Submitted:
19 January 2025
Posted:
20 January 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Results
Data Cleaning and Traditional Bibliometric Analysis
Literature Embedding and Clustering
Hybrid RAG of Literature
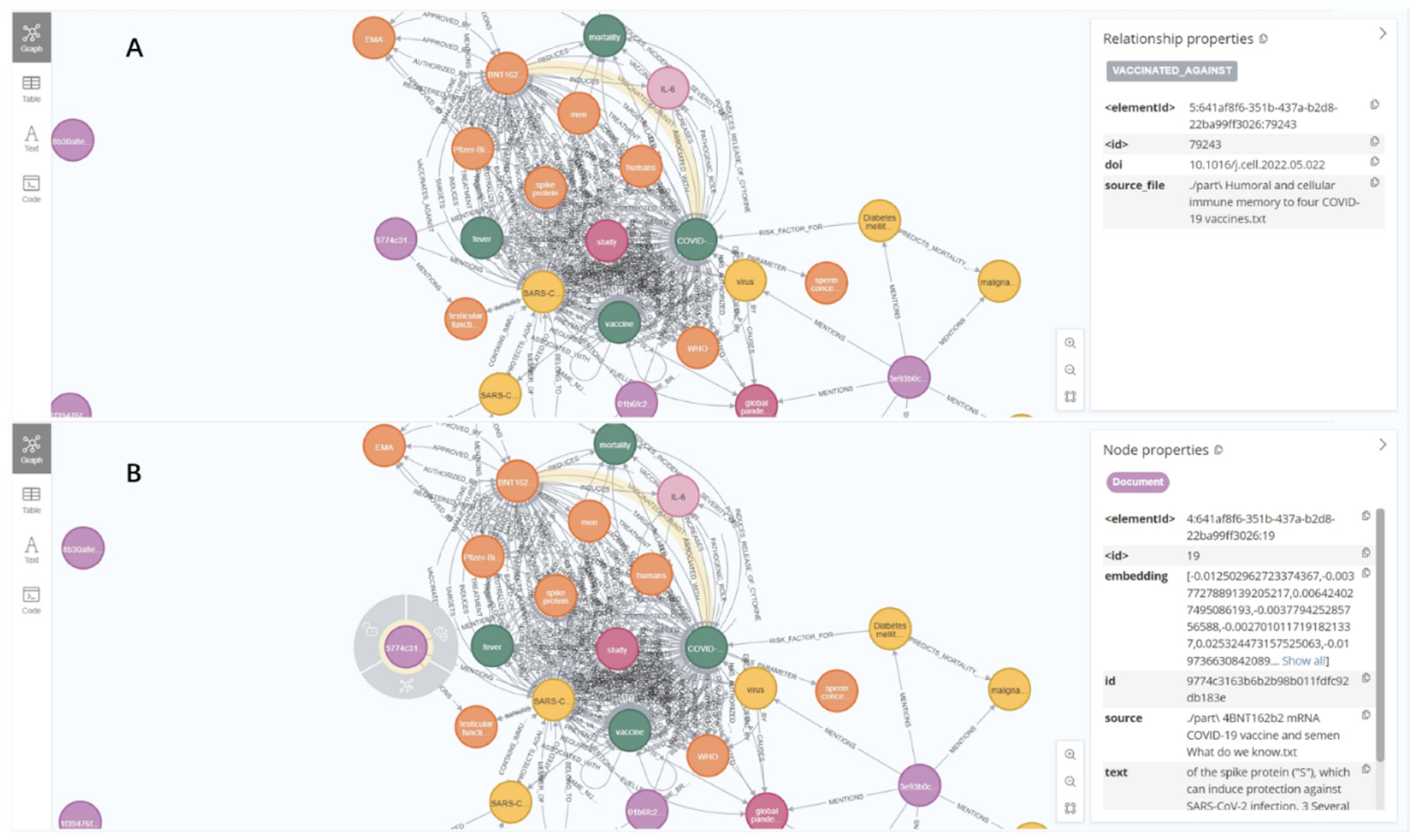
Literature Map
Discussion
Methods
Literature Collection, Cleaning and Traditional Bibliometric Analysis
Literature Embedding and Clustering
Hybrid Retrieval Augmented Generation (RAG) of Literature
Literature Map
Author Contributions
Data Availability Statement
Code Availability:
References
- Azamfirei, R., Kudchadkar, S. R. & Fackler, J. Large language models and the perils of their hallucinations. Crit Care 27, 120 (2023).
