Submitted:
23 January 2025
Posted:
24 January 2025
Read the latest preprint version here
Abstract
A major challenge in aging research is identifying interventions that can improve lifespan and health and minimize toxicity. Clinical studies cannot consider decades-long follow-up periods, and therefore, in-silico evaluations using omics-based surrogate biomarkers are emerging as key tools. However, many current approaches train predictive models on observational data, rather than on intervention data, which can lead to biased conclusions. Yet, the first classifiers for lifespan extension by compounds are now available, learned on intervention data. Here, we review evaluation methodologies and we prioritize training on intervention data whenever available, highlight the importance of safety and toxicity assessments, discuss the role of standardized benchmarks, and present a range of feature processing and predictive modeling approaches. We consider linear and non-linear methods, and automated machine learning workflows. We conclude by emphasizing the need for explainable and reproducible strategies, the integration of safety metrics, and the careful validation of predictors based on interventional benchmarks.
Keywords:
Introduction
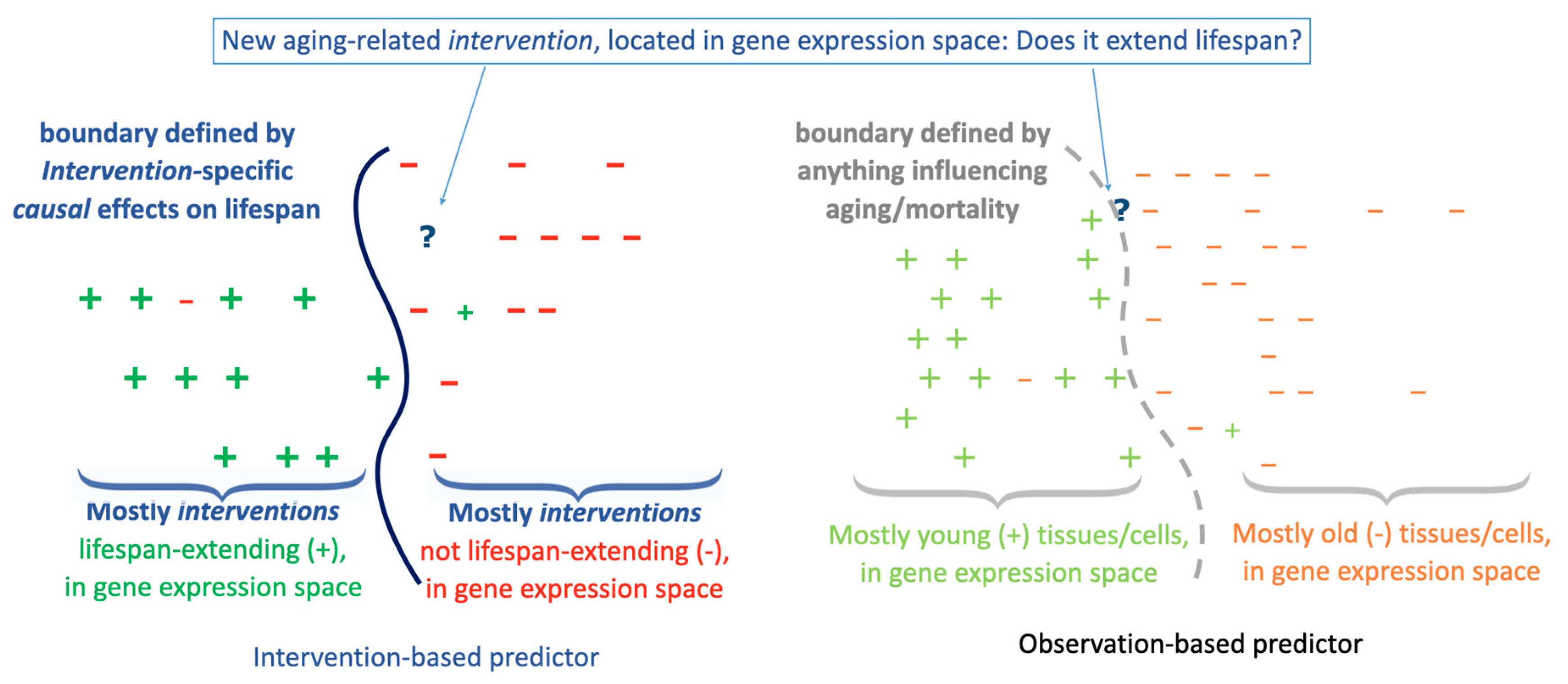
The Challenge of Predicting Intervention Outcomes
The Importance of Safety and Toxicity Assessments
Establishing Benchmarks for Model Comparison
| Benchmark source | Data Type | Intervention Examples | Outcomes | Reference |
|---|---|---|---|---|
