1. Introduction
With globalization of the pig industry, porcine pathogens are emerging more frequently and are spreading worldwide. Enteric diseases that present with severe diarrhea are the predominant cause of morbidity and mortality in piglets [
1]. Although several measures have been taken to prevent swine diarrhea, it remains the most common health problem in the pig industry because of the complexity and unrestrained pathogenic factors [
2]. Most of the viruses causing diarrhea outbreaks in neonatal piglets are RNA viruses, including Coronaviruses, Rotaviruses, Picornaviruses, Astroviruses and Pestiviruses [
3]. Large-scale outbreaks of severe virus-associated diarrhea have been reported in pig populations since 2010 [
4]. In 2014, the porcine epidemic diarrhea virus (PEDV) was identified as the main pathogen causing piglet diarrhea. This was confirmed by metagenome sequencing, which revealed that the PEDV viral content in the diarrheal feces of piglets was more than 50%, and the proportion of other coronaviruses was approximately 3% [
5]. However, in 2024, members of
the Picornaviridae family accounted for the majority of the viral communities in piglet diarrheal feces, whereas the presence of coronaviruses was less than 1.7% [
6]. This was also consistent with the metagenome sequence data of piglet diarrhea stools collected in our laboratory in 2023 [
7].
It has been speculated that the infection pattern of porcine diarrheal pathogens is changing. Reports have shown that the status of PEDV as a major pathogen is decreasing, while the positive rates of emerging diarrheal pathogens are increasing. However, commercially available vaccines are mainly PEDV single vaccines and PEDV/TGEV/PoRV or PEDV/TGEV polyvalent vaccines, which are not suitable or fully protective against current pathogen infection patterns. This may be a key reason for the unsatisfactory prevention and control of diarrheal diseases in swine. Therefore, the identification of pathogenic infection patterns in swine diarrhea is currently an important area of research.
Logistic regression is a type of multiple regression method used to analyze the relationship between a binary or categorical outcome and multiple influencing factors [
8]. In veterinary epidemiology, logistic regression analysis has been used in numerous research investigations involving the study of risk factors, including lambs, goats, minks, poultry, cows, sows, pig [
9,
10,
11]. In this study, logistic regression analysis was used to explore the relationship between emerging pathogens and porcine diarrheal disease. This study will provide scientific data for the prevention and control of porcine emerging diarrheal pathogens.
2. Materials and Methods
2.1. Samples and Multiplex PCR Assay
In this study, 3256 diarrheal fecal samples from piglets were collected from several pig herds in Shanghai, China between 2015 and 2023. All samples were diluted with PBS and stored at -80℃ in sterile plastic tubes until detection.
According to the manufacturer’s instructions, total RNA of the samples were extracted using TIANamp virus RNA Kit (Tiangen Biotech, Beijing, China) followed by transcription using the PrimeScript Double Strand cDNA Synthesis Kit (Takara, Dalian, China). Then, the cDNA was subjected to multiplex PCR assay to detect 11 porcine diarrhea pathogens (PEDV, TGEV, PoRV, BVDV, CSFV, PSV, PAstV, PoSaV, PKoV, PTV, and PToV) [
12]. 2 µL cDNA, 25 µL 2×PCR Mix, 0.5 µL each primers (total eleven pairs of primers), 12 µL H
2O were mixed in a PCR tube with the amplification program of 95 ℃ 5min; 95 ℃ 30s, 55℃ 30s, 72℃ 30s, 25 cycles; 72℃ 5min. All assays were performed using positive and negative controls. Finally, the detection results were analyzed comprehensively.
2.2. Statistical Analysis
The infection status of porcine diarrheal pathogens was defined for each sample as a binary outcome. Tabular methods were used to calculate and map the correlation between emerging diarrheal pathogens and piglet diarrhea using R packages maps [
13]. Furthermore, their correlation with co-infection was analyzed statistically. p-value <0.05 were used to declare statistical significances of factors.
