Submitted:
27 January 2025
Posted:
28 January 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Metabolite Extraction Methods
2.3. UPLC-MS/MS Conditions
2.4. Metabolite Profiling
2.5. RNA-seq
2.6. Differential Gene Analysis
2.7. Weighted Gene Co-Expression Network Analysis
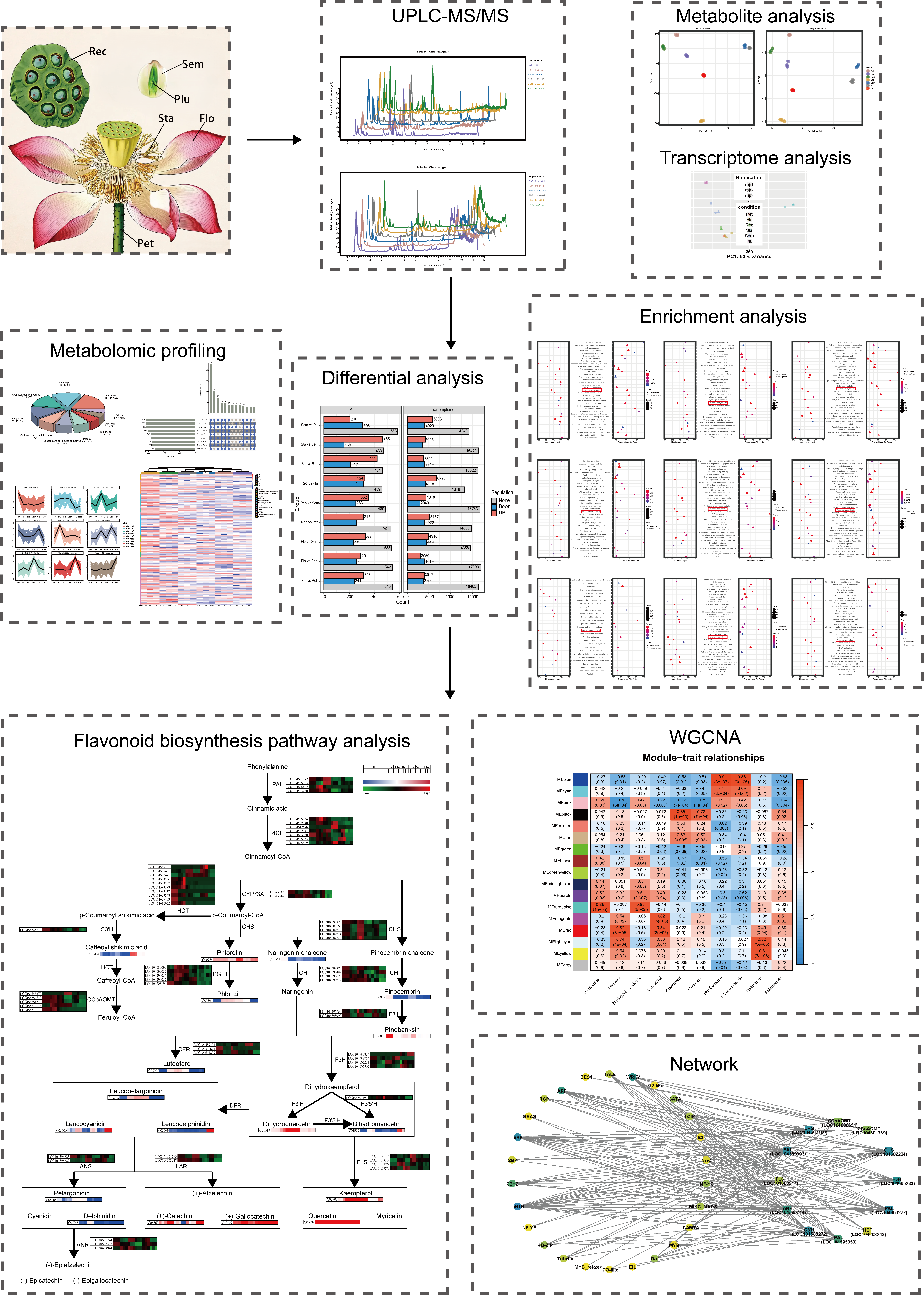
3. Results
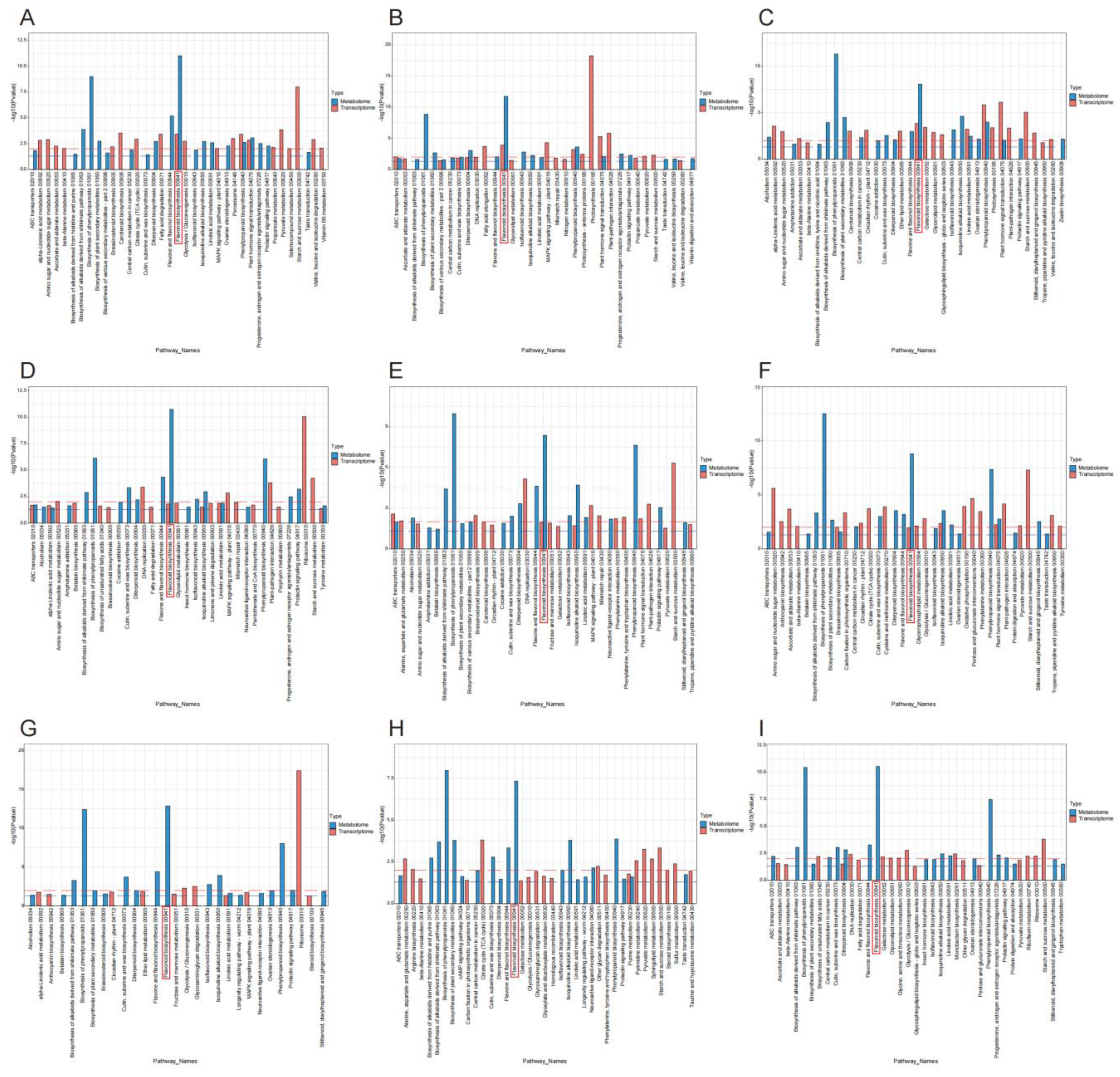
3.1. Differential Analysis of Secondary Metabolism in Lotus Tissues
3.2. Transcriptome Analysis of Lotus Tissues
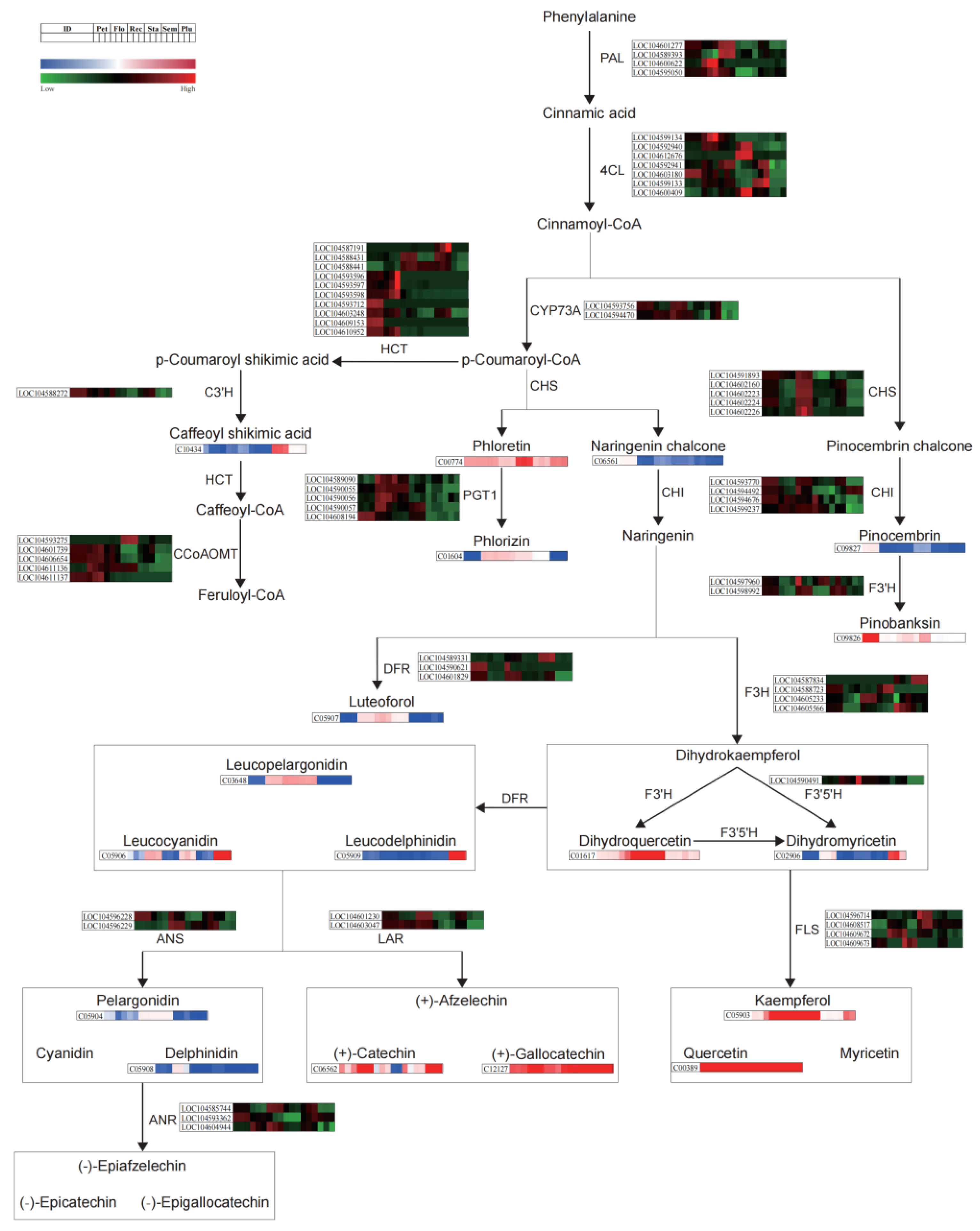
3.3. Joint Metabolomic and Transcriptomic Analysis of Lotus Tissues
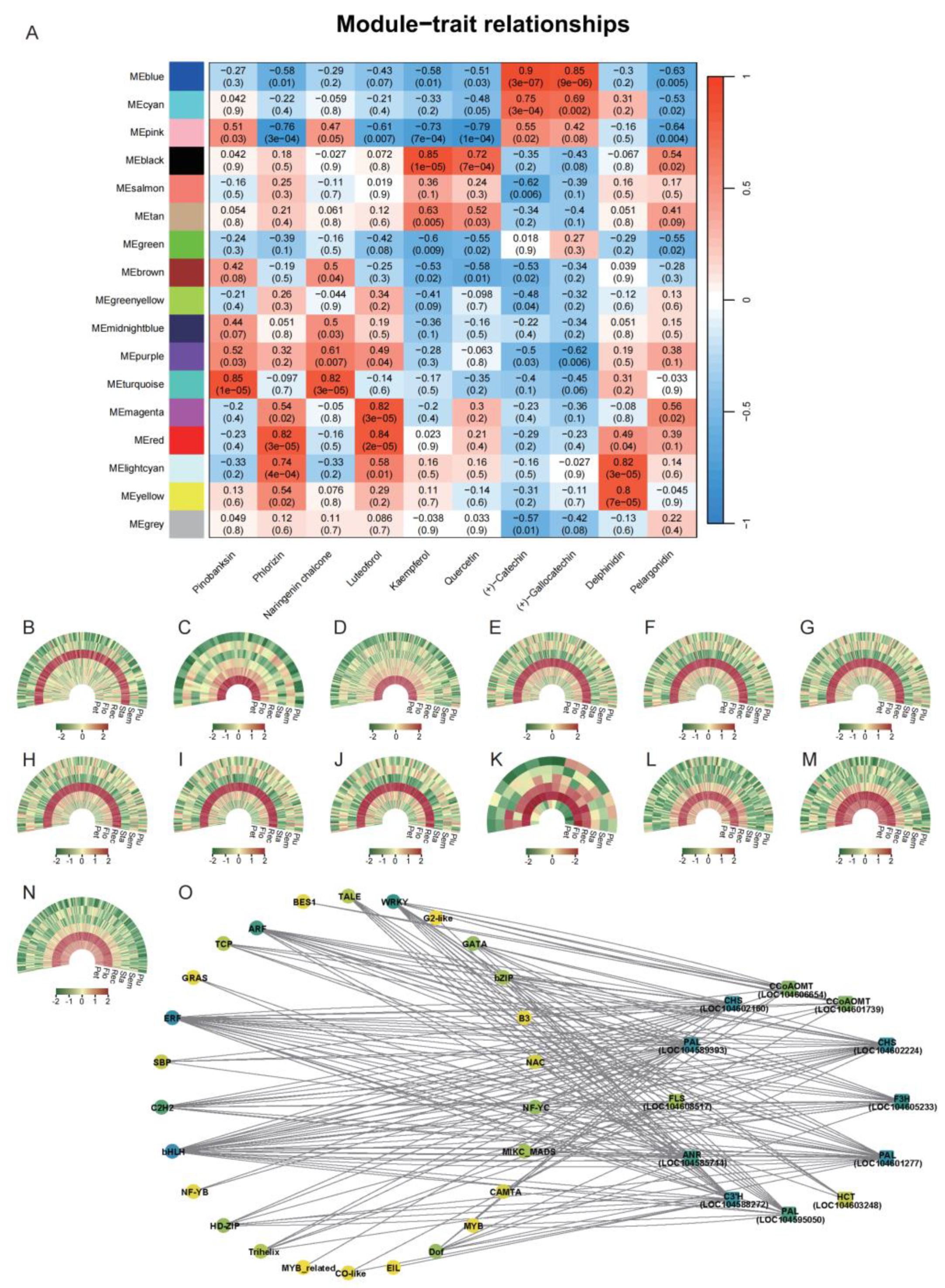
3.4. WGCNA of DEGs in Lotus Tissues
Discussion
4.1. Relationship Between Tissue Specificity and Pharmacological Effects
4.2. Tissue-Specific Regulation of Flavonoid Biosynthetic Pathway
4.3. Regulatory Role of Transcription Factors in Flavonoid Synthesis
4.4. Future Research Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, H. B. Cultivation of Lotus (Nelumbo Nucifera Gaertn. Ssp. Nucifera) and Its Utilization in China. Genet Resour Crop Evol 2009, 56 (3), 323–330. [CrossRef]
- Liu, J.; Chen, S.; Liu, A. Material Basis Analysis of “Homologous and Different Effect” of Lotus Leaf and Lotus Plumule. Chinese Journal of Experimental Traditional Medical Formulae 2020, 131–139. [Google Scholar] [CrossRef]
- National Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China, 2020th ed.; China Medical Science Press, 2020.
- On-nom, N.; Thangsiri, S.; Inthachat, W.; Temviriyanukul, P.; Sahasakul, Y.; Chupeerach, C.; Pruesapan, K.; Trisonthi, P.; Siriwan, D.; Suttisansanee, U. Seasonal Effects on Phenolic Contents and In Vitro Health-Promoting Bioactivities of Sacred Lotus (Nelumbo Nucifera). Plants 2023, 12(7), 1441. [Google Scholar] [CrossRef] [PubMed]
- State Administration of Traditional Chinese Medicine of the People’s Republic of China. Chinese Materia Medica, 1999th ed.; Shanghai Scientific & Technical Publishers, 1999.
- Chen, G.; Zhu, M.; Guo, M. Research Advances in Traditional and Modern Use of Nelumbo Nucifera : Phytochemicals, Health Promoting Activities and Beyond. Critical Reviews in Food Science and Nutrition 2019, 59 (sup1), S189–S209. [Google Scholar] [CrossRef]
- Zheng, H.; Han, L.; Shi, W.; Fang, X.; Hong, Y.; Cao, Y. Research Advances in Lotus Leaf as Chinese Dietary Herbal Medicine. Am. J. Chin. Med. 2022, 50(06), 1423–1445. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, X.; Wu, T.; Li, B.; Liu, T.; Wang, R.; Liu, Q.; Liu, Z.; Gong, Y.; Shao, C. Isoliensinine Induces Apoptosis in Triple-Negative Human Breast Cancer Cells through ROS Generation and P38 MAPK/JNK Activation. Sci Rep 2015, 5(1), 12579. [Google Scholar] [CrossRef]
- Temviriyanukul, P.; Sritalahareuthai, V.; Promyos, N.; Thangsiri, S.; Pruesapan, K.; Srinuanchai, W.; Nuchuchua, O.; Siriwan, D.; On-nom, N.; Suttisansanee, U. The Effect of Sacred Lotus (Nelumbo Nucifera) and Its Mixtures on Phenolic Profiles, Antioxidant Activities, and Inhibitions of the Key Enzymes Relevant to Alzheimer’s Disease. Molecules 2020, 25(16), 3713. [Google Scholar] [CrossRef]
- Kashiwada, Y.; Aoshima, A.; Ikeshiro, Y.; Chen, Y.-P.; Furukawa, H.; Itoigawa, M.; Fujioka, T.; Mihashi, K.; Cosentino, L. M.; Morris-Natschke, S. L.; Lee, K.-H. Anti-HIV Benzylisoquinoline Alkaloids and Flavonoids from the Leaves of Nelumbo Nucifera, and Structure–Activity Correlations with Related Alkaloids. Bioorganic & Medicinal Chemistry 2005, 13 (2), 443–448. [CrossRef]
- Borgi, W.; Ghedira, K.; Chouchane, N. Antiinflammatory and Analgesic Activities of Zizyphus Lotus Root Barks. Fitoterapia 2007, 78(1), 16–19. [Google Scholar] [CrossRef]
- Gao, Z.; Liang, Y.; Wang, Y.; Xiao, Y.; Chen, J.; Yang, X.; Shi, T. Genome-Wide Association Study of Traits in Sacred Lotus Uncovers MITE-Associated Variants Underlying Stamen Petaloid and Petal Number Variations. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef]
- Zheng, P.; Sun, H.; Liu, J.; Lin, J.; Zhang, X.; Qin, Y.; Zhang, W.; Xu, X.; Deng, X.; Yang, D.; Wang, M.; Zhang, Y.; Song, H.; Huang, Y.; Orozco-Obando, W.; Ming, R.; Yang, M. Comparative Analyses of American and Asian Lotus Genomes Reveal Insights into Petal Color, Carpel Thermogenesis and Domestication. The Plant Journal 2022, 110(5), 1498–1515. [Google Scholar] [CrossRef] [PubMed]
- Mao, T.-Y.; Liu, Y.-Y.; Zhu, H.-H.; Zhang, J.; Yang, J.-X.; Fu, Q.; Wang, N.; Wang, Z. Genome-Wide Analyses of the bHLH Gene Family Reveals Structural and Functional Characteristics in the Aquatic Plant Nelumbo Nucifera. PeerJ 2019, 7, e7153. [Google Scholar] [CrossRef]
- Qi, H.; Yu, F.; Lü, S.; Damaris, R. N.; Dong, G.; Yang, P. Exploring Domestication Pattern in Lotus: Insights from Dispensable Genome Assembly. Front. Plant Sci. 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Vasilev, N.; Boccard, J.; Lang, G.; Grömping, U.; Fischer, R.; Goepfert, S.; Rudaz, S.; Schillberg, S. Structured Plant Metabolomics for the Simultaneous Exploration of Multiple Factors. Sci Rep 2016, 6(1), 37390. [Google Scholar] [CrossRef] [PubMed]