| TOXRIC | Transcriptomics + toxicity data | ~2,800 compounds from LINCS/ DrugMatrix/TG-GATEs | Toxicity (acute toxicity such as LD50; genotoxicity such as mutagenicity) | (Wu et al., 2023) |
| DrugAge-based | Gene expression (LINCS) + drug annotations | 56 compounds | Lifespan extension (mouse data) | (Belikov et al., 2024) |
| SIDER-based | Transcriptomics (LINCS) + side effects | 251 compounds | Drug side effects | (Kuhn et al., 2016; Uner et al., 2023; Wang et al., 2016) |
| Tyshkovskiy | Transcriptomics + lifespan | 40 interventions in mice, from ITP and Gene Expression Omnibus | Lifespan effects (extension or shortening) | (Tyshkovskiy et al., 2024) |
| Reprogramming data | Human or mouse data, partial reprogramming | Various partial reprogramming protocols | Various health-related outcomes | (Browder et al., 2022; Hishida et al., 2022; Sarkar et al., 2020) |
| Seno- therapeutic data | Cellular and organismal senotherapy data | Various senotherapeutic/ senolytic compounds | Seno- therapeutic/ senolytic action | (Smer-Barreto et al., 2023) |
| Nutritional interventions | Nutritional data | Various nutritional interventions with mild effects | Various health-related outcomes | (Ford et al., 2023) |
| Gwinn | Rat in-vivo intervention data | Toxic (and non-toxic) compounds | Toxicity outcomes | (Gwinn et al., 2020) |
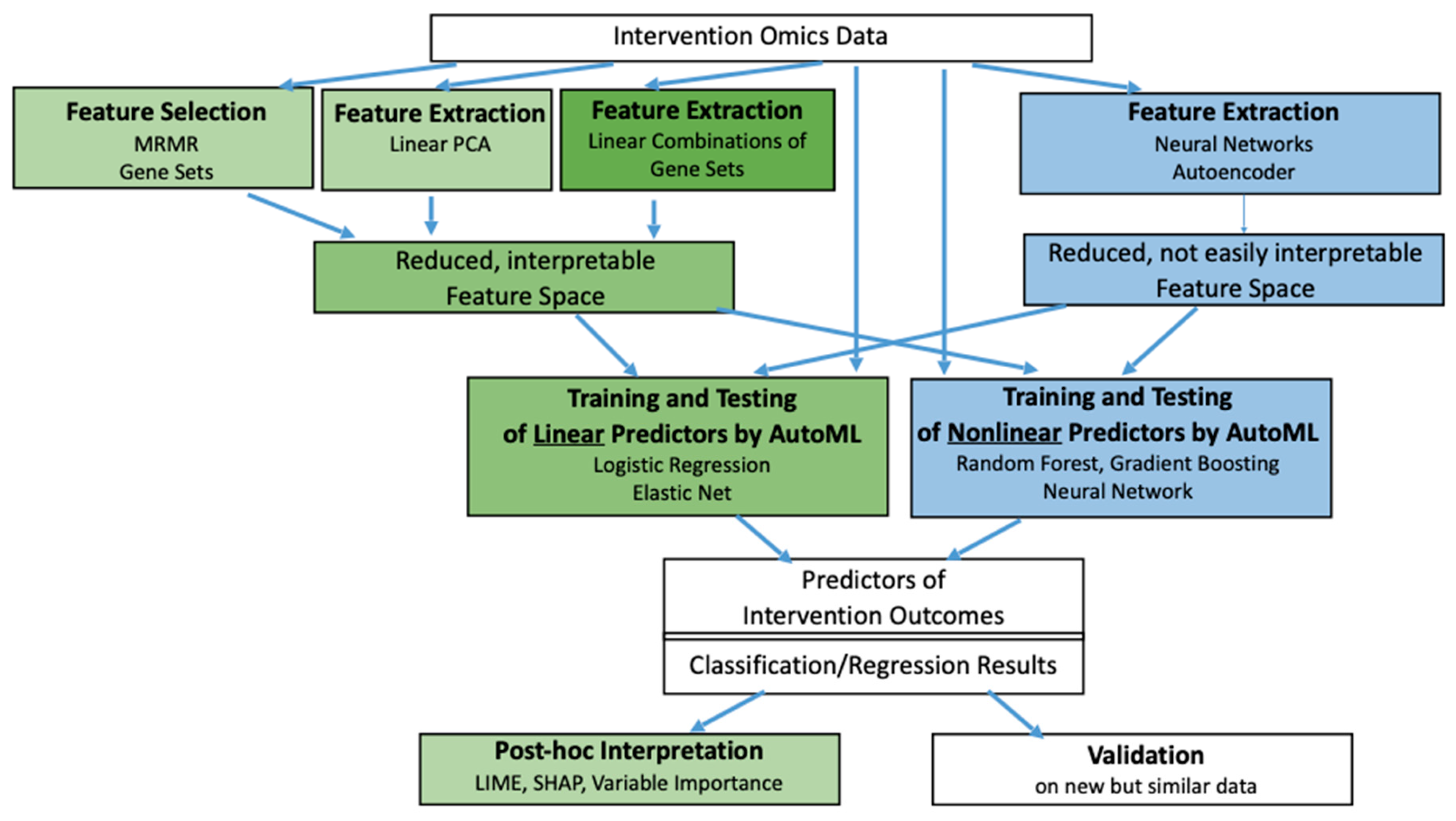
Feature Selection/Extraction and Predictor Learning
Feature Selection/Extraction
Predictor Learning
Early Experiences with Generative AI and LLMs
Perspectives and Future Directions
Conclusions
References
- Abid, A., Zhang, M.J., Bagaria, V.K., Zou, J., 2018. Exploring patterns enriched in a dataset with contrastive principal component analysis. Nat Commun 9, 2134. [CrossRef]
- Basili, D., Reynolds, J., Houghton, J., Malcomber, S., Chambers, B., Liddell, M., Muller, I., White, A., Shah, I., Everett, L.J., Middleton, A., Bender, A., 2022. Latent Variables Capture Pathway-Level Points of Departure in High-Throughput Toxicogenomic Data. Chem Res Toxicol 35, 670-683. [CrossRef]
- Belikov, A.V., Ribeiro, C., Farmer, C.K., de Magalhaes, J.P., Freitas, A.A., 2024. Predicting Mouse Lifespan-Extending Chemical Compounds with Machine Learning. bioRxiv.
- Boileau, P., Hejazi, N.S., Dudoit, S., 2020. Exploring high-dimensional biological data with sparse contrastive principal component analysis. Bioinformatics 36, 3422-3430. [CrossRef]
- Browder, K.C., Reddy, P., Yamamoto, M., Haghani, A., Guillen, I.G., Sahu, S., Wang, C., Luque, Y., Prieto, J., Shi, L., Shojima, K., Hishida, T., Lai, Z., Li, Q., Choudhury, F.K., Wong, W.R., Liang, Y., Sangaraju, D., Sandoval, W., Esteban, C.R., Delicado, E.N., Garcia, P.G., Pawlak, M., Vander Heiden, J.A., Horvath, S., Jasper, H., Izpisua Belmonte, J.C., 2022. In vivo partial reprogramming alters age-associated molecular changes during physiological aging in mice. Nat Aging 2, 243-253. [CrossRef]
- Campager, A., Ciucci, D., Cabitza, F., 2023. Aggregation models in ensemble learning: A large-scale comparison. Information Fusion 90, 241-252. [CrossRef]
- de Oliveira, E.F., Garg, P., Hjerling-Leffler, J., Batista-Brito, R., Sjulson, L., 2024. Identifying patterns differing between high-dimensional datasets with generalized contrastive PCA. bioRxiv.
- Eckhart, L., Lenhof, K., Rolli, L.M., Lenhof, H.P., 2024. A comprehensive benchmarking of machine learning algorithms and dimensionality reduction methods for drug sensitivity prediction. Brief Bioinform 25. [CrossRef]
- Ford, M.L., Cooley, J.M., Sripada, V., Xu, Z., Erickson, J.S., Bennett, K.P., Crawford, D.R., 2023. Eat4Genes: a bioinformatic rational gene targeting app and prototype model for improving human health. Front Nutr 10, 1196520. [CrossRef]
- Fuellen, G., Jansen, L., Cohen, A.A., Luyten, W., Gogol, M., Simm, A., Saul, N., Cirulli, F., Berry, A., Antal, P., Kohling, R., Wouters, B., Moller, S., 2019. Health and Aging: Unifying Concepts, Scores, Biomarkers and Pathways. Aging Dis 10, 883-900. [CrossRef]
- Fuellen, G., Kulaga, A., Lobentanzer, S., Unfried, M., Avelar, R.A., Palmer, D., Kennedy, B.K., 2024. Validation Requirements for AI-based Intervention-Evaluation in Aging and Longevity Research and Practice. Ageing Res Rev 104, 102617. [CrossRef]
- Gwinn, W.M., Auerbach, S.S., Parham, F., Stout, M.D., Waidyanatha, S., Mutlu, E., Collins, B., Paules, R.S., Merrick, B.A., Ferguson, S., Ramaiahgari, S., Bucher, J.R., Sparrow, B., Toy, H., Gorospe, J., Machesky, N., Shah, R.R., Balik-Meisner, M.R., Mav, D., Phadke, D.P., Roberts, G., DeVito, M.J., 2020. Evaluation of 5-day In Vivo Rat Liver and Kidney With High-throughput Transcriptomics for Estimating Benchmark Doses of Apical Outcomes. Toxicol Sci 176, 343-354. [CrossRef]
- Hanzelmann, S., Castelo, R., Guinney, J., 2013. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7. [CrossRef]
- Hartmann, A., Hartmann, C., Secci, R., Hermann, A., Fuellen, G., Walter, M., 2021. Ranking Biomarkers of Aging by Citation Profiling and Effort Scoring. Front Genet 12, 686320. [CrossRef]
- Hartmann, C., Herling, L., Hartmann, A., Kockritz, V., Fuellen, G., Walter, M., Hermann, A., 2023. Systematic estimation of biological age of in vitro cell culture systems by an age-associated marker panel. Front Aging 4, 1129107. [CrossRef]
- Hishida, T., Yamamoto, M., Hishida-Nozaki, Y., Shao, C., Huang, L., Wang, C., Shojima, K., Xue, Y., Hang, Y., Shokhirev, M., Memczak, S., Sahu, S.K., Hatanaka, F., Ros, R.R., Maxwell, M.B., Chavez, J., Shao, Y., Liao, H.K., Martinez-Redondo, P., Guillen-Guillen, I., Hernandez-Benitez, R., Esteban, C.R., Qu, J., Holmes, M.C., Yi, F., Hickey, R.D., Garcia, P.G., Delicado, E.N., Castells, A., Campistol, J.M., Yu, Y., Hargreaves, D.C., Asai, A., Reddy, P., Liu, G.H., Izpisua Belmonte, J.C., 2022. In vivo partial cellular reprogramming enhances liver plasticity and regeneration. Cell Rep 39, 110730. [CrossRef]
- Iturria-Medina, Y., Adewale, Q., Khan, A.F., Ducharme, S., Rosa-Neto, P., O’Donnell, K., Petyuk, V.A., Gauthier, S., De Jager, P.L., Breitner, J., Bennett, D.A., 2022. Unified epigenomic, transcriptomic, proteomic, and metabolomic taxonomy of Alzheimer’s disease progression and heterogeneity. Sci Adv 8, eabo6764.