- Edge, D. et al. From Local to Global: A Graph RAG Approach to Query-Focused Summarization. arXiv.org https://arxiv.org/abs/2404.16130v1 (2024).
- Peng, B. et al. Graph Retrieval-Augmented Generation: A Survey. Preprint at http://arxiv.org/abs/2408.08921 (2024).
- Sarmah, B. et al. HybridRAG: Integrating Knowledge Graphs and Vector Retrieval Augmented Generation for Efficient Information Extraction. Preprint at http://arxiv.org/abs/2408.04948 (2024).
- Chen, C. Searching for intellectual turning points: Progressive knowledge domain visualization. Proceedings of the National Academy of Sciences 101, 5303–5310 (2004).
- Xiao, S., Liu, Z., Zhang, P. & Muennighoff, N. C-Pack: Packaged Resources To Advance General Chinese Embedding. (2023).
- Steinley, D. Properties of the Hubert-Arabie adjusted Rand index. Psychol Methods 9, 386–396 (2004).
- Romano, S., Bailey, J., Nguyen, X. & Verspoor, K. M. Standardized Mutual Information for Clustering Comparisons: One Step Further in Adjustment for Chance. in (2014).
- Guia, J., Gonçalves Soares, V. & Bernardino, J. Graph Databases: Neo4j Analysis: in Proceedings of the 19th International Conference on Enterprise Information Systems 351–356 (SCITEPRESS—Science and Technology Publications, Porto, Portugal, 2017). [CrossRef]
- Reimers, N. & Gurevych, I. Sentence-BERT: Sentence Embeddings using Siamese BERT-Networks. in Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing (Association for Computational Linguistics, 2019).
- Cer, D. et al. Universal Sentence Encoder. arXiv.org https://arxiv.org/abs/1803.11175v2 (2018).
- Řehůřek, R. & Sojka, P. Software Framework for Topic Modelling with Large Corpora. in Proceedings of the LREC 2010 Workshop on New Challenges for NLP Frameworks 45–50 (ELRA, Valletta, Malta, 2010).
- Ester, M., Kriegel, H.-P. & Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise.
- Murtagh, F. & Contreras, P. Algorithms for hierarchical clustering: an overview. WIREs Data Mining and Knowledge Discovery 2, 86–97 (2012).
- Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
- Gu, Y. et al. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. (2020).
- Efficient and Robust Approximate Nearest Neighbor Search Using Hierarchical Navigable Small World Graphs | IEEE Journals & Magazine | IEEE Xplore. https://ieeexplore.ieee.org/document/8594636.
- Tech, Z. Feder: A Federated Learning Library. (2023).
- da Costa, L. S., Oliveira, I. L. & Fileto, R. Text classification using embeddings: a survey. Knowl Inf Syst 65, 2761–2803 (2023).
- Petukhova, A., Matos-Carvalho, J. P. & Fachada, N. Text Clustering with LLM Embeddings. Preprint at https://doi.org/10.48550/arXiv.2403.15112 (2024).
- Feng, F., Yang, Y., Cer, D., Arivazhagan, N. & Wang, W. Language-agnostic BERT Sentence Embedding. Preprint at https://doi.org/10.48550/arXiv.2007.01852 (2022).
- Frey, B. J. & Dueck, D. Clustering by Passing Messages Between Data Points. Science 315, 972–976 (2007).
- GROBID. (2008).
- Dubey, A. et al. The Llama 3 Herd of Models. arXiv.org https://arxiv.org/abs/2407.21783v2 (2024).
- Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17, 261–272 (2020).
- Hagberg, A., Swart, P. & Chult, D. Exploring Network Structure, