Submitted:
31 January 2025
Posted:
03 February 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Novel DNA Repair Components Required for Survival of BRCA1-Deficient, but Not BRCA1-Proficient Cancers
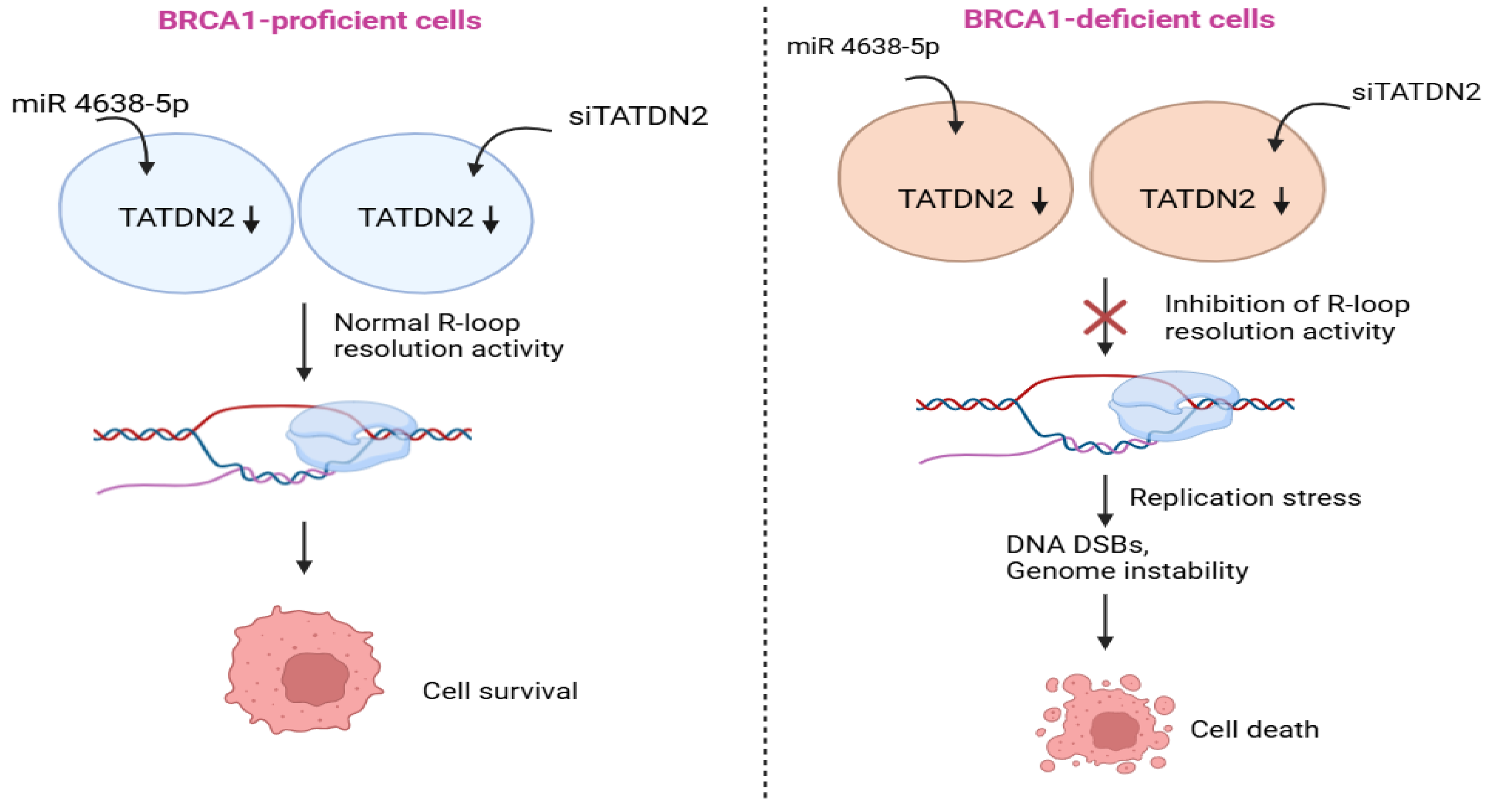
2.1. Role of TATDN2 in BRCA1-Deficient Cancers
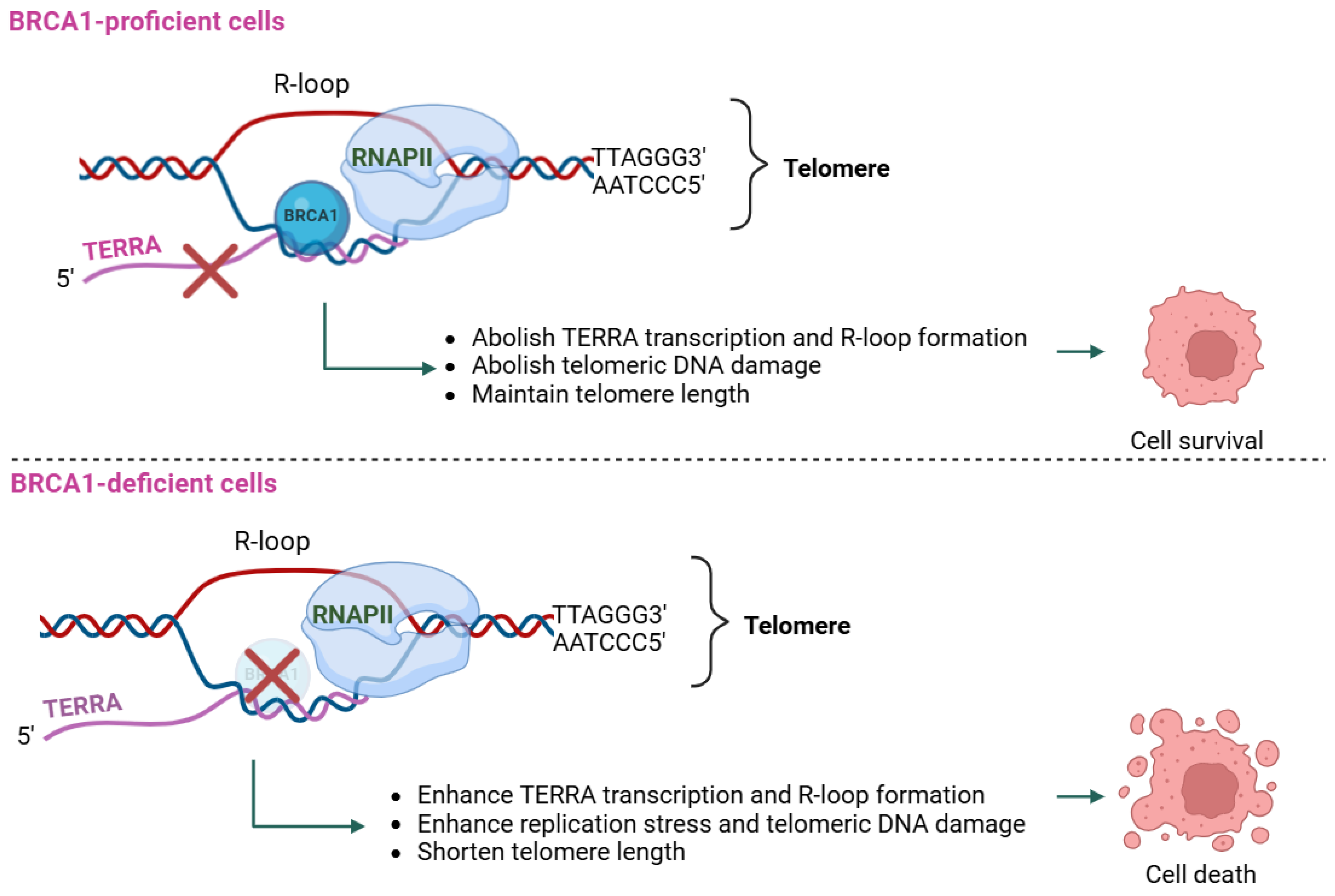
2.2. Role of TERRA in BRCA1-Deficient Cancers
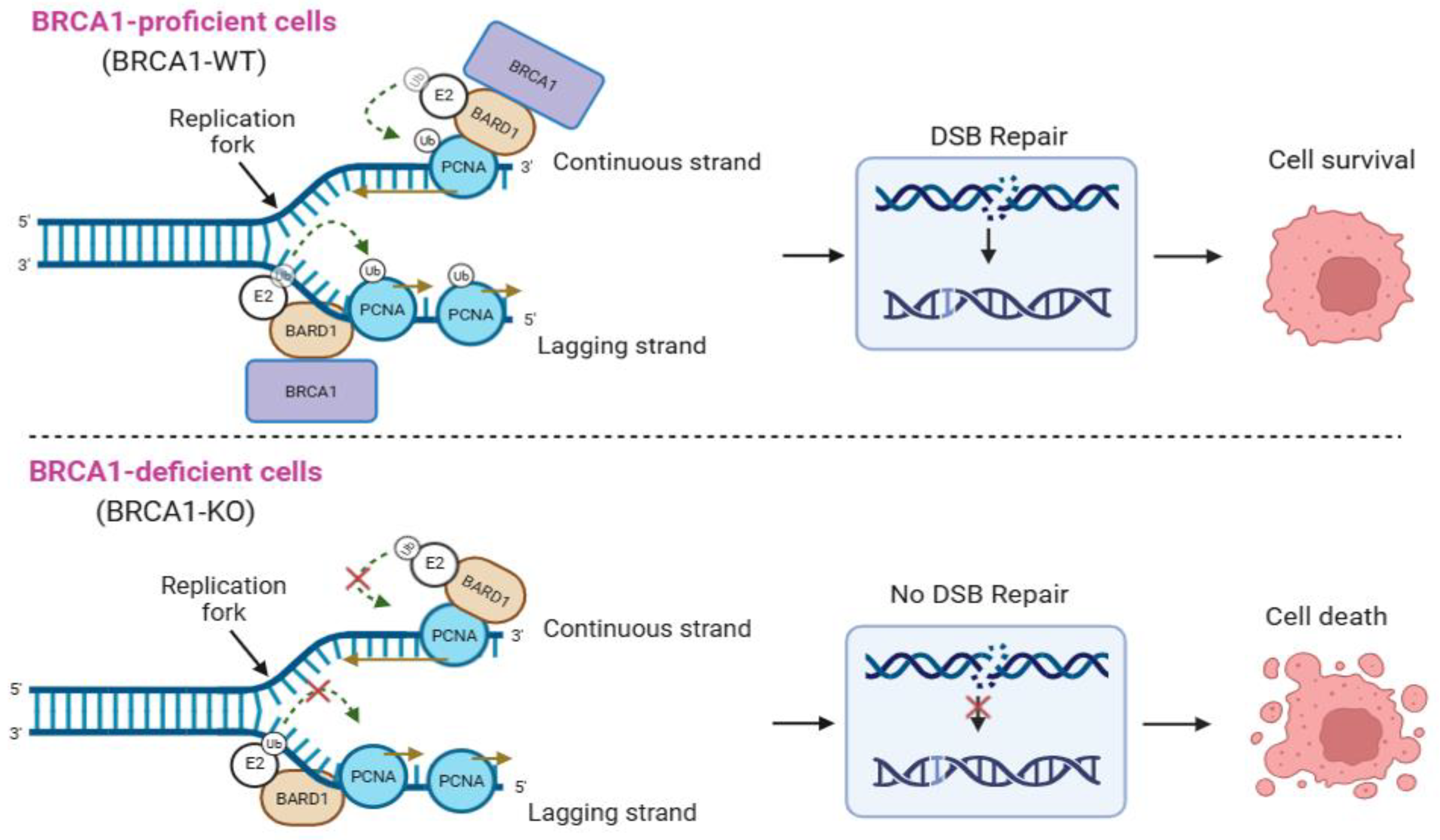
2.3. Role of BARD1 in BRCA1-Deficient Cancers
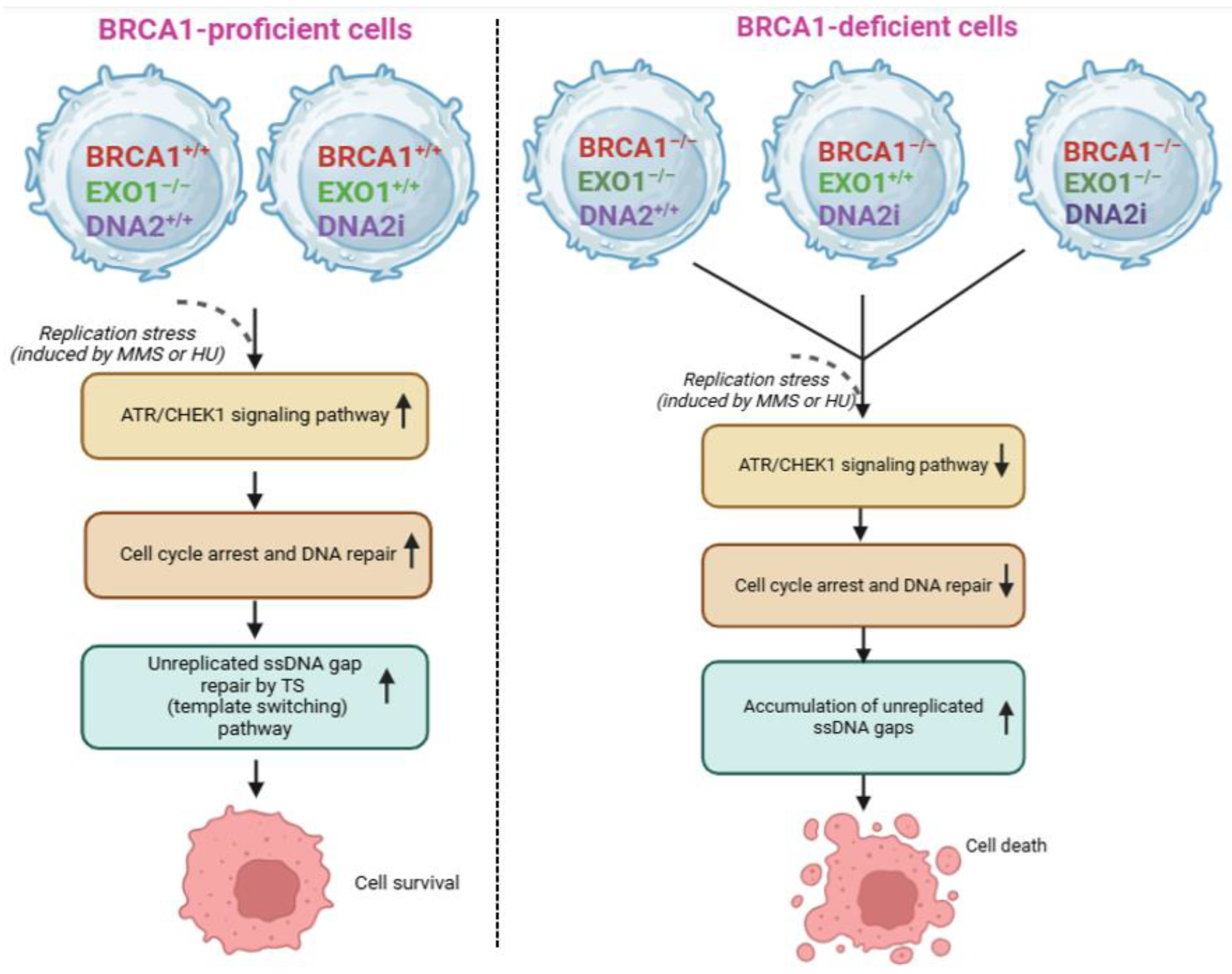
2.4. Role of EXO1 in BRCA1-Deficient Cancers
3. Other Potential Components
3.1. Role of EEPD1 in BRCA1-Deficient Cancers
3.2. Role of FANCJ in BRCA1-Deficient Cancers
3.3. Role of USP1 in BRCA1-Deficient Cancers
4. Discussion
Conflicts of Interests
Author Contributions
Funding
Data Availability Statement
References
- Ceccaldi, R.; Rondinelli, B.; D'Andrea, A.D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol 2016, 26, 52-64. [CrossRef]
- Yu, W.; Lescale, C.; Babin, L.; Bedora-Faure, M.; Lenden-Hasse, H.; Baron, L.; Demangel, C.; Yelamos, J.; Brunet, E.; Deriano, L. Repair of G1 induced DNA double-strand breaks in S-G2/M by alternative NHEJ. Nature Communications 2020, 11, 5239. [CrossRef]
- Pannunzio, N.R.; Watanabe, G.; Lieber, M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. Journal of Biological Chemistry 2018, 293, 10512-10523. [CrossRef]
- Hussain, S.S.; Majumdar, R.; Moore, G.M.; Narang, H.; Buechelmaier, Erika S.; Bazil, M.J.; Ravindran, P.T.; Leeman, Jonathan E.; Li, Y.; Jalan, M.; et al. Measuring nonhomologous end-joining, homologous recombination and alternative end-joining simultaneously at an endogenous locus in any transfectable human cell. Nucleic Acids Research 2021, 49, e74-e74. [CrossRef]
- Hanscom, T.; McVey, M. Regulation of Error-Prone DNA Double-Strand Break Repair and Its Impact on Genome Evolution. Cells 2020, 9, 1657.
- Dueva, R.; Iliakis, G. Alternative pathways of non-homologous end joining (NHEJ) in genomic instability and cancer. Translational Cancer Research 2013, 2, 163-177.
- Bennardo, N.; Cheng, A.; Huang, N.; Stark, J.M. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 2008, 4, e1000110. [CrossRef]
- Sfeir, A.; Symington, L.S. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci 2015, 40, 701-714. [CrossRef]
- Paul, A.; Paul, S. The breast cancer susceptibility genes (BRCA) in breast and ovarian cancers. Front Biosci (Landmark Ed) 2014, 19, 605-618. [CrossRef]
- Yamamoto, H.; Hirasawa, A. Homologous Recombination Deficiencies and Hereditary Tumors. Int J Mol Sci 2021, 23. [CrossRef]
- Wang, M.; Chen, S.; Ao, D. Targeting DNA repair pathway in cancer: Mechanisms and clinical application. MedComm 2021, 2, 654-691. [CrossRef]
- Nickoloff, J.A.; Jaiswal, A.S.; Sharma, N.; Williamson, E.A.; Tran, M.T.; Arris, D.; Yang, M.; Hromas, R. Cellular Responses to Widespread DNA Replication Stress. International Journal of Molecular Sciences 2023, 24, 16903.
- Huang, R.; Zhou, P.K. DNA damage repair: historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct Target Ther 2021, 6, 254. [CrossRef]
- Deng, C.X. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res 2006, 34, 1416-1426. [CrossRef]
- Rodgers, K.; McVey, M. Error-Prone Repair of DNA Double-Strand Breaks. J Cell Physiol 2016, 231, 15-24. [CrossRef]
- Torgovnick, A.; Schumacher, B. DNA repair mechanisms in cancer development and therapy. Front Genet 2015, 6, 157. [CrossRef]
- Sishc, B.J.; Davis, A.J. The Role of the Core Non-Homologous End Joining Factors in Carcinogenesis and Cancer. Cancers (Basel) 2017, 9. [CrossRef]
- Schiewer, M.J.; Knudsen, K.E. DNA Damage Response in Prostate Cancer. Cold Spring Harb Perspect Med 2019, 9. [CrossRef]
- Wu, W.; Koike, A.; Takeshita, T.; Ohta, T. The ubiquitin E3 ligase activity of BRCA1 and its biological functions. Cell Div 2008, 3, 1. [CrossRef]
- Clark, S.L.; Rodriguez, A.M.; Snyder, R.R.; Hankins, G.D.; Boehning, D. Structure-Function Of The Tumor Suppressor BRCA1. Comput Struct Biotechnol J 2012, 1. [CrossRef]
- Leung, C.C.; Glover, J.N. BRCT domains: easy as one, two, three. Cell Cycle 2011, 10, 2461-2470. [CrossRef]
- Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat Rev Mol Cell Biol 2020, 21, 284-299. [CrossRef]
- Rosen, E.M.; Fan, S.; Ma, Y. BRCA1 regulation of transcription. Cancer Letters 2006, 236, 175-185. [CrossRef]
- Pan, H.; He, Z.; Ling, L.; Ding, Q.; Chen, L.; Zha, X.; Zhou, W.; Liu, X.; Wang, S. Reproductive factors and breast cancer risk among BRCA1 or BRCA2 mutation carriers: Results from ten studies. Cancer Epidemiology 2014, 38, 1-8. [CrossRef]
- Shao, F.; Sun, H.; Deng, C.X. Potential therapeutic targets of triple-negative breast cancer based on its intrinsic subtype. Oncotarget 2017, 8, 73329-73344. [CrossRef]
- Peshkin, B.N.; Alabek, M.L.; Isaacs, C. BRCA1/2 mutations and triple negative breast cancers. Breast Dis 2010, 32, 25-33. [CrossRef]
- Burga, L.N.; Hu, H.; Juvekar, A.; Tung, N.M.; Troyan, S.L.; Hofstatter, E.W.; Wulf, G.M. Loss of BRCA1 leads to an increase in epidermal growth factor receptor expression in mammary epithelial cells, and epidermal growth factor receptor inhibition prevents estrogen receptor-negative cancers in BRCA1-mutant mice. Breast Cancer Research 2011, 13, R30. [CrossRef]
- Mersch, J.; Jackson, M.A.; Park, M.; Nebgen, D.; Peterson, S.K.; Singletary, C.; Arun, B.K.; Litton, J.K. Cancers associated with 1 and 2 mutations other than breast and ovarian. Cancer 2015, 121, 269-275. [CrossRef]
- Castro, E.; Eeles, R. The role of BRCA1 and BRCA2 in prostate cancer. Asian J Androl 2012, 14, 409-414. [CrossRef]
- Narod, S.A.; Metcalfe, K.; Finch, A.; Chan, A.-W.; Armel, S.R.; Aeilts, A.; Eisen, A.; Karlan, B.; Bordeleau, L.; Tung, N.; et al. The risk of skin cancer in women who carry BRCA1 or BRCA2 mutations. Hereditary Cancer in Clinical Practice 2024, 22, 7. [CrossRef]
- Devico Marciano, N.; Kroening, G.; Dayyani, F.; Zell, J.A.; Lee, F.C.; Cho, M.; Valerin, J.G. BRCA-Mutated Pancreatic Cancer: From Discovery to Novel Treatment Paradigms. Cancers (Basel) 2022, 14. [CrossRef]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 2011, 12, 68-78. [CrossRef]
- Patel, A.G.; Sarkaria, J.N.; Kaufmann, S.H. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A 2011, 108, 3406-3411. [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 2017, 18, 495-506. [CrossRef]
- Liang, S.; Blundell, T.L. Human DNA-dependent protein kinase activation mechanism. Nature Structural & Molecular Biology 2023, 30, 140-147. [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 2017, 18, 610-621. [CrossRef]
- Couto, C.A.; Wang, H.Y.; Green, J.C.; Kiely, R.; Siddaway, R.; Borer, C.; Pears, C.J.; Lakin, N.D. PARP regulates nonhomologous end joining through retention of Ku at double-strand breaks. J Cell Biol 2011, 194, 367-375. [CrossRef]
- Dale Rein, I.; Solberg Landsverk, K.; Micci, F.; Patzke, S.; Stokke, T. Replication-induced DNA damage after PARP inhibition causes G2 delay, and cell line-dependent apoptosis, necrosis and multinucleation. Cell Cycle 2015, 14, 3248-3260. [CrossRef]
- Wang, S.S.Y.; Jie, Y.E.; Cheng, S.W.; Ling, G.L.; Ming, H.V.Y. PARP Inhibitors in Breast and Ovarian Cancer. Cancers (Basel) 2023, 15. [CrossRef]