- Janssens, G.E., Houtkooper, R.H., 2020. Identification of longevity compounds with minimized probabilities of side effects. Biogerontology 21, 709-719. [CrossRef]
- Joachimiak, M.P., Caufield, J.H., Harris, N.L., Kim, H., Mungall, C.J., 2024. Gene Set Summarization Using Large Language Models. ArXiv.
- Kistler-Fischbacher, M., Armbrecht, G., Gangler, S., Theiler, R., Rizzoli, R., Dawson-Hughes, B., Kanis, J.A., Hofbauer, L.C., Schimmer, R.C., Vellas, B., Da Silva, J.A.P., John, O.E., Kressig, R.W., Andreas, E., Lang, W., Wanner, G.A., Bischoff-Ferrari, H.A., Group, D.-H.R., 2024. Effects of vitamin D3, omega-3s, and a simple strength training exercise program on bone health: the DO-HEALTH randomized controlled trial. J Bone Miner Res 39, 661-671.
- Kriukov, D., Kuzmina, E., Efimov, E., Dylov, D.V., Khrameeva, E.E., 2024. Epistemic uncertainty challenges aging clock reliability in predicting rejuvenation effects. Aging Cell, e14283. [CrossRef]
- Kuhn, M., Letunic, I., Jensen, L.J., Bork, P., 2016. The SIDER database of drugs and side effects. Nucleic Acids Res 44, D1075-1079. [CrossRef]
- Lewis, C.J., de Grey, A.D., 2024. Combining rejuvenation interventions in rodents: a milestone in biomedical gerontology whose time has come. Expert Opin Ther Targets 28, 501-511.
- Liu, J., Yang, M., Yu, Y., Xu, H., Li, K., Zhou, X., 2024. Large language models in bioinformatics: applications and perspectives. ArXiv arXiv:2401.04155.
- Lopez-Otin, C., Blasco, M.A., Partridge, L., Serrano, M., Kroemer, G., 2013. The hallmarks of aging. Cell 153, 1194-1217.
- Lopez-Otin, C., Kroemer, G., 2021. Hallmarks of Health. Cell 184, 33-63.
- Merico, D., Isserlin, R., Stueker, O., Emili, A., Bader, G.D., 2010. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984. [CrossRef]
- Moqri, M., Herzog, C., Poganik, J.R., Justice, J., Belsky, D.W., Higgins-Chen, A., Moskalev, A., Fuellen, G., Cohen, A.A., Bautmans, I., Widschwendter, M., Ding, J., Fleming, A., Mannick, J., Han, J.J., Zhavoronkov, A., Barzilai, N., Kaeberlein, M., Cummings, S., Kennedy, B.K., Ferrucci, L., Horvath, S., Verdin, E., Maier, A.B., Snyder, M.P., Sebastiano, V., Gladyshev, V.N., 2023. Biomarkers of aging for the identification and evaluation of longevity interventions. Cell 186, 3758-3775. [CrossRef]
- Moqri, M., Herzog, C., Poganik, J.R., Ying, K., Justice, J.N., Belsky, D.W., Higgins-Chen, A., Chen, B.H., Cohen, A.A., Fuellen, G., Hagg, S., Marioni, R.E., Widschwendter, M., Fortney, K., Fedichev, P.O., Zhavoronkov, A., Barzilai, N., Lasky-Su, J., Kiel, D.P., Kennedy, B.K., Cummings, S., Slagboom, P.E., Verdin, E., Maier, A.B., Sebastiano, V., Snyder, M.P., Gladyshev, V.N., Horvath, S., Ferrucci, L., 2024. Validation of biomarkers of aging. Nat Med 30, 360-372. [CrossRef]
- Nadon, N.L., Strong, R., Miller, R.A., Nelson, J., Javors, M., Sharp, Z.D., Peralba, J.M., Harrison, D.E., 2008. Design of aging intervention studies: the NIA interventions testing program. Age (Dordr) 30, 187-199. [CrossRef]
- Nguyen, H., Tran, D., Galazka, J.M., Costes, S.V., Beheshti, A., Petereit, J., Draghici, S., Nguyen, T., 2021. CPA: a web-based platform for consensus pathway analysis and interactive visualization. Nucleic Acids Res 49, W114-W124.
- Ortigossa, E.S., Gonçalves, T., Nonato, L.G., 2024. EXplainable Artificial Intelligence (XAI)—From Theory to Methods and Applications. IEEE Access 24, 80799-80846. [CrossRef]
- Pantazis, Y., Tselas, C., Lakiotaki, K., Lagani, V., Tsamardinos, I., 2020. Latent Feature Representations for Human Gene Expression Data Improve Phenotypic Predictions, 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE, pp. 2505-2512.
- Piccolo, S.R., Mecham, A., Golightly, N.P., Johnson, J.L., Miller, D.B., 2022. The ability to classify patients based on gene-expression data varies by algorithm and performance metric. PLoS Comput Biol 18, e1009926. [CrossRef]
- Pickard, J., Choi, M.A., Oliven, N., Stansbury, C., Cwycyshyn, J., Galioto, N., Gorodetsky, A., Velasquez, A., Rajapakse, I., 2024. Bioinformatics Retrieval Augmentation Data (BRAD) Digital Assistant. ArXiv arXiv:2409.02864.
- Pun, F.W., Leung, G.H.D., Leung, H.W., Liu, B.H.M., Long, X., Ozerov, I.V., Wang, J., Ren, F., Aliper, A., Izumchenko, E., Moskalev, A., de Magalhaes, J.P., Zhavoronkov, A., 2022. Hallmarks of aging-based dual-purpose disease and age-associated targets predicted using PandaOmics AI-powered discovery engine. Aging (Albany NY) 14, 2475-2506. [CrossRef]
- Ringner, M., 2008. What is principal component analysis? Nat Biotechnol 26, 303-304.
- Ryan, C.P., Corcoran, D.L., Banskota, N., Eckstein Indik, C., Floratos, A., Friedman, R., Kobor, M.S., Kraus, V.B., Kraus, W.E., MacIsaac, J.L., Orenduff, M.C., Pieper, C.F., White, J.P., Ferrucci, L., Horvath, S., Huffman, K., Belsky, D.W., 2024. The CALERIE™ Genomic Data Resource. BioArxiv 10.1101/2024.05.17.594714. [CrossRef]
- Saarimaki, L.A., Fratello, M., Pavel, A., Korpilahde, S., Leppanen, J., Serra, A., Greco, D., 2023a. A curated gene and biological system annotation of adverse outcome pathways related to human health. Sci Data 10, 409. [CrossRef]
- Saarimaki, L.A., Morikka, J., Pavel, A., Korpilahde, S., Del Giudice, G., Federico, A., Fratello, M., Serra, A., Greco, D., 2023b. Toxicogenomics Data for Chemical Safety Assessment and Development of New Approach Methodologies: An Adverse Outcome Pathway-Based Approach. Adv Sci (Weinh) 10, e2203984. [CrossRef]
- Sarkar, T.J., Quarta, M., Mukherjee, S., Colville, A., Paine, P., Doan, L., Tran, C.M., Chu, C.R., Horvath, S., Qi, L.S., Bhutani, N., Rando, T.A., Sebastiano, V., 2020. Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat Commun 11, 1545. [CrossRef]
- Simm, A., Grosskopf, A., Fuellen, G., 2024. Detailing the biomedical aspects of geroscience by molecular data and large-scale "deep" bioinformatics analyses. Z Gerontol Geriatr 57, 355-360.