3. Results
3.1. Analysis on the Prevalence of the Eleven Porcine Diarrhea Pathogens
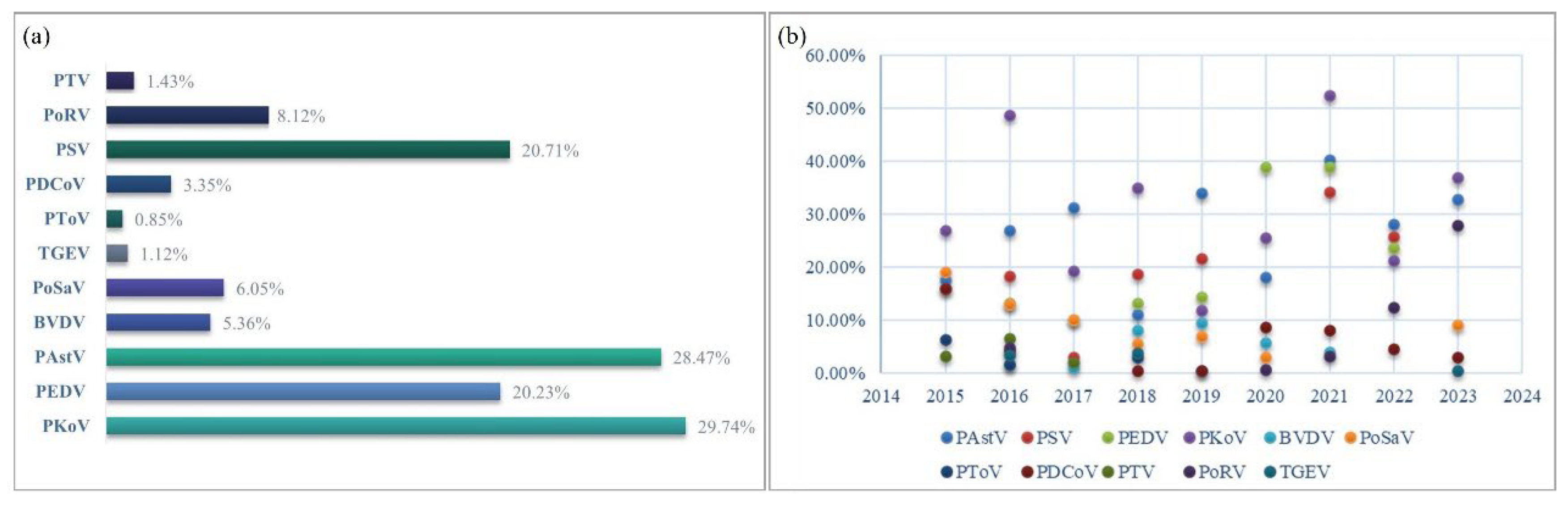
Based on an early investigation, 11 diarrhea pathogens, including PEDV, TGEV, PoRV, BVDV, CSFV, PSV, PAstV, PoSaV, PKoV, PTV, and PToV, were selected for detecting piglet fecal samples. Continuous pathogen monitoring from 2015 to 2023 revealed that PKoV had the highest positivity rate (29.74%), followed by PAstV (28.47%), PSV (20.71%), and PEDV (20.23%) (
Figure 1a). In previous years, PEDV, TGEV, and PoRV were the three most important pathogens causing porcine diarrhea. However, their detection rates have decreased. Compared to the low detection rate of TGEV (1.12%), PoRV had a higher rate of 8.12%. Since its discovery in 2012, PDCoV has been prevalent for several years and was once considered to have the same important status as PEDV. However, the total positive rate was only 3.35%, which was even lower than the BVDV (5.36%) and PoSaV (6.05%) detection rates.
Annual trend analysis showed that at least six types of porcine diarrheal pathogens were present in clinical samples, and up to nine pathogens were detected in 2018 (
Figure 1b). It can be clearly seen that PKoV (light purple ball) and PAstV (light blue ball) were the two most dominant pathogens in piglet diarrhea samples with high positive rates in each year (
Figure 1b). PSV and PEDV have similar infection advantages, ranking third or fourth, respectively.
