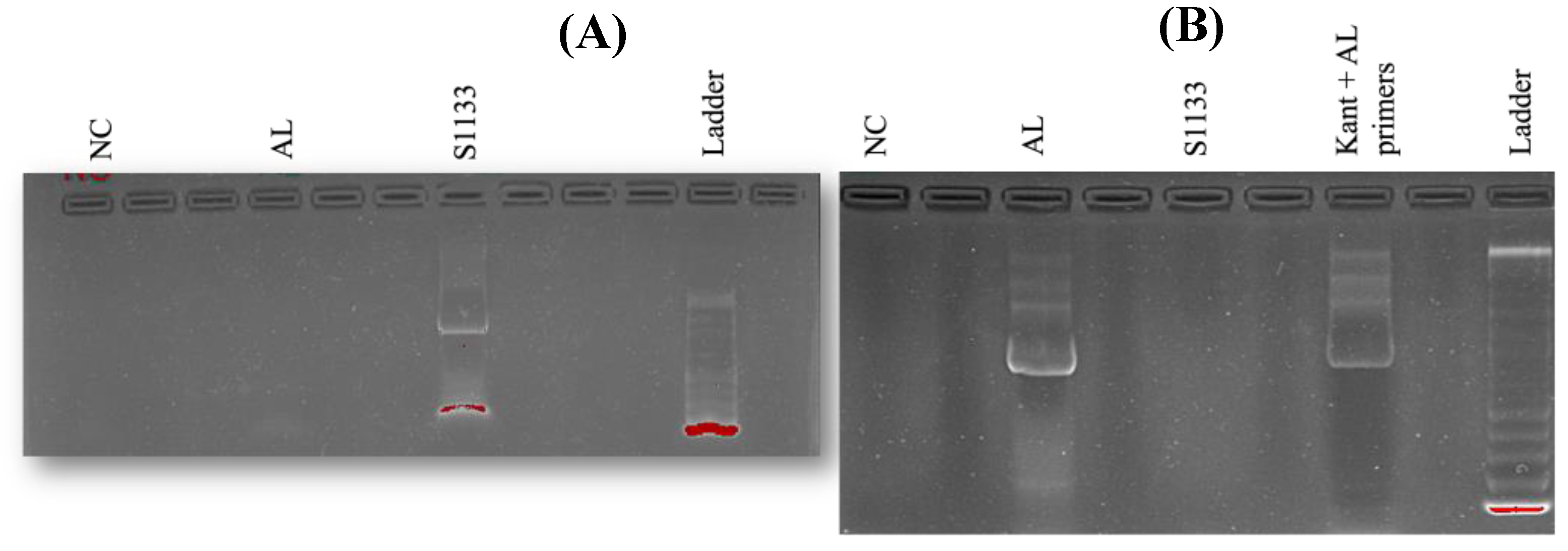
The conventional PCR targeting σC gene with Kant et al. primers yielded amplicons only for ARV S1133 and not for AL isolate, as visualized by gel electrophoresis (
Figure 2-A). The raw chromatograms of forward and reverse reads were visualized, and ambiguous bases were removed. The read length reduced to 563 bases for the reverse and 750 bases for the forward reads.
The consensus sequences obtained by Sanger sequencing were 976 bases in length for S1133. σC sequences S1133-isolate obtained by Sanger sequencing had pairwise identities of 99.6%, when aligned against the σC region of reference S1133 from GenBank. Across the length of gene, ARV S1133 had 8 mutations compared to the reference, most of which were synonymous. An overview of the results is shown in
Table 1.
Whole Genome Sequencing: Read Quality
Following de novo assembly of the trimmed reads, a total of 35 scaffolds were analyzed, ranging from 213 bases to 3895 bp in length. The assembly had an N50 value (the length of the shortest contig or scaffold at which 50% of the total assembled genome length is contained in contigs of equal or greater length) of 1619 bp with the lower, median and upper quartile nodes were around 225 bp, 443 bp and 1217 bp, respectively. The longest node was found to comprise 3895 bp.
WGS: Contigs Similar and Divergent from the Reference S1133 Were Found
Upon alignment of the contigs to reference S1133, divergent contigs (<90% pairwise identity to the reference) as well as similar (95% or greater pairwise identity) were found for most of the segments obtained from presumptively plaque-purified AL isolate. An example of presence of contigs similar to and divergent from reference S1133 is shown in (
Figure 3).
As shown in
Table 2, up to 3 contigs of AL isolate varying in pairwise identity to the reference were identified. The most striking difference among the contigs was observed for S1 segment with contig 1 and 3 having 55.8% and 93.3% similarity to the reference S1133. The other segments had either only one contigs or multiple contigs but with little divergence.
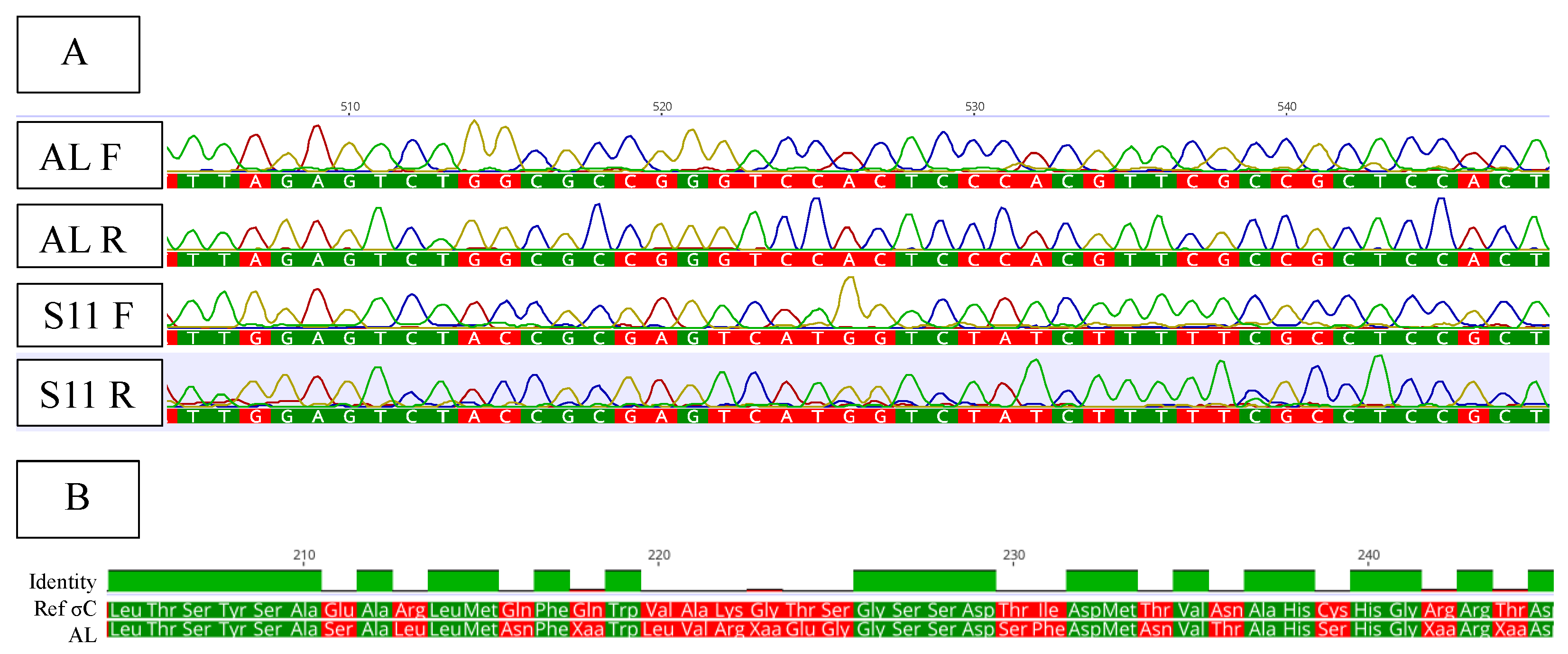
On the other hand, σC sequences of AL- and S1133-isolates obtained by Sanger sequencing had pairwise identities of 99.6% and 54.5% respectively, when aligned against the σC region of reference S1133 from GenBank. An analysis of translation of the σC nucleotide sequences from AL isolate upon alignment with reference σC protein suggested a drastically different amino acid composition (
Figure 4-B), with a pairwise identity of ~44% with the reference.