- Smer-Barreto, V., Quintanilla, A., Elliott, R.J.R., Dawson, J.C., Sun, J., Campa, V.M., Lorente-Macias, A., Unciti-Broceta, A., Carragher, N.O., Acosta, J.C., Oyarzun, D.A., 2023. Discovery of senolytics using machine learning. Nat Commun 14, 3445. [CrossRef]
- Smith, A.M., Walsh, J.R., Long, J., Davis, C.B., Henstock, P., Hodge, M.R., Maciejewski, M., Mu, X.J., Ra, S., Zhao, S., Ziemek, D., Fisher, C.K., 2020. Standard machine learning approaches outperform deep representation learning on phenotype prediction from transcriptomics data. BMC Bioinformatics 21, 119. [CrossRef]
- Subramanian, A., Tamayo, P., Mootha, V.K., Mukherjee, S., Ebert, B.L., Gillette, M.A., Paulovich, A., Pomeroy, S.L., Golub, T.R., Lander, E.S., Mesirov, J.P., 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545-15550. [CrossRef]
- Tang, X., Qian, B., Gao, R., Chen, J., Chen, X., Gerstein, M.B., 2024. BioCoder: a benchmark for bioinformatics code generation with large language models. Bioinformatics 40, i266-i276. [CrossRef]
- Tsamardinos, I., Charonyktakis, P., Papoutsoglou, G., Borboudakis, G., Lakiotaki, K., Zenklusen, J.C., Juhl, H., Chatzaki, E., Lagani, V., 2022. Just Add Data: automated predictive modeling for knowledge discovery and feature selection. NPJ Precis Oncol 6, 38. [CrossRef]
- Tyshkovskiy, A., Kholdina, D., Ying, K., Davitadze, M., Molière, A., Tongu, Y., Kasahara, T., Kats, L.M., Vladimirova, A., Moldakozhyev, A., Liu, H., Zhang, B., Khasanova, U., Moqri, M., Van Raamsdonk, J.M., Harrison, D.E., Strong, R., Abe, T., Dmitriev, S.E., Gladyshev, V.N., 2024. Transcriptomic Hallmarks of Mortality Reveal Universal and Specific Mechanisms of Aging, Chronic Disease, and Rejuvenation. BioRxiv 10.1101/2024.07.04.601982.
- Uner, O.C., Kuru, H.I., Cinbis, R.G., Tastan, O., Cicek, A.E., 2023. DeepSide: A Deep Learning Approach for Drug Side Effect Prediction. IEEE/ACM Trans Comput Biol Bioinform 20, 330-339. [CrossRef]
- Vyas, C.M., Manson, J.E., Sesso, H.D., Rist, P.M., Weinberg, A., Kim, E., Moorthy, M.V., Cook, N.R., Okereke, O.I., 2024. Effect of cocoa extract supplementation on cognitive function: results from the clinic subcohort of the COSMOS trial. Am J Clin Nutr 119, 39-48. [CrossRef]
- Wang, J., Wang, J., Athiwaratkun, B., Zheng, C., Zou, J., 2024. Mixture-of-Agents Enhances Large Language Model Capabilities. ArXiv arXiv:2406.04692.
- Wang, Z., Clark, N.R., Ma’ayan, A., 2016. Drug-induced adverse events prediction with the LINCS L1000 data. Bioinformatics 32, 2338-2345. [CrossRef]
- Wu, L., Yan, B., Han, J., Li, R., Xiao, J., He, S., Bo, X., 2023. TOXRIC: a comprehensive database of toxicological data and benchmarks. Nucleic Acids Res 51, D1432-D1445. [CrossRef]
- Xin, Q., Kong, Q., Ji, H., Shen, Y., Liu, Y., Y, S., Zhang, Z., Li, Z., Xia, X., Deng, B., Bai, Y., 2024. BioInformatics Agent (BIA): Unleashing the Power of Large Language Models to Reshape Bioinformatics Workflow. bioRxiv 2024.05.22.595240.
- Yang, Y., Sun, H., Zhang, Y., Zhang, T., Gong, J., Wei, Y., Duan, Y.G., Shu, M., Yang, Y., Wu, D., Yu, D., 2021. Dimensionality reduction by UMAP reinforces sample heterogeneity analysis in bulk transcriptomic data. Cell Rep 36, 109442. [CrossRef]
- Zhou, J., Zhang, B., Li, G., Chen, X., Li, H., Xu, X., Chen, S., He, W., Xu, C., Liu, L., Gao, X., 2024. An AI Agent for Fully Automated Multi-Omic Analyses. Adv Sci (Weinh), e2407094. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