3.2. Analysis of the Mixed-Infection in Porcine Diarrhea Samples
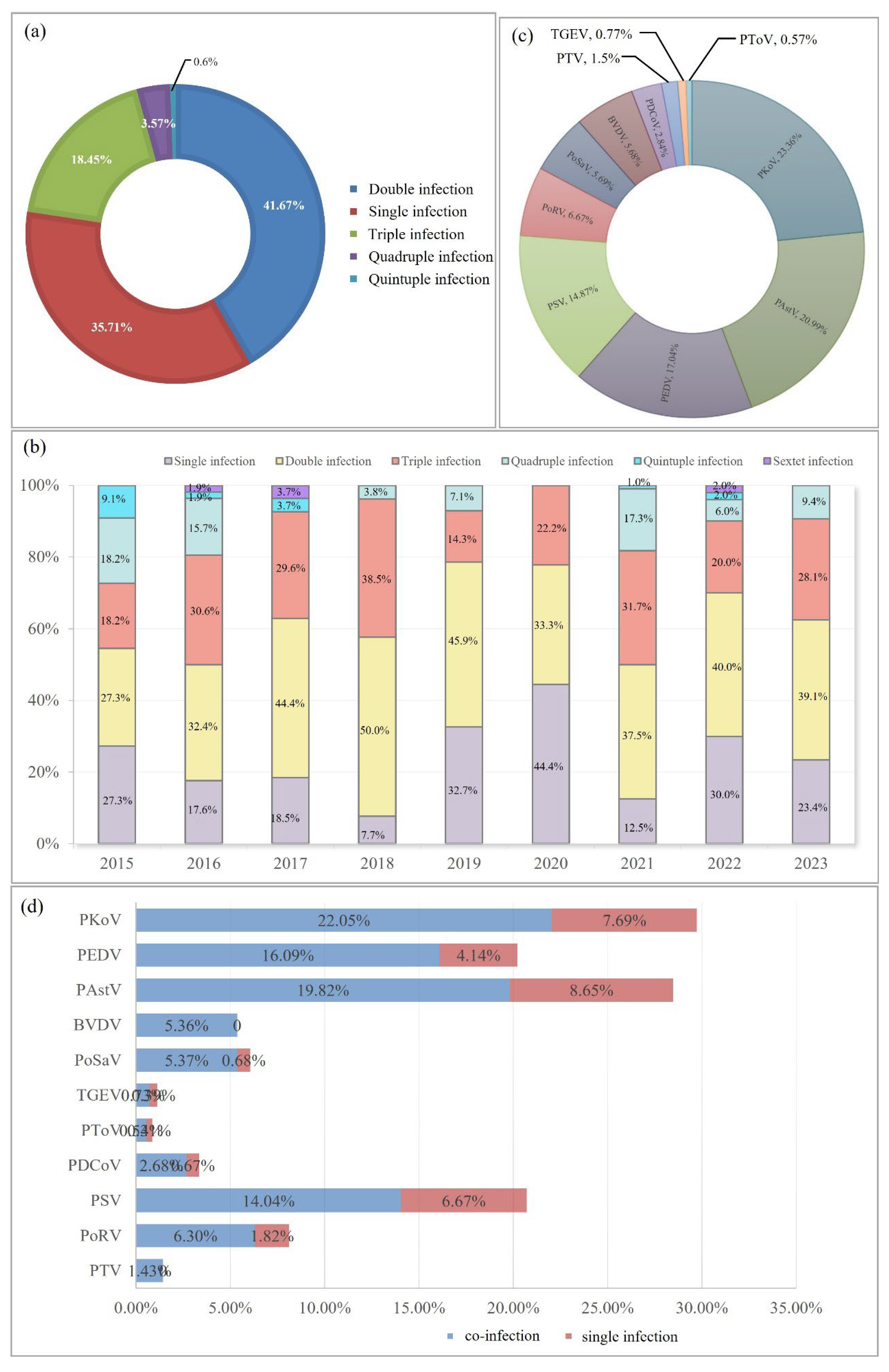
Subsequently, infection models of porcine diarrheal pathogens were explored. The proportion of single, double, triple, quadruple, and quintuple infections was 35.71%, 41.67%, 18.45%, 3.57%, and 0.6%, respectively (
Figure 2a). Double infection has a higher rate than single infection each year from 2015 to 2023, except for 2020 (
Figure 2b). In 2018, double and triple infection rates reached 50% and 38.5%, respectively, which were the highest percentage in nine years. This was the only time that a single infection (44.4%) was higher than the double infection in 2020 (
Figure 2b). This may be related to the COVID-19 pandemic. Improved quarantine measures on farms and stricter controls on personnel, vehicles, and logistics may result in relatively few outbreaks of pathogens.
Among these mixed infection agents, PKoV (23.36%), PAstV (20.99%), PEDV (17.04%), and PSV (14.87%) were the top four pathogens identified in high proportions in the co-infected samples (
Figure 2c).