Primers Designed Based on WGS Amplified σC Gene of AL Isolate
The read length reduced from around 900 bases to around 750 bases post-trimming for each of forward and reverse reads of AL isolate. The consensus sequences obtained by de novo assembly were 855 in length for AL isolate. The pairwise identity of σC gene obtained by Sanger sequencing of AL isolate and corresponding σC sequence obtained by WGS was 99.1%.
Phylogenetic Analysis Confirmed a Mixture of Divergent Sequences
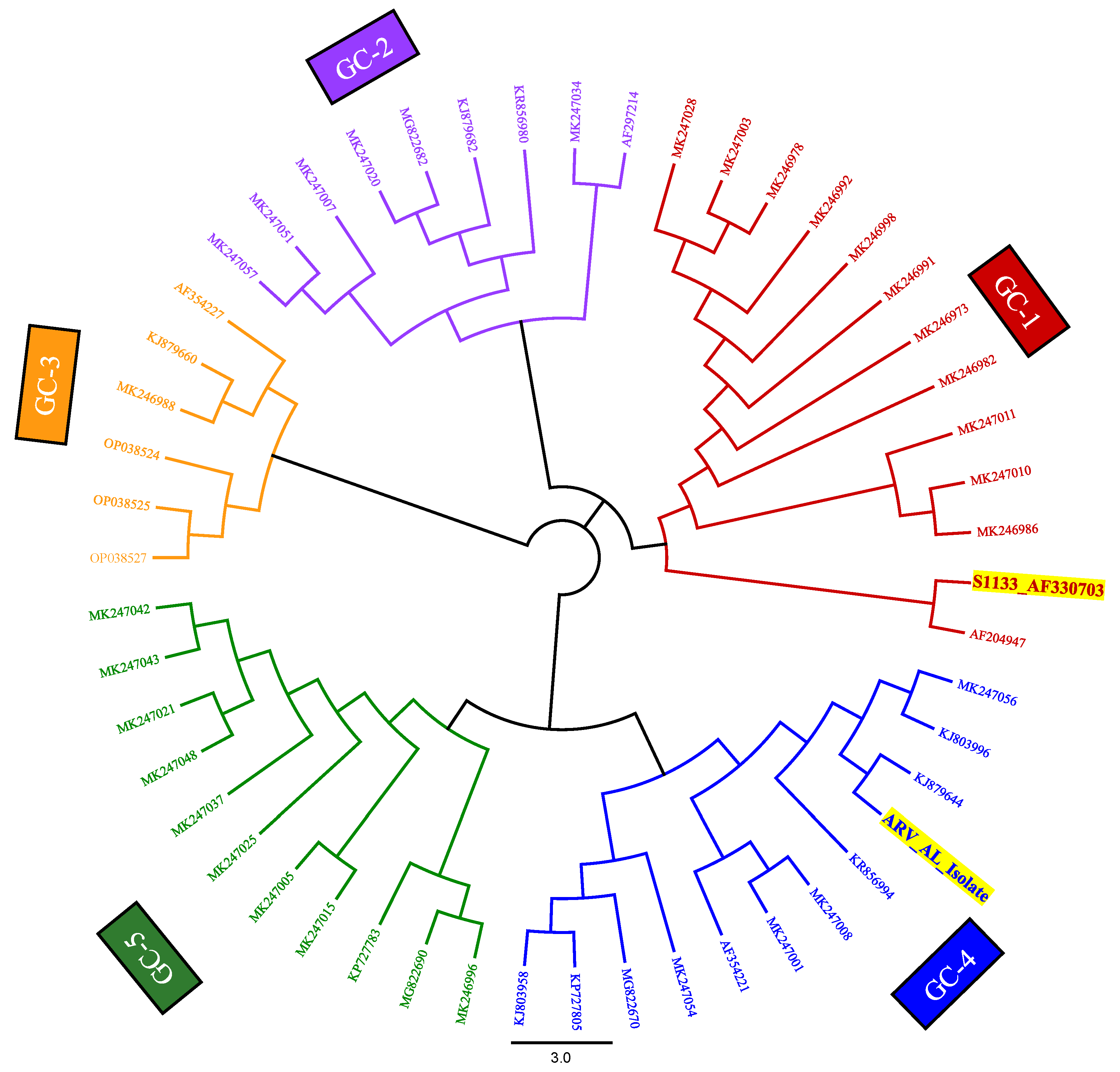
The phylogenetic analysis revealed that the AL isolate-like sequences clustered in GC4 while S1133 belonged to GC1 which contained other vaccine strains as well as pathogenic ARVs (
Figure 5).