- Want, E. J.; Masson, P.; Michopoulos, F.; Wilson, I. D.; Theodoridis, G.; Plumb, R. S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J. K. Global Metabolic Profiling of Animal and Human Tissues via UPLC-MS. Nat Protoc 2013, 8(1), 17–32. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, D. A.; Piccolo, B. D.; Vaziri, N. D.; Liu, S.; Lau, W. L.; Khazaeli, M.; Nazertehrani, S.; Moore, M. E.; Marco, M. L.; Martin, R. J.; Adams, S. H. Resistant Starch Alters Gut Microbiome and Metabolomic Profiles Concurrent with Amelioration of Chronic Kidney Disease in Rats. American Journal of Physiology-Renal Physiology 2016, 310 (9), F857–F871. [CrossRef]
- He, X.-Y.; Wu, L.-J.; Wang, W.-X.; Xie, P.-J.; Chen, Y.-H.; Wang, F. Amygdalin - A Pharmacological and Toxicological Review. Journal of Ethnopharmacology 2020, 254, 112717. [Google Scholar] [CrossRef] [PubMed]
- Tauber, J. P.; Tozkar, C. Ö.; Schwarz, R. S.; Lopez, D.; Irwin, R. E.; Adler, L. S.; Evans, J. D. Colony-Level Effects of Amygdalin on Honeybees and Their Microbes. Insects 2020, 11(11), 783. [Google Scholar] [CrossRef]
- London-Shafir, I.; Shafir, S.; Eisikowitch, D. Amygdalin in Almond Nectar and Pollen – Facts and Possible Roles. Plant Syst Evol 2003, 238(1), 87–95. [Google Scholar] [CrossRef]
- Li, R.; Zhang, Q.; Liu, J.; He, L.; Huang, Q.; Peng, W.; Wu, C. Processing Methods and Mechanisms for Alkaloid-Rich Chinese Herbal Medicines: A Review - ScienceDirect. [CrossRef]
- Zhao, C.; Liu, X.; Gong, Q.; Cao, J.; Shen, W.; Yin, X.; Grierson, D.; Zhang, B.; Xu, C.; Li, X.; Chen, K.; Sun, C. Three AP2/ERF Family Members Modulate Flavonoid Synthesis by Regulating Type IV Chalcone Isomerase in Citrus. Plant Biotechnology Journal 2021, 19(4), 671–688. [Google Scholar] [CrossRef] [PubMed]
- Guodong, R.; Jianguo, Z.; Xiaoxia, L.; Ying, L. Identification of Putative Genes for Polyphenol Biosynthesis in Olive Fruits and Leaves Using Full-Length Transcriptome Sequencing. Food Chemistry 2019, 300, 125246. [Google Scholar] [CrossRef]
- Burin, T.; Grohar, M. C.; Jakopic, J.; Veberic, R.; Stajner, N.; Cesar, T.; Kunej, U.; Hudina, M. Interplay of Phenolic Compounds and Gene Expression in Phenylpropanoid and Flavonoid Pathways during Olive (Olea Europaea L. ) Ripening of ‘Leccino’ Cultivar. Scientia Horticulturae 2024, 338, 113640. [Google Scholar] [CrossRef]
- Nabavi, S. M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; Giampieri, F.; Battino, M.; Sobarzo-Sanchez, E.; Nabavi, S. F.; Yousefi, B.; Jeandet, P.; Xu, S.; Shirooie, S. Flavonoid Biosynthetic Pathways in Plants: Versatile Targets for Metabolic Engineering. Biotechnology Advances 2020, 38, 107316. [Google Scholar] [CrossRef] [PubMed]
- Jariani, P.; Shahnejat-Bushehri, A.-A.; Naderi, R.; Zargar, M.; Naghavi, M. R. Characterization of Key Genes in Anthocyanin and Flavonoid Biosynthesis during Floral Development in Rosa Canina L. International Journal of Biological Macromolecules 2024, 276, 133937. [Google Scholar] [CrossRef] [PubMed]
- Silva, G. dos S. e; Marques, J. N. de J.; Linhares, E. P. M.; Bonora, C. M.; Costa, É. T.; Saraiva, M. F. Review of Anticancer Activity of Monoterpenoids: Geraniol, Nerol, Geranial and Neral. Chemico-Biological Interactions 2022, 362, 109994. [Google Scholar] [CrossRef]
- Hong, P.; Liu, Q.-W.; Xie, Y.; Zhang, Q.-H.; Liao, L.; He, Q.-Y.; Li, B.; Xu, W. W. Echinatin Suppresses Esophageal Cancer Tumor Growth and Invasion through Inducing AKT/mTOR-Dependent Autophagy and Apoptosis. Cell Death Dis 2020, 11(7), 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Liu, C.; Jiang, H.-L.; Zhang, Y.; Han, J.; Liu, J.; Chen, M. Cardioprotection Provided by Echinatin against Ischemia/Reperfusion in Isolated Rat Hearts. BMC Cardiovasc Disord 2016, 16(1), 119. [Google Scholar] [CrossRef]
- Zhang, C.; Lin, J.; Zhen, C.; Wang, F.; Sun, X.; Kong, X.; Gao, Y. Amygdalin Protects against Acetaminophen-Induced Acute Liver Failure by Reducing Inflammatory Response and Inhibiting Hepatocyte Death. Biochemical and Biophysical Research Communications 2022, 602, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Chen, Q.; Xu, M.; Xia, Q.; Zheng, T.; Teng, J.; Li, M.; Fan, L. Recent Updates and Future Perspectives about Amygdalin as a Potential Anticancer Agent: A Review. Cancer Medicine 2019, 8(6), 3004–3011. [Google Scholar] [CrossRef]
- Liang, M.; Li, X.; Ouyang, X.; Xie, H.; Chen, D. Antioxidant Mechanisms of Echinatin and Licochalcone A. Molecules 2019, 24(1), 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Y.; Shen, Y.; Zhu, C.; Qin, X.; Gao, Y. Liquidambaric Acid Inhibits Cholangiocarcinoma Progression by Disrupting the STAMBPL1/NRF2 Positive Feedback Loop. Phytomedicine 2024, 156303. [Google Scholar] [CrossRef]
- Zhao, X.; Zhu, X.; Tao, H.; Zou, H.; Cao, J.; Chen, Y.; Zhang, Z.; Zhu, Y.; Li, Q.; Li, M. Liquidambaric Acid Inhibits the Proliferation of Hepatocellular Carcinoma Cells by Targeting PPARα-RXRα to down-Regulate Fatty Acid Metabolism. Toxicology and Applied Pharmacology 2024, 490, 117042. [Google Scholar] [CrossRef]
- Baczewska-Dąbrowska, A. H.; Dmuchowski, W.; Gozdowski, D.; Gworek, B.; Jozwiak, A.; Swiezewska, E.; Dąbrowski, P.; Suwara, I. The Importance of Prenol Lipids in Mitigating Salt Stress in the Leaves of Tilia × Euchlora Trees. Trees 2022, 36(1), 393–404. [Google Scholar] [CrossRef]
- Ren, C.; Wang, J.; Xian, B.; Tang, X.; Liu, X.; Hu, X.; Hu, Z.; Wu, Y.; Chen, C.; Wu, Q.; Chen, J.; Pei, J. Transcriptome Analysis of Flavonoid Biosynthesis in Safflower Flowers Grown under Different Light Intensities. PeerJ 2020, 8, e8671. [Google Scholar] [CrossRef]
- Elbatreek, M. H.; Mahdi, I.; Ouchari, W.; Mahmoud, M. F.; Sobeh, M. Current Advances on the Therapeutic Potential of Pinocembrin: An Updated Review. Biomedicine & Pharmacotherapy 2023, 157, 114032. [Google Scholar] [CrossRef]
- Kim, Y. W.; Zhao, R. J.; Park, S. J.; Lee, J. R.; Cho, I. J.; Yang, C. H.; Kim, S. G.; Kim, S. C. Anti-Inflammatory Effects of Liquiritigenin as a Consequence of the Inhibition of NF-κB-Dependent iNOS and Proinflammatory Cytokines Production. British Journal of Pharmacology 2008, 154(1), 165–173. [Google Scholar] [CrossRef] [PubMed]
- Gaur, R.; Yadav, K. S.; Verma, R. K.; Yadav, N. P.; Bhakuni, R. S. In Vivo Anti-Diabetic Activity of Derivatives of Isoliquiritigenin and Liquiritigenin. Phytomedicine 2014, 21(4), 415–422. [Google Scholar] [CrossRef]
- Ramalingam, M.; Kim, H.; Lee, Y.; Lee, Y.-I. Phytochemical and Pharmacological Role of Liquiritigenin and Isoliquiritigenin From Radix Glycyrrhizae in Human Health and Disease Models. Front. Aging Neurosci. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Huang, H.; Zhao, M.; Sun-Waterhouse, D.; Lin, L.; Xiao, C. Mechanisms Underlying the Xanthine Oxidase Inhibitory Effects of Dietary Flavonoids Galangin and Pinobanksin. Journal of Functional Foods 2016, 24, 26–36. [Google Scholar] [CrossRef]
- Elangovan, B. A Review on Pharmacological Studies of Natural Flavanone: Pinobanksin. 3 Biotech 2024, 14(4), 111. [Google Scholar] [CrossRef] [PubMed]
- Escribano-Ferrer, E.; Queralt Regué, J.; Garcia-Sala, X.; Boix Montañés, A.; Lamuela-Raventos, R. M. In Vivo Anti-Inflammatory and Antiallergic Activity of Pure Naringenin, Naringenin Chalcone, and Quercetin in Mice. J. Nat. Prod. 2019, 82(2), 177–182. [Google Scholar] [CrossRef]
- Sunil, C.; Xu, B. An Insight into the Health-Promoting Effects of Taxifolin (Dihydroquercetin). Phytochemistry 2019, 166, 112066. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qiu, D.; Yang, T.; Su, J.; Liu, C.; Su, X.; Li, A.; Sun, P.; Li, J.; Yan, L.; Ding, C.; Zhang, S. Research Progress of Dihydroquercetin in the Treatment of Skin Diseases. Molecules 2023, 28(19), 6989. [Google Scholar] [CrossRef]
- Shevelev, A. B.; La Porta, N.; Isakova, E. P.; Martens, S.; Biryukova, Y. K.; Belous, A. S.; Sivokhin, D. A.; Trubnikova, E. V.; Zylkova, M. V.; Belyakova, A. V.; Smirnova, M. S.; Deryabina, Y. I. In Vivo Antimicrobial and Wound-Healing Activity of Resveratrol, Dihydroquercetin, and Dihydromyricetin against Staphylococcus Aureus, Pseudomonas Aeruginosa, and Candida Albicans. Pathogens 2020, 9(4), 296. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, R.; Shi, W.; Li, L.; Liu, H.; Liu, Z.; Wu, L. The Multifunctional Benefits of Naturally Occurring Delphinidin and Its Glycosides. J. Agric. Food Chem. 2019, 67(41), 11288–11306. [Google Scholar] [CrossRef]