- Valabrega, G.; Scotto, G.; Tuninetti, V.; Pani, A.; Scaglione, F. Differences in PARP Inhibitors for the Treatment of Ovarian Cancer: Mechanisms of Action, Pharmacology, Safety, and Efficacy. Int J Mol Sci 2021, 22. [CrossRef]
- Bhamidipati, D.; Haro-Silerio, J.I.; Yap, T.A.; Ngoi, N. PARP inhibitors: enhancing efficacy through rational combinations. British Journal of Cancer 2023, 129, 904-916. [CrossRef]
- Dilmac, S.; Ozpolat, B. Mechanisms of PARP-Inhibitor-Resistance in BRCA-Mutated Breast Cancer and New Therapeutic Approaches. Cancers (Basel) 2023, 15. [CrossRef]
- Dhillon, K.K.; Swisher, E.M.; Taniguchi, T. Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci 2011, 102, 663-669. [CrossRef]
- Giudice, E.; Gentile, M.; Salutari, V.; Ricci, C.; Musacchio, L.; Carbone, M.V.; Ghizzoni, V.; Camarda, F.; Tronconi, F.; Nero, C.; et al. PARP Inhibitors Resistance: Mechanisms and Perspectives. Cancers (Basel) 2022, 14. [CrossRef]
- Konstantinopoulos, P.A.; Lheureux, S.; Moore, K.N. PARP Inhibitors for Ovarian Cancer: Current Indications, Future Combinations, and Novel Assets in Development to Target DNA Damage Repair. American Society of Clinical Oncology Educational Book 2020, e116-e131. [CrossRef]
- Revythis, A.; Limbu, A.; Mikropoulos, C.; Ghose, A.; Sanchez, E.; Sheriff, M.; Boussios, S. Recent Insights into PARP and Immuno-Checkpoint Inhibitors in Epithelial Ovarian Cancer. Int J Environ Res Public Health 2022, 19. [CrossRef]
- Veneris, J.T.; Matulonis, U.A.; Liu, J.F.; Konstantinopoulos, P.A. Choosing wisely: Selecting PARP inhibitor combinations to promote anti-tumor immune responses beyond BRCA mutations. Gynecologic Oncology 2020, 156, 488-497. [CrossRef]
- Yazinski, S.A.; Comaills, V.; Buisson, R.; Genois, M.M.; Nguyen, H.D.; Ho, C.K.; Todorova Kwan, T.; Morris, R.; Lauffer, S.; Nussenzweig, A.; et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev 2017, 31, 318-332. [CrossRef]
- Nolan, E.; Savas, P.; Policheni, A.N.; Darcy, P.K.; Vaillant, F.; Mintoff, C.P.; Dushyanthen, S.; Mansour, M.; Pang, J.B.; Fox, S.B.; et al. Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci Transl Med 2017, 9. [CrossRef]
- Chen, Y.C.; Li, C.L.; Hsiao, Y.Y.; Duh, Y.; Yuan, H.S. Structure and function of TatD exonuclease in DNA repair. Nucleic Acids Res 2014, 42, 10776-10785. [CrossRef]
- Petrů, M.; Wideman, J.; Moore, K.; Alcock, F.; Palmer, T.; Doležal, P. Evolution of mitochondrial TAT translocases illustrates the loss of bacterial protein transport machines in mitochondria. BMC Biology 2018, 16, 141. [CrossRef]
- Kudva, R.; Denks, K.; Kuhn, P.; Vogt, A.; Müller, M.; Koch, H.G. Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res Microbiol 2013, 164, 505-534. [CrossRef]
- Dorival, J.; Eichman, B.F. Human and bacterial TatD enzymes exhibit apurinic/apyrimidinic (AP) endonuclease activity. Nucleic Acids Res 2023, 51, 2838-2849. [CrossRef]
- Yang, H.; Liu, C.; Jamsen, J.; Wu, Z.; Wang, Y.; Chen, J.; Zheng, L.; Shen, B. The DNase domain-containing protein TATDN1 plays an important role in chromosomal segregation and cell cycle progression during zebrafish eye development. Cell Cycle 2012, 11, 4626-4632. [CrossRef]
- Jaiswal, A.S.; Dutta, A.; Srinivasan, G.; Yuan, Y.; Zhou, D.; Shaheen, M.; Sadideen, D.T.; Kirby, A.; Williamson, E.A.; Gupta, Y.K.; et al. TATDN2 resolution of R-loops is required for survival of BRCA1-mutant cancer cells. Nucleic Acids Res 2023, 51, 12224-12241. [CrossRef]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol Cell 2019, 73, 398-411. [CrossRef]
- García-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604-618. [CrossRef]
- Chang, E.Y.; Stirling, P.C. Replication Fork Protection Factors Controlling R-Loop Bypass and Suppression. Genes (Basel) 2017, 8. [CrossRef]
- Bettin, N.; Oss Pegorar, C.; Cusanelli, E. The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability. Cells 2019, 8. [CrossRef]
- Porro, A.; Feuerhahn, S.; Reichenbach, P.; Lingner, J. Molecular Dissection of Telomeric Repeat-Containing RNA Biogenesis Unveils the Presence of Distinct and Multiple Regulatory Pathways. Molecular and Cellular Biology 2010, 30, 4808-4817. [CrossRef]
- Redon, S.; Reichenbach, P.; Lingner, J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Research 2010, 38, 5797-5806. [CrossRef]