4. Discussion
The study aimed to determine the effectiveness of plaque purification to obtain clonal population of ARV. Multiple rounds of plaque purification can theoretically yield clonal viral populations of a virus of interest or help remove contamination by unwanted agents, as demonstrated for influenza viruses [
39]. Two [
21,
22,
40,
41] or three [
24,
25] rounds of plaque purification have been employed to obtain clonal viral populations from ARV field samples. However, unsuccessful plaque purification of ARV has been previously suggested based on electrophoretograms analyses of various gene segments [
23,
42]. In the current study, the combination of
de novo assembly of full-length sequences and alignment of the contigs to the reference genome allowed for a high-resolution analysis of viral homogeneity. We performed three rounds of plaque purification on CELi cells and demonstrated the presence of two divergent types of contigs in the genome assembly of the AL isolate. This demonstrates a failure of the approach to obtain pure sequences. A possibility of coinfection of a monolayer by multiple ARV particles in larger plaque sizes has been previously speculated [
40]. Moreover, such an observation has been made for polioviruses even at low multiplicity of infection, wherein chimeric plaques containing more than one parental viruses was reported due to aggregation of the particles [
43]. One potential explanation for the failure to obtain clonal population of AL isolate could lie in the three-dimensional nature of CELi cells used for the purpose. Such a cell culture morphology could contribute to the problem of isolating divergent subpopulations from different layers of the culture in a single plaque. Therefore, the possibility of successful ARV purification with other cell types cannot be ruled out.
Before the extensive utilization of WGS, several indicators including plaque morphology [
44], size and diameter of the viral particles [
45], tissue tropism [
44] and restriction fragment length polymorphisms [
23,
46] have been utilized to differentiate various ARV strains. To organize these strains, ARVs have been classified into various genotypes using σC gene sequences due to its antigenic relevance and variability being a surface protein [
5,
9,
47,
48]. The primers described previously [
5] are widely utilized to target σC gene for detection of ARV or Sanger sequencing. Since the gene is highly variable, there remains a possibility of false negative PCR outcomes because of the inability of primers to bind to the significantly altered σC sequences. A similar observation was made in the present study where the σC gene of the ARV isolate AL did not amplify with the conventionally utilized primers. Upon WGS, several mismatches preventing the binding of the reverse primer were found. Therefore,
de novo primers exclusively binding to σC gene of the AL isolate had to be designed based on the WGS results, which yielded amplicons for Sanger sequencing. It remains ambiguous as to why the primers by Kant et al. [
5] unexpectedly failed to amplify the S1133-like sequences in the AL isolate. One possible explanation could be an insufficient sensitivity of the PCR as the length of the amplicon decreases the sensitivity of the PCR and since the sequences similar to S1133 were less abundant in AL isolate (results not shown), the amplification of S1133-like sequences might have been too little to be detected by electrophoresis. Collectively, these findings highlight the shortcomings associated with targeting variable genomic regions for characterization of field isolates using a given set of primers.