Analysis from the perspective of a single pathogen showed that the number of mixed-infection samples was 3.88 times that of single-infection samples in PEDV-positive samples (
Figure 2d). Except BVDV and PTV with no single infection, this data of PoSaV was biggest (7.88) followed by PDCoV (4) and PoRV (3.46).
3.3. Identification of Mixed Infection Models of PEDV and Emerging Diarrhea Pathogens
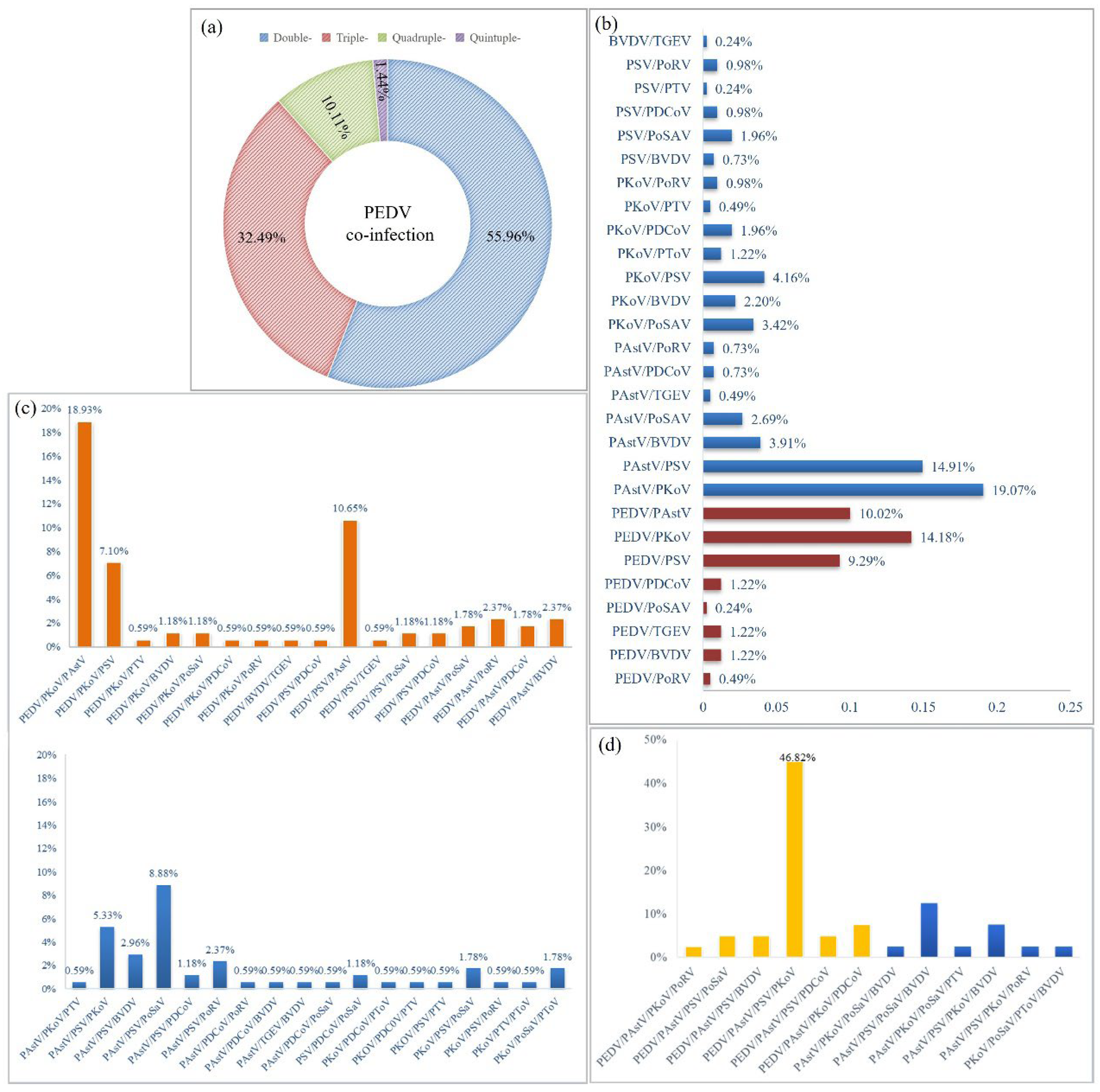
PEDV is a classic and important diarrheal agent involved in porcine diarrheal disease; however, it is always mixed with other diarrheal pathogens. Therefore, mixed infection models of PEDV and emerging diarrheal pathogens were identified. The results confirmed that the PEDV-based double, triple, quadruple and quintuple infection rates were 55.96%, 32.49%, 10.11%, and 1.44%, respectively (
Figure 3a). Over half of the PEDV-positive samples were double-infected (55.96%). In addition, double and triple infections were dominant in PEDV-positive samples, with rates of 32.49% and 10.11%, respectively.