- Chebly, A.; Ropio, J.; Baldasseroni, L.; Prochazkova-Carlotti, M.; Idrissi, Y.; Ferrer, J.; Farra, C.; Beylot-Barry, M.; Merlio, J.P.; Chevret, E. Telomeric Repeat-Containing RNA (TERRA): A Review of the Literature and First Assessment in Cutaneous T-Cell Lymphomas. Genes (Basel) 2022, 13. [CrossRef]
- Cusanelli, E.; Romero, C.A.; Chartrand, P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol Cell 2013, 51, 780-791. [CrossRef]
- Chu, H.P.; Cifuentes-Rojas, C.; Kesner, B.; Aeby, E.; Lee, H.G.; Wei, C.; Oh, H.J.; Boukhali, M.; Haas, W.; Lee, J.T. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell 2017, 170, 86-101.e116. [CrossRef]
- Rivosecchi, J.; Jurikova, K.; Cusanelli, E. Telomere-specific regulation of TERRA and its impact on telomere stability. Seminars in Cell & Developmental Biology 2024, 157, 3-23. [CrossRef]
- MacKenzie, D., Jr.; Watters, A.K.; To, J.T.; Young, M.W.; Muratori, J.; Wilkoff, M.H.; Abraham, R.G.; Plummer, M.M.; Zhang, D. ALT Positivity in Human Cancers: Prevalence and Clinical Insights. Cancers (Basel) 2021, 13. [CrossRef]
- Gong, Y.; Liu, Y. R-Loops at Chromosome Ends: From Formation, Regulation, and Cellular Consequence. Cancers (Basel) 2023, 15. [CrossRef]
- Al-Turki, T.M.; Maranon, D.G.; Nelson, C.B.; Lewis, A.M.; Luxton, J.J.; Taylor, L.E.; Altina, N.; Wu, F.; Du, H.; Kim, J.; et al. Telomeric RNA (TERRA) increases in response to spaceflight and high-altitude climbing. Communications Biology 2024, 7, 698. [CrossRef]
- Cusanelli, E.; Chartrand, P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet 2015, 6, 143. [CrossRef]
- Vohhodina, J.; Goehring, L.J.; Liu, B.; Kong, Q.; Botchkarev Jr, V.V.; Huynh, M.; Liu, Z.; Abderazzaq, F.O.; Clark, A.P.; Ficarro, S.B.; et al. BRCA1 binds TERRA RNA and suppresses R-Loop-based telomeric DNA damage. Nature Communications 2021, 12, 3542. [CrossRef]
- Flynn, R.L.; Cox, K.E.; Jeitany, M.; Wakimoto, H.; Bryll, A.R.; Ganem, N.J.; Bersani, F.; Pineda, J.R.; Suvà, M.L.; Benes, C.H.; et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 2015, 347, 273-277. [CrossRef]
- Deng, Z.; Wang, Z.; Stong, N.; Plasschaert, R.; Moczan, A.; Chen, H.S.; Hu, S.; Wikramasinghe, P.; Davuluri, R.V.; Bartolomei, M.S.; et al. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. Embo j 2012, 31, 4165-4178. [CrossRef]
- Monk, B.J.; Lorusso, D.; Italiano, A.; Kaye, S.B.; Aracil, M.; Tanović, A.; D'Incalci, M. Trabectedin as a chemotherapy option for patients with BRCA deficiency. Cancer Treat Rev 2016, 50, 175-182. [CrossRef]
- Cruz, C.; Llop-Guevara, A.; Garber, J.E.; Arun, B.K.; Pérez Fidalgo, J.A.; Lluch, A.; Telli, M.L.; Fernández, C.; Kahatt, C.; Galmarini, C.M.; et al. Multicenter Phase II Study of Lurbinectedin in BRCA-Mutated and Unselected Metastatic Advanced Breast Cancer and Biomarker Assessment Substudy. J Clin Oncol 2018, 36, 3134-3143. [CrossRef]
- Shakya, R.; Szabolcs, M.; McCarthy, E.; Ospina, E.; Basso, K.; Nandula, S.; Murty, V.; Baer, R.; Ludwig, T. The basal-like mammary carcinomas induced by Brca1 or Bard1 inactivation implicate the BRCA1/BARD1 heterodimer in tumor suppression. Proc Natl Acad Sci U S A 2008, 105, 7040-7045. [CrossRef]
- Edwards, R.A.; Lee, M.S.; Tsutakawa, S.E.; Williams, R.S.; Nazeer, I.; Kleiman, F.E.; Tainer, J.A.; Glover, J.N. The BARD1 C-terminal domain structure and interactions with polyadenylation factor CstF-50. Biochemistry 2008, 47, 11446-11456. [CrossRef]
- Zhao, W.; Steinfeld, J.B.; Liang, F.; Chen, X.; Maranon, D.G.; Jian Ma, C.; Kwon, Y.; Rao, T.; Wang, W.; Sheng, C.; et al. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature 2017, 550, 360-365. [CrossRef]
- Mallery, D.L.; Vandenberg, C.J.; Hiom, K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. Embo j 2002, 21, 6755-6762. [CrossRef]
- Densham, R.M.; Garvin, A.J.; Stone, H.R.; Strachan, J.; Baldock, R.A.; Daza-Martin, M.; Fletcher, A.; Blair-Reid, S.; Beesley, J.; Johal, B.; et al. Human BRCA1–BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nature Structural & Molecular Biology 2016, 23, 647-655. [CrossRef]
- Zhong, Q.; Chen, C.-F.; Li, S.; Chen, Y.; Wang, C.-C.; Xiao, J.; Chen, P.-L.; Sharp, Z.D.; Lee, W.-H. Association of BRCA1 with the hRad50-hMre11-p95 Complex and the DNA Damage Response. Science 1999, 285, 747-750, doi:doi:10.1126/science.285.5428.747.