Upon Sanger sequencing, the observed low (~44%) pairwise amino acid identity of the σC protein between the AL isolate and the S1133 strain suggests drastic divergence from the reference similar to a previous observation on the differences between the American and Australian isolates [
49]. Sequence variations, especially within regions corresponding to immunodominant epitopes, could alter the structural conformation or accessibility of these epitopes, potentially leading to immune evasion. The σC protein is a critical determinant of host immune response, as it mediates receptor binding and the major antigen to which neutralizing antibodies bind [
50]. Most commercial vaccines utilize a σC protein sequence similar to S1133, and such a radical change in protein structure results in a change in the antigenicity index impacting vaccine efficacy [
18,
19].
Phylogenetic analyses of the sequences provide significant insights into genetic relatedness of the circulating field strains with the vaccine strains. Various genes including σC [
1,
4,
5,
51,
52], σNS [
51], p10- and p17-encoding genes [
53] and full-length genes or gene segments [
8,
9,
54] have been utilized to study ARV phylogenetics. A global study categorized over 200 ARV isolates into five major genetic clusters and suggested notable discrepancies in genotyping systems [
1]. In California, six distinct genotypic clusters were observed based on S1 with various degrees of sequence homology between clusters [
9]. In brief, a universally accepted system of genotypic classification is lacking at present and the association of genetic cluster with disease outcomes has not been established [
1,
55]. Based on the classification criteria of these studies, representative sequences of σC gene belonging to various genotypic clusters were retrieved from GenBank and the phylogenetic tree was constructed to classify ARV AL isolate. Owing to the divergence of its σC sequence, the AL isolate was classified in the farthest cluster GC4, away from the reference S1133 which belonged to GC1, a cluster representing commercially available vaccine strains. Presence of GC1 (vaccine-like) as well as GC4 (field-like) sequences after plaque purification could also be suggestive of the potential persistence of vaccine virus in the chickens (from which AL isolate was collected) followed by superinfection with the field virus.
Briefly, our results suggest that three rounds of plaque purification are insufficient to ensure homogeneity of ARV populations. Moreover, the retrieval of full-length divergent contigs highlights the advantage of WGS over traditional methods like PCR and Sanger sequencing for detection and characterization of mixed populations. We recommend that WGS should be utilized to determine the purity of an ARV isolate prior to experimentation.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Supplementary file S1.
Author Contributions
Conceptualization, R.H., Z.K. and S.C.; methodology, Z.K., S.A., T.H. and E.C.; software, Z.K., and S.A.; formal analysis, Z.K., S. A.; data curation, Z.K., R.H. and S.A.; writing—original draft preparation, Z.K.; writing—review and editing, R.H., S.A., S.C.; supervision, R.H. and S.C; funding acquisition, R.H., S.C. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Acknowledgments
We thank Ms. Emily Handley from the State Veterinary Diagnostic Lab at Auburn and Dr. Teresa Dormitorio for technical assistance with SPF embryos. We are grateful to Dr. Haroldo Toro for permissions to use their laboratory for plaque purification and Cassandra Kitchens for logistical assistance.
Conflicts of Interest
“The authors declare no conflicts of interest.”
References
- Kovács, E.; Varga-Kugler, R.; Mató, T.; Homonnay, Z.; Tatár-Kis, T.; Farkas, S.; Kiss, I.; Bányai, K.; Palya, V. Identification of the Main Genetic Clusters of Avian Reoviruses from a Global Strain Collection. Front. Vet. Sci. 2023, 9, 1094761. [Google Scholar] [CrossRef] [PubMed]
- French, D. Incidence and Economic Impact of Reovirus in the Poultry Industries in the United States. Avian Dis. 2022, 66, 432–434. [Google Scholar] [CrossRef] [PubMed]