Furthermore, we statistically analyzed all the co-infection models. In double infections, PAstV/PKoV (19.07%) and PAstV/PSV(14.91%) had the highest rates. PEDV/PKoV (14.18%), PEDV/PAstV (10.02%), and PEDV/PSV (9.29%) were the three most dominant double-infection models of PEDV (
Figure 3b). In triple infection, PEDV/PKoV/PAstV had the highest proportion (18.93%), followed by PEDV/PSV/PAstV (10.65%), and PAstV/PSV/PoSaV (8.88%), while PEDV/PKoV/PSV also presented a relatively high proportion of 7.10% (
Figure 3c). PEDV/PKoV/PAstV, PEDV/PSV/PAstV, and PEDV/PKoV/PSV are the predominant triple infection models for PEDV. In quadruple infections, PEDV/PAstV/PSV/PKoV was the only dominant infection model (46.82%) (
Figure 3d). In conclusion, PEDV is mainly co-infected with PAstV, PKoV, and PSV in clinical diarrhea samples, which may be the dominant infection model in pig herds.
3.4. Logistic Regression Analysis of the Procine Diarrhea Pathogens
To explore the correlation between these pathogens and diarrhea, both single-factor and multiple-factor logistic regression analyses were performed. Single-factor regression analysis showed that all 11 pathogens were associated with porcine diarrhea with significant
p-values (
Table 1). Because the interference of other factors was not controlled for during the single-factor regression analysis, the results may not be reliable. Therefore, it is necessary to conduct further multiple-factor regression analyses. In this study, multiple-factor regression analysis using the maximum likelihood method revealed that the
p-values of PAstV, PKoV, BVDV, and PEDV were lower than 0.05 and Exp (B) values were higher than 1, indicating that these five pathogens had a significant relationship with porcine diarrhea (
Table 2). The regression equation was logit (P)=-46.439+0.734PAstV(1)+1.554PKoV(1)+18.787PoSaV(1)+18.499PDCoV(1)+1.747BVDV(1)+3.709PEDV(1), with a prediction accuracy of 89.5%. Furthermore, statistical analysis showed that co-infection correlations existed among PKoV, PoSaV, PDCoV, BVDV, and PEDV (
Figure 4,
Table 3).
4. Discussion
Diarrheic diseases in swine have strong economic impacts on production units around the world. In recent years, porcine coronaviruses have represented some of the principal causes of these diseases [
14]. PEDV has been described as the major pathogen that causes diarrheal outbreaks in pigs. However, reports of naturally occurring coinfection of coronaviruses with other viral agents highlight the importance of less characterized viruses on disease severity and outcome [
15]. It has been confirmed that PKoV, PSV and PAstV are also responsible for gastrointestinal diseases, although they have been detected in fecal samples of animals that did not exhibit any clinical signs [
16,
17,
18]. The close association between PKoV, PSV and PAstV with neonatal piglet diarrhea has been previously reported [
19,
20,
21] suggesting that these viruses may play synergistic roles in causing diarrheal disease. Wu et al. confirmed that PKoV enhanced PEDV pathogenicity and altered the number of intestinal lymphocytes in piglets [
22]. Our group also identified that PSV and PEDV coinfections could aggravate the clinical symptoms observed in piglets (data not published). This could be attributed to an ineffective piglet immune response towards the viruses causing porcine diarrheal disease. At present, commercial vaccines comprise mainly inactivated or live viruses to induce an immune response against PEDV, TGEV and PoRV and these vaccines are not effective against the current swine epidemic pathogens. Findings from this study propose that the development of effective vaccines against a combination of PEDV and the novel co-infecting diarrheal pathogens (PSV, PKoV, PAstV, etc.) is an important direction to pursue.