- Chen, L.; Nievera, C.J.; Lee, A.Y.-L.; Wu, X. Cell Cycle-dependent Complex Formation of BRCA1·CtIP·MRN Is Important for DNA Double-strand Break Repair*. Journal of Biological Chemistry 2008, 283, 7713-7720. [CrossRef]
- Zong, D.; Adam, S.; Wang, Y.; Sasanuma, H.; Callén, E.; Murga, M.; Day, A.; Kruhlak, M.J.; Wong, N.; Munro, M.; et al. BRCA1 Haploinsufficiency Is Masked by RNF168-Mediated Chromatin Ubiquitylation. Mol Cell 2019, 73, 1267-1281.e1267. [CrossRef]
- Sherker, A.; Chaudhary, N.; Adam, S.; Heijink, A.M.; Noordermeer, S.M.; Fradet-Turcotte, A.; Durocher, D. Two redundant ubiquitin-dependent pathways of BRCA1 localization to DNA damage sites. EMBO reports 2021, 22, e53679. [CrossRef]
- Salas-Lloret, D.; García-Rodríguez, N.; Soto-Hidalgo, E.; González-Vinceiro, L.; Espejo-Serrano, C.; Giebel, L.; Mateos-Martín, M.L.; de Ru, A.H.; van Veelen, P.A.; Huertas, P.; et al. BRCA1/BARD1 ubiquitinates PCNA in unperturbed conditions to promote continuous DNA synthesis. Nat Commun 2024, 15, 4292. [CrossRef]
- Salas-Lloret, D.; García-Rodríguez, N.; Soto-Hidalgo, E.; González-Vinceiro, L.; Espejo-Serrano, C.; Giebel, L.; Mateos-Martín, M.L.; de Ru, A.H.; van Veelen, P.A.; Huertas, P.; et al. BRCA1/BARD1 ubiquitinates PCNA in unperturbed conditions to promote continuous DNA synthesis. Nature Communications 2024, 15, 4292. [CrossRef]
- van de Kooij, B.; Schreuder, A.; Pavani, R.S.; Garzero, V.; Van Hoeck, A.; San Martin Alonso, M.; Koerse, D.; Wendel, T.J.; Callen, E.; Boom, J.; et al. EXO1-mediated DNA repair by single-strand annealing is essential for BRCA1-deficient cells. bioRxiv 2023, 2023.2002.2024.529205. [CrossRef]
- García-Rodríguez, N.; Domínguez-García, I.; Domínguez-Pérez, M.D.C.; Huertas, P. EXO1 and DNA2-mediated ssDNA gap expansion is essential for ATR activation and to maintain viability in BRCA1-deficient cells. Nucleic Acids Res 2024, 52, 6376-6391. [CrossRef]
- Howard, S.M.; Ceppi, I.; Anand, R.; Geiger, R.; Cejka, P. The internal region of CtIP negatively regulates DNA end resection. Nucleic Acids Res 2020, 48, 5485-5498. [CrossRef]
- Patterson-Fortin, J.; D'Andrea, A.D. Exploiting the Microhomology-Mediated End-Joining Pathway in Cancer Therapy. Cancer Res 2020, 80, 4593-4600. [CrossRef]
- Zhao, F.; Kim, W.; Kloeber, J.A.; Lou, Z. DNA end resection and its role in DNA replication and DSB repair choice in mammalian cells. Experimental & Molecular Medicine 2020, 52, 1705-1714. [CrossRef]
- Halder, S.; Sanchez, A.; Ranjha, L.; Reginato, G.; Ceppi, I.; Acharya, A.; Anand, R.; Cejka, P. Double-stranded DNA binding function of RAD51 in DNA protection and its regulation by BRCA2. Molecular Cell 2022, 82, 3553-3565.e3555. [CrossRef]
- Bunting, S.F.; Callén, E.; Kozak, M.L.; Kim, J.M.; Wong, N.; López-Contreras, A.J.; Ludwig, T.; Baer, R.; Faryabi, R.B.; Malhowski, A.; et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell 2012, 46, 125-135. [CrossRef]
- Nimonkar, A.V.; Genschel, J.; Kinoshita, E.; Polaczek, P.; Campbell, J.L.; Wyman, C.; Modrich, P.; Kowalczykowski, S.C. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 2011, 25, 350-362. [CrossRef]
- Xue, C.; Greene, E.C. DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing. Trends Genet 2021, 37, 639-656. [CrossRef]
- Bhargava, R.; Onyango, D.O.; Stark, J.M. Regulation of Single-Strand Annealing and its Role in Genome Maintenance. Trends Genet 2016, 32, 566-575. [CrossRef]
- van de Kooij, B.; Schreuder, A.; Pavani, R.; Garzero, V.; Uruci, S.; Wendel, T.J.; van Hoeck, A.; San Martin Alonso, M.; Everts, M.; Koerse, D.; et al. EXO1 protects BRCA1-deficient cells against toxic DNA lesions. Molecular Cell 2024, 84, 659-674.e657. [CrossRef]
- Jaiswal, A.S.; Kim, H.S.; Schärer, O.D.; Sharma, N.; Williamson, E.A.; Srinivasan, G.; Phillips, L.; Kong, K.; Arya, S.; Misra, A.; et al. EEPD1 promotes repair of oxidatively-stressed replication forks. NAR Cancer 2023, 5, zcac044. [CrossRef]
- Wu, Y.; Lee, S.H.; Williamson, E.A.; Reinert, B.L.; Cho, J.H.; Xia, F.; Jaiswal, A.S.; Srinivasan, G.; Patel, B.; Brantley, A.; et al. EEPD1 Rescues Stressed Replication Forks and Maintains Genome Stability by Promoting End Resection and Homologous Recombination Repair. PLoS Genet 2015, 11, e1005675. [CrossRef]
- Teleanu, D.M.; Niculescu, A.G.; Lungu, II; Radu, C.I.; Vladâcenco, O.; Roza, E.; Costăchescu, B.; Grumezescu, A.M.; Teleanu, R.I. An Overview of Oxidative Stress, Neuroinflammation, and Neurodegenerative Diseases. Int J Mol Sci 2022, 23. [CrossRef]
- Fleming, A.M.; Zhu, J.; Ding, Y.; Burrows, C.J. 8-Oxo-7,8-dihydroguanine in the Context of a Gene Promoter G-Quadruplex Is an On–Off Switch for Transcription. ACS Chemical Biology 2017, 12, 2417-2426. [CrossRef]
- Sugden, K.D.; Martin, B.D. Guanine and 7,8-dihydro-8-oxo-guanine-specific oxidation in DNA by chromium(V). Environ Health Perspect 2002, 110 Suppl 5, 725-728. [CrossRef]
- Andrs, M.; Stoy, H.; Boleslavska, B.; Chappidi, N.; Kanagaraj, R.; Nascakova, Z.; Menon, S.; Rao, S.; Oravetzova, A.; Dobrovolna, J.; et al. Excessive reactive oxygen species induce transcription-dependent replication stress. Nat Commun 2023, 14, 1791. [CrossRef]
- Yoshizawa-Sugata, N.; Masai, H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem 2007, 282, 2729-2740. [CrossRef]
- Maynard, S.; Schurman, S.H.; Harboe, C.; de Souza-Pinto, N.C.; Bohr, V.A. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis 2009, 30, 2-10. [CrossRef]
- Thompson, P.S.; Cortez, D. New insights into abasic site repair and tolerance. DNA Repair (Amst) 2020, 90, 102866. [CrossRef]
- Rajan, A.; Varghese, G.R.; Yadev, I.; Anandan, J.; Latha, N.R.; Patra, D.; Krishnan, N.; Kuppusamy, K.; Warrier, A.V.; Bhushan, S.; et al. Modulation of BRCA1 mediated DNA damage repair by deregulated ER-α signaling in breast cancers. Am J Cancer Res 2022, 12, 17-47.