- Spandidos, D. and; Graham, A.F. Physical and Chemical Characterization of an Avian Reovirus. J. Virol. 1976, 19, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Ayalew, L.E.; Ahmed, K.A.; Mekuria, Z.H.; Lockerbie, B.; Popowich, S.; Tikoo, S.K.; Ojkic, D.; Gomis, S. The Dynamics of Molecular Evolution of Emerging Avian Reoviruses through Accumulation of Point Mutations and Genetic Re-Assortment. Virus Evol. 2020, 6, veaa025. [Google Scholar] [CrossRef]
- Kant, A.; Balk, F.; Born, L.; van Roozelaar, D.; Heijmans, J.; Gielkens, A.; ter Huurne, A. Classification of Dutch and German Avian Reoviruses by Sequencing the Sigma C Protein. Vet. Res. 2003, 34, 203–212. [Google Scholar] [CrossRef]
- Goldenberg, D.; Pasmanik-Chor, M.; Pirak, M.; Kass, N.; Lublin, A.; Yeheskel, A.; Heller, D.; Pitcovski, J. Genetic and Antigenic Characterization of Sigma C Protein from Avian Reovirus. Avian Pathol 2010, 39, 189–199. [Google Scholar] [CrossRef]
- Narvaez, S.A.; Harrell, T.L.; Oluwayinka, O.L.T.D.; Sellers, H.S.; Khalid, Z.; Hauck, R.; Chowdhury, E.U.; Conrad, S.J. Optimizing the Conditions for Whole-Genome Sequencing of Avian Reoviruses. Viruses 2023, 15, 1938. [Google Scholar] [CrossRef]
- Nour, I.; Alvarez-Narvaez, S.; Harrell, T.L.; Conrad, S.J.; Mohanty, S.K. Whole Genomic Constellation of Avian Reovirus Strains Isolated from Broilers with Arthritis in North Carolina, USA. Viruses 2023, 15. [Google Scholar] [CrossRef]
- Egaña-Labrin, S.; Hauck, R.; Figueroa, A.; Stoute, S.; Shivaprasad, H.L.; Crispo, M.; Corsiglia, C.; Zhou, H.; Kern, C.; Crossley, B.; et al. Genotypic Characterization of Emerging Avian Reovirus Genetic Variants in California. Sci. Rep. 2019, 9, 9351. [Google Scholar] [CrossRef]
- Rafique, S.; Rashid, F.; Wei, Y.; Zeng, T.T.; Xie, L.J.; Xie, Z.X. Avian Orthoreoviruses: A Systematic Review of Their Distribution, Dissemination Patterns, and Genotypic Clustering. Viruses 2024, 16, 1056. [Google Scholar] [CrossRef]
- Meanger, J.; Wickramasinghe, R.; Enriquez, C.E.; Robertson, M.D.; Wilcox, G.E. Type-specific Antigenicity of Avian Reoviruses. Avian Pathol. 1995, 24, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, D.; Lublin, A.; Rosenbluth, E.; Heller, E.D.; Pitcovski, J. Differentiating Infected from Vaccinated Animals, and among Virulent Prototypes of Reovirus. J. Virol. Methods 2011, 177, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Lee, L.H.; Shih, W.L.; Hu, Y.C.; Liu, H.J. Baculovirus Surface Display of σC and σB Proteins of Avian Reovirus and Immunogenicity of the Displayed Proteins in a Mouse Model. Vaccine 2008, 26, 6361–6367. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.-H.; Hsu, A.-P.; Shien, J.-H.; Chang, T.-J.; Liao, J.-W.; Chen, J.-R.; Lin, C.-F.; Hsu, W.-L. Avian Reovirus Sigma C Enhances the Mucosal and Systemic Immune Responses Elicited by Antigen-Conjugated Lactic Acid Bacteria. Vaccine 2012, 30, 5019–5029. [Google Scholar] [CrossRef]
- Goldenberg, D.; Lublin, A.; Rosenbluth, E.; Heller, E.D.; Pitcovski, J. Optimized Polypeptide for a Subunit Vaccine against Avian Reovirus. Vaccine 2016, 34, 3178–3183. [Google Scholar] [CrossRef]
- Saikia, D.P.; Yadav, K.; Pathak, D.C.; Ramamurthy, N.; D’Silva, A.L.; Marriappan, A.K.; Ramakrishnan, S.; Vakharia, V.N.; Chellappa, M.M.; Dey, S. Recombinant Newcastle Disease Virus (NDV) Expressing Sigma C Protein of Avian Reovirus (ARV) Protects against Both ARV and NDV in Chickens. Pathogens 2019, 8. [Google Scholar] [CrossRef]
- Clark, F.D.; Ni, Y.; Collisson, E.W.; Kemp, M.C. Characterization of Avian Reovirus Strain-Specific Polymorphisms. Avian Dis. 1990, 34, 304–314. [Google Scholar] [CrossRef]
- Vasserman, Y.; Eliahoo, D.; Hemsani, E.; Kass, N.; Ayali, G.; Pokamunski, S.; Pitcovski, J. The Influence of Reovirus Sigma C Protein Diversity on Vaccination Efficiency. Avian Dis. 2004, 48, 271–278. [Google Scholar] [CrossRef]
- Dawe, W.H.; Kapczynski, D.R.; Linnemann, E.G.; Gauthiersloan, V.R.; Sellers, H.S. Analysis of the Immune Response and Identification of Antibody Epitopes against the Sigma C Protein of Avian Orthoreovirus Following Immunization with Live or Inactivated Vaccines. Avian Dis. 2023, 66, 465–475. [Google Scholar] [CrossRef]
- Egaña-Labrin, S.; Gallardo, R.A. Concomitant Virus Detection on Avian Reovirus Isolates. In Proceedings of the Proceedings of Western Poultry Disease Conference.
- Tran, A.; Berard, A.; Coombs, K.M. Avian Reoviruses: Propagation, Quantification, and Storage. In Current Protocols in Microbiology; Coico, R., McBride, A., Quarles, J.M., Stevenson, B., Taylor, R.K., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; ISBN 978-0-471-72925-9. [Google Scholar]
- Rekik, M.R.; Silim, A.; Elazhary, M.A.S.Y. Characteristics and Analysis of Electropherotypes of Avian Reovirus Field Isolates. Spec. Issue: Adv. Vet. Virol. 1990, 23, 273–281. [Google Scholar] [CrossRef]
- Wu, W.Y.; Shien, J.H.; Lee, L.H.; Shieh, H.K. Analysis of the Double-Stranded RNA Genome Segments among Avian Reovirus Field Isolates. J. Virol. Methods 1994, 48, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Guneratne, J.R.M.; Jones, R.C.; Georgiou, K. Some Observations on the Isolation and Cultivation of Avian Reoviruses. Avian Pathol. 1982, 11, 453–462. [Google Scholar] [CrossRef] [PubMed]
- van Loon, A.A.W.M.; Koopman, H.C.; Kosman, W.; Mumczur, J.; Szeleszczuk, O.; Karpinska, E.; Kosowska, G.; Lütticken, D. Virology: Isolation of a New Serotype of Avian Reovirus Associated with Malabsorption Syndrome in Chickens. Vet. Q 2001, 23, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Lu, H.G. Whole Genome Alignment Based One-Step Real-Time RT-PCR for Universal Detection of Avian Orthoreoviruses of Chicken, Pheasant and Turkey Origins. Infect., Genet. Evol. 2016, 39, 120–126. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinf. 2004, 5, 113. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Alvarez-Narvaez, S.; Harrell, T.L.; Conrad, S.J. Protocol: Optimized Conditions for Whole Genome Sequencing of Avian Reoviruses 2024.
- Parris, D.J.; Kariithi, H.; Suarez, D.L. Non-Target RNA Depletion Strategy to Improve Sensitivity of next-Generation Sequencing for the Detection of RNA Viruses in Poultry. J. Vet. Diagn. Invest. 2022, 34, 638–645. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data 2010.
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive Visualization of de Novo Genome Assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Murata, H.; Macauley, J.; Lewis, A.M.; Peden, K. Plaque Purification as a Method to Mitigate the Risk of Adventitious-Agent Contamination in Influenza Vaccine Virus Seeds. Vaccine 2011, 29, 3155–3161. [Google Scholar] [CrossRef]
- Markis, M. In Vivo and in Vitro Characterization of Virus Isolates Involved in Viral Enteritis/Enteropathy, Also Known as Runting Stunting Syndrome (Rss), in Broiler Chickens. Ph.D., University of Delaware, 2013.