This study demonstrated the dominant infection patterns of porcine diarrheal pathogens in Shanghai of China. The increasing epidemic trends of novel diarrheal viruses are not limited to China but have also been reported in other countries, including Korea [
23,
24], Thailand [
25], France [
26], Japan [
27], USA [
28,
29,
30], Italy [
31], Switzerland [
17], India [
32,
33], and Croatia [
34]. Therefore, it is imperative that greater attention is paid to these novel diarrheal pathogens. PEDV can cause serious clinical symptoms while most of the novel diarrheal pathogens also exist in healthy pigs albeit with higher rates detected in co-infection clinical samples. This suggests that these viruses may be synergistically interacting with PEDV, which makes clinical diarrhea prevention and control difficult.
Logistic regression model has been a routine technique for epidemic risk assessment. Though the emerging diarrheal pathogens were prevalenced in pig herds with high detection rates, their risk of infection and epidemic is still unknown. In this study, two kinds of logistic regression models were compared. Single-factor logistic regression model showed that all the 11 pathogens were closly related with porcine diarrhea while multiple-factor logistic regression model revealed that only PAstV, PKoV, BVDV, and PEDV were closly related with porcine diarrhea. And PEDV/PKoV, PEDV/PoSaV, PKoV/BVDV, PoSaV/BVDV, and PDCoV/PoSaV had great co-infection dominance. This may provide pathogens models for the researches on the co-infection pathogenesis.
In conclusion, the dominant co-infection models of porcine diarrheal pathogens were explored in this study and found that PEDV mainly mix-infected with the emerging diarrheal pathogens (PKoV, PAstV, PSV). Further logistic regression analysis confirmed that emerging pathogens of PAstV and PKoV were significant related with porcine diarrhea. Enhanced warnging should be raised. However, additional techniques will be applied to conduct more comprehensive co-infection risk assessments.
Author Contributions
Conceptualization, H.L. (Huili Liu) and J.T. (Jie Tao); methodology, J.C. (Jinghua Cheng) and Y.S. (Ying Shi); investigation, B.L. (Benqiang Li); resources, B.L. and P.T. (Pan Tang); data curation, J.J. (Jiajie Jiao) and P.T.; writing—original draft preparation, H.L. and J.T.; writing—review and editing, J.C. and Y.S.; project administration, H.L. and J.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Shanghai Agricultural Technology Development Program, China (Grant No. 2022-02-08-00-12-F01167), and the SAAS Program for Excellent Research Team (Grant No. [2022]012).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors do not have any conflicts of interest.
References
- Zhang, Q.; Hu, R.; Tang, X.; Wu, C.; He, Q.; Zhao, Z.; Chen, H.; Wu, B. Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch Virol 2013, 158, 1631–1636. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zheng, H.; Wang, S.W.; Luo, H.Q.; Li, Z.W.; Song, X.Z.; Xu, H.X.; Li, P.D.; Sun, S.Y.; Wang, Y.; Yuan, Z.J. Comparative analysis of changes in diarrhea and gut microbiota in Beigang pigs. Microb Pathog 2023, 185, 106441. [Google Scholar] [CrossRef] [PubMed]
- Cortey, M.; Díaz, I.; Vidal, A.; Martín-Valls, G.; Franzo, G.; Gómez de Nova, P.J.; Darwich, L.; Puente, H.; Carvajal, A.; Martín, M.; Mateu, E. High levels of unreported intraspecific diversity among RNA viruses in faeces of neonatal piglets with diarrhoea. BMC Vet Res 2019, 15, 441. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Pan, Y.; Xi, Y.; Wang, M.; Zeng, Q. Insights and progress on epidemic characteristics, genotyping, and preventive measures of PEDV in China: A review. Microb Pathog 2023, 181, 106185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Tang, C.; Yue, H.; Ren, Y.; Song, Z. Viral metagenomics analysis demonstrates the diversity of viral flora in piglet diarrhoeic faeces in China. J Gen Virol 2014, 95, 1603–1611. [Google Scholar] [CrossRef]