- Kim, H.S.; Nickoloff, J.A.; Wu, Y.; Williamson, E.A.; Sidhu, G.S.; Reinert, B.L.; Jaiswal, A.S.; Srinivasan, G.; Patel, B.; Kong, K.; et al. Endonuclease EEPD1 Is a Gatekeeper for Repair of Stressed Replication Forks. J Biol Chem 2017, 292, 2795-2804. [CrossRef]
- Awate, S.; Sommers, J.A.; Datta, A.; Nayak, S.; Bellani, M.A.; Yang, O.; Dunn, C.A.; Nicolae, C.M.; Moldovan, G.L.; Seidman, M.M.; et al. FANCJ compensates for RAP80 deficiency and suppresses genomic instability induced by interstrand cross-links. Nucleic Acids Res 2020, 48, 9161-9180. [CrossRef]
- Awate, S.; Sommers, J.A.; Datta, A.; Nayak, S.; Bellani, M.A.; Yang, O.; Dunn, C.A.; Nicolae, C.M.; Moldovan, G.-L.; Seidman, M.M.; et al. FANCJ compensates for RAP80 deficiency and suppresses genomic instability induced by interstrand cross-links. Nucleic Acids Research 2020, 48, 9161-9180. [CrossRef]
- Clark, D.W.; Tripathi, K.; Dorsman, J.C.; Palle, K. FANCJ protein is important for the stability of FANCD2/FANCI proteins and protects them from proteasome and caspase-3 dependent degradation. Oncotarget 2015, 6, 28816-28832. [CrossRef]
- Bogliolo, M.; Surrallés, J. The Fanconi Anemia/BRCA Pathway: FANCD2 at the Crossroad between Repair and Checkpoint Responses to DNA Damage. 2006.
- Fang, C.B.; Wu, H.T.; Zhang, M.L.; Liu, J.; Zhang, G.J. Fanconi Anemia Pathway: Mechanisms of Breast Cancer Predisposition Development and Potential Therapeutic Targets. Front Cell Dev Biol 2020, 8, 160. [CrossRef]
- Cong, K.; MacGilvary, N.; Lee, S.; MacLeod, S.G.; Calvo, J.; Peng, M.; Nedergaard Kousholt, A.; Day, T.A.; Cantor, S.B. FANCJ promotes PARP1 activity during DNA replication that is essential in BRCA1 deficient cells. Nat Commun 2024, 15, 2599. [CrossRef]
- Cong, K.; MacGilvary, N.; Lee, S.; MacLeod, S.G.; Calvo, J.; Peng, M.; Kousholt, A.N.; Day, T.; Cantor, S.B. FANCJ promotes PARP1 activity during DNA replication that is essential in BRCA1 deficient cells. bioRxiv 2024. [CrossRef]
- Yu, W.; Lescale, C.; Babin, L.; Bedora-Faure, M.; Lenden-Hasse, H.; Baron, L.; Demangel, C.; Yelamos, J.; Brunet, E.; Deriano, L. Repair of G1 induced DNA double-strand breaks in S-G2/M by alternative NHEJ. Nat Commun 2020, 11, 5239. [CrossRef]
- Lim, K.S.; Li, H.; Roberts, E.A.; Gaudiano, E.F.; Clairmont, C.; Sambel, L.A.; Ponnienselvan, K.; Liu, J.C.; Yang, C.; Kozono, D.; et al. USP1 Is Required for Replication Fork Protection in BRCA1-Deficient Tumors. Mol Cell 2018, 72, 925-941.e924. [CrossRef]
- García-Santisteban, I.; Peters, G.J.; Giovannetti, E.; Rodríguez, J.A. USP1 deubiquitinase: cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Molecular Cancer 2013, 12, 91. [CrossRef]
- Dharadhar, S.; Clerici, M.; van Dijk, W.J.; Fish, A.; Sixma, T.K. A conserved two-step binding for the UAF1 regulator to the USP12 deubiquitinating enzyme. J Struct Biol 2016, 196, 437-447. [CrossRef]
- Murai, J.; Yang, K.; Dejsuphong, D.; Hirota, K.; Takeda, S.; D'Andrea, A.D. The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Mol Cell Biol 2011, 31, 2462-2469. [CrossRef]
- Edmunds, C.E.; Simpson, L.J.; Sale, J.E. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell 2008, 30, 519-529. [CrossRef]
- Ghosal, G.; Chen, J. DNA damage tolerance: a double-edged sword guarding the genome. Translational Cancer Research 2013, 2, 107-129.
- Nicolae, C.M.; Aho, E.R.; Vlahos, A.H.S.; Choe, K.N.; De, S.; Karras, G.I.; Moldovan, G.-L. The ADP-ribosyltransferase PARP10/ARTD10 Interacts with Proliferating Cell Nuclear Antigen (PCNA) and Is Required for DNA Damage Tolerance*. Journal of Biological Chemistry 2014, 289, 13627-13637. [CrossRef]
- Venkadakrishnan, J.; Lahane, G.; Dhar, A.; Xiao, W.; Bhat, K.M.; Pandita, T.K.; Bhat, A. Implications of Translesion DNA Synthesis Polymerases on Genomic Stability and Human Health. Mol Cell Biol 2023, 43, 401-425. [CrossRef]
- Dexheimer, T.S.; Rosenthal, A.S.; Liang, Q.; Chen, J.; Villamil, M.A.; Kerns, E.H.; Simeonov, A.; Jadhav, A.; Zhuang, Z.; Maloney, D.J. Discovery of ML323 as a Novel Inhibitor of the USP1/UAF1 Deubiquitinase Complex. In Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information (US): Bethesda (MD), 2010.
- Tung, N.M.; Garber, J.E. BRCA1/2 testing: therapeutic implications for breast cancer management. British Journal of Cancer 2018, 119, 141-152. [CrossRef]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nature Reviews Cancer 2012, 12, 68-78. [CrossRef]
- Arun, B.; Couch, F.J.; Abraham, J.; Tung, N.; Fasching, P.A. BRCA-mutated breast cancer: the unmet need, challenges and therapeutic benefits of genetic testing. British Journal of Cancer 2024, 131, 1400-1414. [CrossRef]
- Petrucelli, N.; Daly, M.B.; Pal, T. BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer. In GeneReviews(®), Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington, Seattle. Copyright © 1993-2024, University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved.: Seattle (WA), 1993.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