- Lamy, S.; Blanchette, M.; Michaud-Levesque, J.; Lafleur, R.; Durocher, Y.; Moghrabi, A.; Barrette, S.; Gingras, D.; Béliveau, R. Delphinidin, a Dietary Anthocyanidin, Inhibits Vascular Endothelial Growth Factor Receptor-2 Phosphorylation. Carcinogenesis 2006, 27(5), 989–996. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, B.; Sun, S.; Liu, Q.; Zhu, J.; Zhou, X.; Zhang, H.; Wu, Q.; Wang, L. Analysis of Proanthocyanidins and Flavonols in the Seedpods of Chinese Antique Lotus: A Rich Source of Antioxidants. Food Chemistry 2023, 415, 135756. [Google Scholar] [CrossRef] [PubMed]
- Proshkina, E.; Plyusnin, S.; Babak, T.; Lashmanova, E.; Maganova, F.; Koval, L.; Platonova, E.; Shaposhnikov, M.; Moskalev, A. Terpenoids as Potential Geroprotectors. Antioxidants 2020, 9(6), 529. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Zhang, X.; Su, Z.; Xiao, J.; Lv, M.; Cao, Y.; Chen, Y. Carnosol Improved Lifespan and Healthspan by Promoting Antioxidant Capacity in Caenorhabditis Elegans. Oxidative Medicine and Cellular Longevity 2019, 2019(1), 5958043. [Google Scholar] [CrossRef]
- Okoro, N. O.; Odiba, A. S.; Osadebe, P. O.; Omeje, E. O.; Liao, G.; Fang, W.; Jin, C.; Wang, B. Bioactive Phytochemicals with Anti-Aging and Lifespan Extending Potentials in Caenorhabditis Elegans. Molecules 2021, 26(23), 7323. [Google Scholar] [CrossRef]
- Arooj, M.; Imran, S.; Inam-ur-Raheem, M.; Rajoka, M. S. R.; Sameen, A.; Siddique, R.; Sahar, A.; Tariq, S.; Riaz, A.; Hussain, A.; Siddeeg, A.; Aadil, R. M. Lotus Seeds (Nelumbinis Semen) as an Emerging Therapeutic Seed: A Comprehensive Review. Food Science & Nutrition 2021, 9 (7), 3971–3987. [CrossRef]
- Wang, Z.; Li, Y.; Ma, D.; Zeng, M.; Wang, Z.; Qin, F.; Chen, J.; Christian, M.; He, Z. Alkaloids from Lotus (Nelumbo Nucifera): Recent Advances in Biosynthesis, Pharmacokinetics, Bioactivity, Safety, and Industrial Applications. Critical Reviews in Food Science and Nutrition 2023, 63(21), 4867–4900. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.-Z.; Chang, Q.; Zhong, Y.; Xiao, B.-X.; Feng, L.; Cao, F.-R.; Pan, R.-L.; Zhang, Z.-S.; Liao, Y.-H.; Liu, X.-M. Lotus Leaf Alkaloid Extract Displays Sedative–Hypnotic and Anxiolytic Effects through GABAA Receptor. J. Agric. Food Chem. 2015, 63(42), 9277–9285. [Google Scholar] [CrossRef]
- Jung, H. A.; Jin, S. E.; Choi, R. J.; Kim, D. H.; Kim, Y. S.; Ryu, J. H.; Kim, D.-W.; Son, Y. K.; Park, J. J.; Choi, J. S. Anti-Amnesic Activity of Neferine with Antioxidant and Anti-Inflammatory Capacities, as Well as Inhibition of ChEs and BACE1. Life Sciences 2010, 87(13), 420–430. [Google Scholar] [CrossRef]