- Qian, L.; Tang, C.; Yue, H.; Ren, Y.; Song, Z. Metagenomic survey of viral diversity obtained from feces of piglets with diarrhea. Heliyon 2024, 10, e25616. [Google Scholar] [CrossRef]
- Tao, J.; Li, B.Q.; Cheng, J.H.; Shi, Y.; Liu, P.H.; He, G.X.; Xu, W.J.; Liu, H.L. Viral metagenome analysis of the viral community composition of the porcine diarrhea feaces. Biodivers Sci 2023, 31, 9. [Google Scholar] [CrossRef]
- Bouyer, J. Logistics regression in epidemiology. II. Rev Epidemiol Sante Publique 1991, 39, 183–196. [Google Scholar]
- Rattenborg, E.; Chriel, M.; Dietz, H.H. Influence of farm, feed-producer and season on incidence of gastrointestinal disorders in Danish farm mink. Prev Vet Med 1999, 38, 231–237. [Google Scholar] [CrossRef]
- Birkegard, A.C.; Halasa, T.; Toft, N. Sampling pig farms at the abattoir in a cross-sectional study - Evaluation of a sampling method. Prev Vet Med 2017, 145, 83–90. [Google Scholar] [CrossRef]
- Herve-Claude, L.P.; Lwanga-Iga, I.; Kroll-Lwanga-Iga, S.; Nyangiwe, N.; Ruddat, I.; Kreienbrock, L. Village livestock population and sampling strategies in communal areas in the Eastern Cape Province, South Africa. Trop Anim Health Prod 2011, 43, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Li, B.Q.; Tao, J.; Cheng, J.H. The Complex Co-infections of Multiple Porcine Diarrhea Viruses in Local Area Based on the Luminex xTAG Multiplex Detection Method. Front Vet Sci 2021, 8, 602866. [Google Scholar] [CrossRef] [PubMed]
- Homwong, N.; Diaz, A.; Rossow, S.; Ciarlet, M.; Marthaler, D. Three-Level Mixed-Effects Logistic Regression Analysis Reveals Complex Epidemiology of Swine Rotaviruses in Diagnostic Samples from North America. PLoS One 2016, 11, e0154734. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Luo, S.; Gu, J.; Li, Z.; Li, K.; Yuan, W.; Ye, Y.; Li, H.; Ding, Z.; Song, D.; Tang, Y. Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet Res 2019, 15, 470. [Google Scholar] [CrossRef]
- Su, M.; Qi, S.; Yang, D.; Guo, D.; Yin, B.; Sun, D. Coinfection and Genetic Characterization of Porcine Astrovirus in Diarrheic Piglets in China From 2015 to 2018. Front Vet Sci 2020, 7, 462. [Google Scholar] [CrossRef]
- Li, Y.; Liang, J.; Wu, S.; Yan, Z.; Zhang, W. Complete genomic sequence analysis and intestinal tissue localization of a porcine Kobuvirus variant in China. Infect Genet Evol 2022, 104, 105362. [Google Scholar] [CrossRef]
- Staubli, T.; Rickli, C.I.; Torgerson, P.R.; Fraefel, C.; Lechmann, J. Porcine teschovirus, sapelovirus, and enterovirus in Swiss pigs: multiplex RT-PCR investigation of viral frequencies and disease association. J Vet Diagn Invest 2021, 33, 864–874. [Google Scholar] [CrossRef]
- Kumthip, K.; Khamrin, P.; Saikruang, W.; Kongkaew, A.; Vachirachewin, R.; Ushijima, H.; Maneekarn, N. Detection and genetic characterization of porcine astroviruses in piglets with and without diarrhea in Thailand. Arch Virol 2018, 163, 1823–1829. [Google Scholar] [CrossRef]
- Nantel-Fortier, N.; Gauthier, M.; L’Homme, Y.; Lachapelle, V.; Fravalo, P.; Brassard, J. The swine enteric virome in a com mercial production system and its association with neonatal diarrhea. Vet Microbiol 2022, 266, 109366. [Google Scholar] [CrossRef]
- Qiu, M.; Li, S.; Xiao, Y.; Li, J.; Zhang, Y.; Li, X.; Feng, B.; Li, C.; Lin, H.; Zhu, J.; Chen, N. Pathogenic and metagenomic evaluations reveal the correlations of porcine epidemic diarrhea virus, porcine kobuvirus and porcine astroviruses with neonatal piglet diarrhea. Microb Pathog 2022, 170, 105703. [Google Scholar] [CrossRef]