- Shapouri, M.R.S.; Arella, M.; Silim, A. Evidence for the Multimeric Nature and Cell Binding Ability of Avian Reovirus Σ3 Protein. J. Gen. Virol. 1996, 77, 1203–1210. [Google Scholar] [CrossRef]
- Rekik, M.R.; Silim, A.; Bernier, G. Serological and Pathogenic Characterization of Avian Reoviruses Isolated in Quebec. Avian Pathol. 1991, 20, 607–617. [Google Scholar] [CrossRef]
- Aguilera, E.R.; Erickson, A.K.; Jesudhasan, P.R.; Robinson, C.M.; Pfeiffer, J.K. Plaques Formed by Mutagenized Viral Populations Have Elevated Coinfection Frequencies. mBio 2017, 8, e02020–16. [Google Scholar] [CrossRef]
- Ni, Y.W.; Kemp, M.C. A Comparative Study of Avian Reovirus Pathogenicity: Virus Spread and Replication and Induction of Lesions. Avian Dis. 1995, 39, 554–566. [Google Scholar] [CrossRef]
- Deshmukh, D.R.; Dutta, S.K.; Pomeroy, B.S. Avian Reoviruses V. Studies of Ultrastructural Morphology by Electron Microscopy. Avian Dis. 1971, 15, 588–595. [Google Scholar] [CrossRef]
- Gouvea, V.S.; Schnitzer, T.J. Polymorphism of the Migration of Double-Stranded RNA Genome Segments of Avian Reoviruses. J. Virol. 1982, 43, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Masaji Mase; Makiko Gotou; Daisuke Inoue; Tsuneyuki Masuda; Satoko Watanabe; Hiroshi Iseki Genetic Analysis of Avian Reovirus Isolated from Chickens in Japan. Avian Dis. 2021, 65, 346–350. [CrossRef]
- Farnoushi, Y.; Heller, D.; Lublin, A. Genetic Characterization of Newly Emerging Avian Reovirus Variants in Chickens with Viral Arthritis/Tenosynovitis in Israel. Virology 2024, 589, 109908. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-J.; Giambrone, J.J. Amplification, Cloning and Sequencing of the σC-Encoded Gene of Avian Reovirus. J. Virol. Methods 1997, 63, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, R.; Meanger, J.; Enriquez, C.E.; Wilcox, G.E. Avian Reovirus Proteins Associated with Neutralization of Virus Infectivity. Virology 1993, 194, 688–696. [Google Scholar] [CrossRef]
- Kim, S.-W.; Choi, Y.-R.; Park, J.-Y.; Wei, B.; Shang, K.; Zhang, J.-F.; Jang, H.-K.; Cha, S.-Y.; Kang, M. Isolation and Genomic Characterization of Avian Reovirus from Wild Birds in South Korea. Front. Vet. Sci. 2022, 9. [Google Scholar] [CrossRef]
- De Carli, S.; Wolf, J.M.; Gräf, T.; Lehmann, F.K.M.; Fonseca, A.S.K.; Canal, C.W.; Lunge, V.R.; Ikuta, N. Genotypic Characterization and Molecular Evolution of Avian Reovirus in Poultry Flocks from Brazil. Avian Pathol. 2020, 49, 611–620. [Google Scholar] [CrossRef]
- Hsu, H.W.; Su, H.Y.; Huang, P.H.; Lee, L.H.; Liu, H.J. Sequence and Phylogenetic Analysis of P10- and P17-Encoding Genes of Avian Reovirus. Avian Dis. 2005, 49, 36–42. [Google Scholar] [CrossRef]
- Teng, L.; Xie, Z.; Xie, L.; Liu, J.; Pang, Y.; Deng, X.; Xie, Z.; Fan, Q.; Luo, S.; Feng, J.; et al. Sequencing and Phylogenetic Analysis of an Avian Reovirus Genome. Virus Genes 2014, 48, 381–386. [Google Scholar] [CrossRef]
- Egaña-Labrin, S.; Jerry, C.; Roh, H.J.; Silva, A.P. da; Corsiglia, C.; Crossley, B.; Rejmanek, D.; Gallardo, R.A. Avian Reoviruses of the Same Genotype Induce Different Pathology in Chickens. Avian Dis. 2021, 65, 529–539. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).