- Martínez-Vázquez, M.; Estrada-Reyes, R.; Araujo Escalona, A. G.; Ledesma Velázquez, I.; Martínez-Mota, L.; Moreno, J.; Heinze, G. Antidepressant-like Effects of an Alkaloid Extract of the Aerial Parts of Annona Cherimolia in Mice. Journal of Ethnopharmacology 2012, 139(1), 164–170. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Hu, T.; Yang, C.; Li, H.; Yang, M.; Ijaz, R.; Ye, Z.; Zhang, Y. Transcriptome Profiling of Tomato Fruit Development Reveals Transcription Factors Associated with Ascorbic Acid, Carotenoid and Flavonoid Biosynthesis. PLOS ONE 2015, 10(7), e0130885. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; Machemer, K.; Braun, E. L.; Grotewold, E. Evolutionary and Comparative Analysis of MYB and bHLH Plant Transcription Factors. The Plant Journal 2011, 66(1), 94–116. [Google Scholar] [CrossRef]
- Pu, Y.; Wang, C.; Jiang, Y.; Wang, X.; Ai, Y.; Zhuang, W. Metabolic Profiling and Transcriptome Analysis Provide Insights into the Accumulation of Flavonoids in Chayote Fruit during Storage. Front. Nutr. 2023, 10. [Google Scholar] [CrossRef]
- Zhou, T.; Xing, Q.; Bu, J.; Han, W.; Shen, Z. Integrated Metabolomic and Transcriptomic Analysis Reveals the Regulatory Mechanisms of Flavonoid and Alkaloid Biosynthesis in the New and Old Leaves of Murraya Tetramera Huang. BMC Plant Biol 2024, 24(1), 499. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Xia, Y.; Jiang, W.; Shen, G.; Pang, Y. GbMYBR1 from Ginkgo Biloba Represses Phenylpropanoid Biosynthesis and Trichome Development in Arabidopsis. Planta 2020, 252(4), 68. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J. F.; Bilbao-Castro, J. R.; Robertson, D. L. Genome-Wide Classification and Evolutionary Analysis of the bHLH Family of Transcription Factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiology 2010, 153(3), 1398–1412. [Google Scholar] [CrossRef]
- Naik, J.; Misra, P.; Trivedi, P. K.; Pandey, A. Molecular Components Associated with the Regulation of Flavonoid Biosynthesis. Plant Science 2022, 317, 111196. [Google Scholar] [CrossRef] [PubMed]
- Nemesio-Gorriz, M.; Blair, P. B.; Dalman, K.; Hammerbacher, A.; Arnerup, J.; Stenlid, J.; Mukhtar, S. M.; Elfstrand, M. Identification of Norway Spruce MYB-bHLH-WDR Transcription Factor Complex Members Linked to Regulation of the Flavonoid Pathway. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, G.; Li, C.; Zhang, C.; Cui, L.; Ai, G.; Wang, X.; Zheng, F.; Zhang, D.; Larkin, R. M.; Ye, Z.; Zhang, J. NF-Y Plays Essential Roles in Flavonoid Biosynthesis by Modulating Histone Modifications in Tomato. New Phytologist 2021, 229(6), 3237–3252. [Google Scholar] [CrossRef] [PubMed]
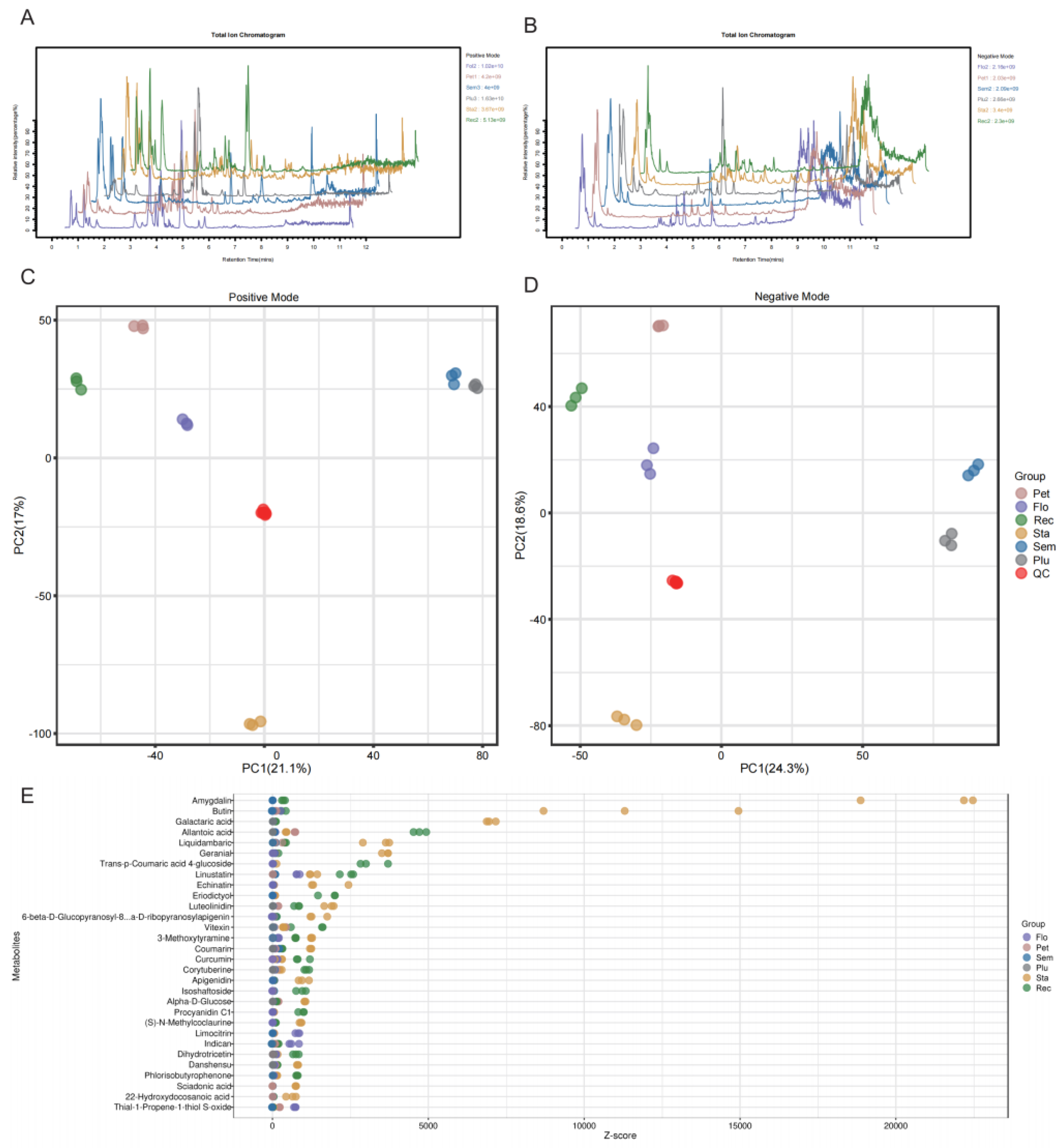
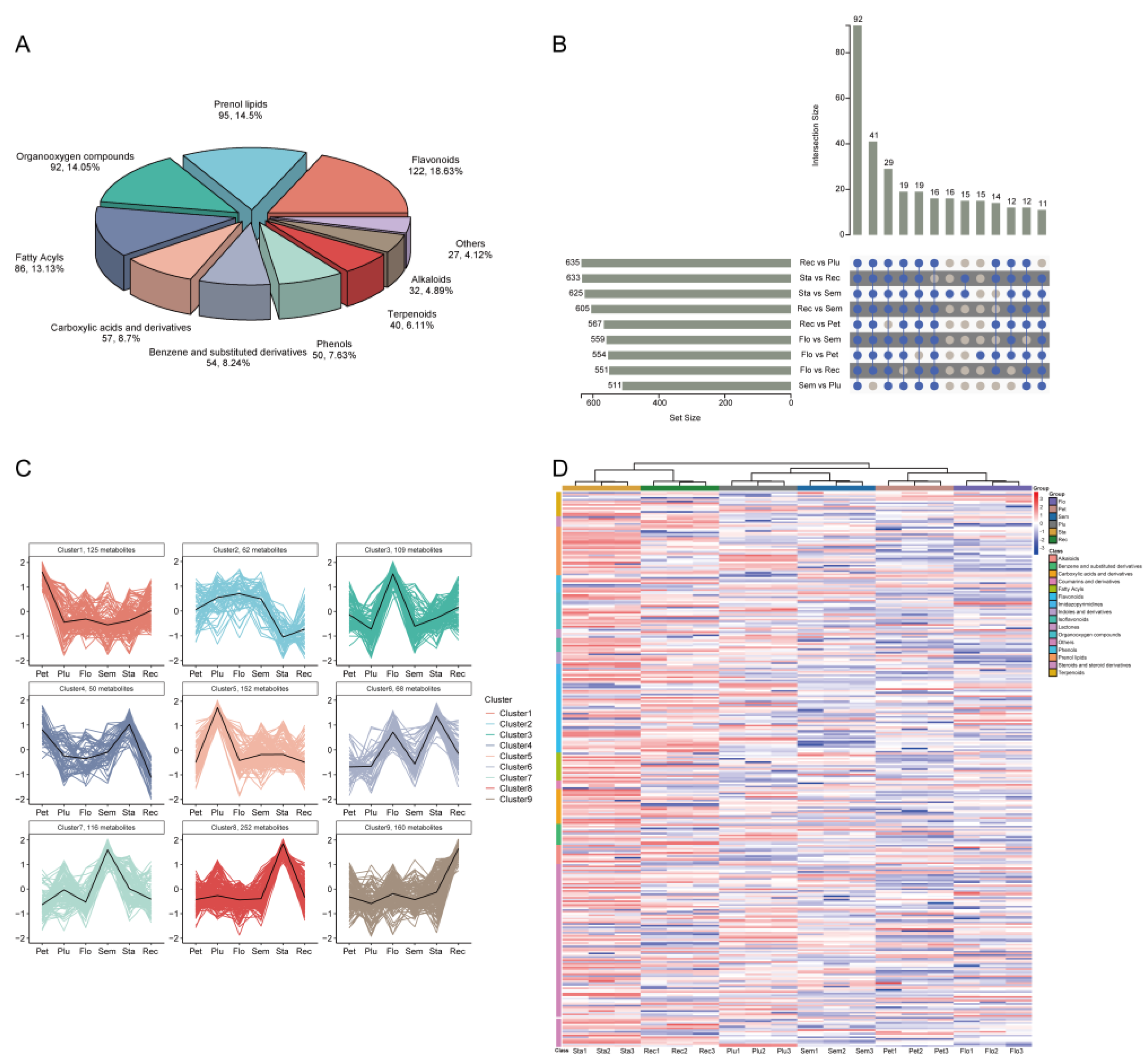
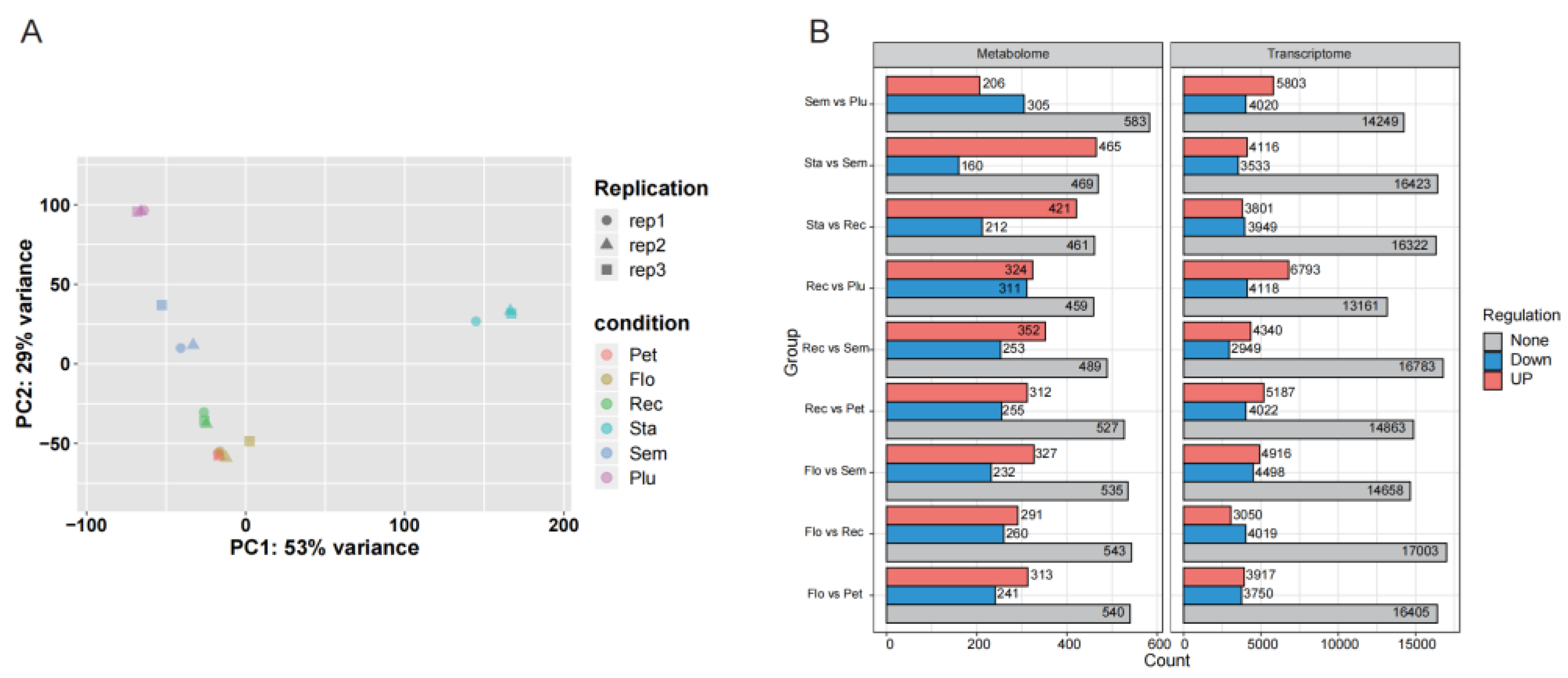

| Sample | Reads No. | Clean Reads No. | Clean Data (bp) | N (%) | Q30 (%) |
| Pet1 | 44796076 | 44195988 | 6663704056 | 0.004196 | 96.85 |
| Pet2 | 45964130 | 45336206 | 6825848171 | 0.004153 | 96.8 |
| Pet3 | 50239706 | 49472494 | 7454090951 | 0.007693 | 96.06 |
| Plu1 | 41582154 | 40895430 | 6151047640 | 0.007237 | 95.59 |
| Plu2 | 46805894 | 46177374 | 6952022595 | 0.007229 | 96.18 |
| Plu3 | 54998098 | 54188514 | 8134266098 | 0.007403 | 95.91 |
| Flo1 | 40858150 | 40235688 | 6066986452 | 0.006309 | 96.87 |
| Flo2 | 41037134 | 40556292 | 6118355186 | 0.006397 | 97.52 |
| Flo3 | 45648060 | 45110000 | 6801259085 | 0.006159 | 97.28 |
| Sem1 | 44918700 | 44284232 | 6680348123 | 0.006197 | 97 |
| Sem2 | 43378918 | 42815524 | 6456737375 | 0.006276 | 97.12 |
| Sem3 | 40897244 | 40409936 | 6090914512 | 0.006272 | 97.33 |
| Sta1 | 38924410 | 38349966 | 5780282858 | 0.006001 | 96.9 |
| Sta2 | 43999936 | 43409676 | 6547853705 | 0.006264 | 97.03 |
| Sta3 | 39146748 | 38586882 | 5817568921 | 0.006328 | 96.9 |
| Rec1 | 39690262 | 39175626 | 5904032544 | 0.006293 | 97.07 |
| Rec2 | 45727084 | 45109178 | 6799917596 | 0.006294 | 97.03 |
| Rec3 | 52071532 | 51358160 | 7742502649 | 0.006194 | 96.98 |
| Total | 800684236 | 789667166 | 1.18988E+11 | - | - |
| Module | Structural genes | Interacting genes number | Strongly correlated flavonoids |
| MEblack | LOC104608517 (FLS) | 154 | kaempferol |
| MEturquoise | LOC104603248 (HCT) | 54 | pinobanksin, naringenin chalcone |
| MEturquoise | LOC104588272 (C3’H) | 199 | pinobanksin, naringenin chalcone |
| MEmagenta | LOC104601277 (PAL) | 189 | luteoforol |
| MEmagenta | LOC104589393 (PAL) | 173 | luteoforol |
| MEmagenta | LOC104602160 (CHS) | 181 | luteoforol |
| MEmagenta | LOC104602224 (CHS) | 178 | luteoforol |
| MEmagenta | LOC104605233 (F3H) | 139 | luteoforol |
| MEmagenta | LOC104585744 (ANR) | 116 | luteoforol |
| MElightcyan | LOC104611136 (CCoAOMT) | 15 | delphinidin |
| MEyellow | LOC104595050 (PAL) | 131 | delphinidin |
| MEyellow | LOC104601739 (CCoAOMT) | 133 | delphinidin |
| MEyellow | LOC104606654 (CCoAOMT) | 198 | delphinidin |
| Structural genes | Interacting transcription factors | Strongly correlated flavonoids |
| LOC104601277 (PAL) | bHLH,ERF,NF-YC | luteoforol |
| LOC104602224 (CHS) | bHLH,ERF,NF-YC | luteoforol |
| LOC104589393 (PAL) | bHLH,ERF,NF-YC | luteoforol |
| LOC104588272 (C3’H) | bHLH,ERF,NF-YC | pinobanksin, naringenin chalcone |
| LOC104585744 (ANR) | bHLH,ERF,NF-YC | luteoforol |
| LOC104595050 (PAL) | bHLH,ERF | delphinidin |
| LOC104608517 (FLS) | bHLH | kaempferol |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