- Chen, J.; Suo, X.; Cao, L.; Yuan, C.; Shi, L.; Duan, Y.; Zheng, H.; Wang, Q. Virome Analysis for Identification of a Novel Porcine Sapelovirus Isolated in Western China. Microbiol Spectr 2022, 10, e0180122. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Gou, F.; Meng, J.; Jin, X.; Liu, W.; Ding, W.; Xu, W.; Gu, C.; Hu, X.; Cheng, G.; Tao, P.; Zhang, W. Porcine kobuvirus enhances porcine epidemic diarrhea virus pathogenicity and alters the number of intestinal lymphocytes in piglets. Vet Microbiol 2024, 293, 110100. [Google Scholar] [CrossRef] [PubMed]
- An, D.J.; Jeoung, H.Y.; Jeong, W.; Lee, H.S.; Park, J.Y.; Kim, B. Porcine kobuvirus from pig stool in Korea. Virus Genes 2011, 42, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Jeong, C.G.; Chae, S.B.; Yang, M.S.; Oh, B.; Lee, S.Y.; Oem, J.K. Porcine Astrovirus Infection in Brains of Pigs in Korea. Viruses 2024, 16, 1372. [Google Scholar] [CrossRef]
- Chuchaona, W.; Khamrin, P.; Yodmeeklin, A.; Kongkaew, A.; Vachirachewin, R.; Kumthip, K.; Ushijima, H.; Maneekarn, N. Detection and molecular characterization of porcine kobuvirus in piglets in 2009-2013 in northern Thailand. Trop Anim Health Prod 2017, 49, 1077–1080. [Google Scholar] [CrossRef]
- Capai, L.; Piorkowski, G.; Maestrini, O.; Casabianca, F.; Masse, S.; de Lamballerie, X.; Charrel, R.N.; Falchi, A. Detection of porcine enteric viruses (Kobuvirus, Mamastrovirus and Sapelovirus) in domestic pigs in Corsica, France. PLoS One 2022, 17, e0260161. [Google Scholar] [CrossRef]
- Okitsu, S.; Khamrin, P.; Thongprachum, A.; Hidaka, S.; Kongkaew, S.; Kongkaew, A.; Maneekarn, N.; Mizuguchi, M.; Hayakawa, S.; Ushijima, H. Sequence analysis of porcine kobuvirus VP1 region detected in pigs in Japan and Thailand. Virus Genes 2012, 44, 253–257. [Google Scholar] [CrossRef]
- Verma, H.; Mor, S.K.; Abdel-Glil, M.Y.; Goyal, S.M. Identification and molecular characterization of porcine kobuvirus in U. S. swine. Virus Genes 2013, 46, 551–553. [Google Scholar] [CrossRef]
- Matias Ferreyra, F.; Harmon, K.; Bradner, L.; Burrough, E.; Derscheid, R.; Magstadt, D.R.; Michael, A.; de Almeida, M.N.; Schumacher, L.; Siepker, C.; Sitthicharoenchai, P.; Stevenson, G.; Arruda, B. Comparative Analysis of Novel Strains of Porcine Astrovirus Type 3 in the USA. Viruses 2021, 13, 1859. [Google Scholar] [CrossRef]
- Mor, S.K.; Chander, Y.; Marthaler, D.; Patnayak, D.P.; Goyal, S.M. Detection and molecular characterization of Porcine astrovirus strains associated with swine diarrhea. J Vet Diagn Invest 2012, 24, 1064–1067. [Google Scholar] [CrossRef]
- Chelli, E.; De Sabato, L.; Vaccari, G.; Ostanello, F.; Di Bartolo, I. Detection and Characterization of Porcine Sapelovirus in Italian Pig Farms. Animals (Basel) 2020, 10, 966. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Saikumar, G.; Desingu, P.A.; Das, T.; Singh, R. Immunohistochemical detection of naturally occurring porcine Sapelovirus infection in Indian pigs. J Immunoassay Immunochem 2019, 40, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Vaishali Gupta, R.; Kumar, M.; Bansal, N.; Vivek Kumar, P.; Kumar, P.; Jindal, N. Coinfection of porcine astrovirus and other porcine viruses in diarrheic pigs in Haryana, India. Arch Virol 2023, 168, 246. [Google Scholar] [CrossRef] [PubMed]
- Brnic, D.; Prpić, J.; Keros, T.; Roić, B.; Starešina, V.; Jemeršić, L. Porcine astrovirus viremia and high genetic variability in pigs on large holdings in Croatia. Infect Genet Evol 2013, 14, 258–264. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).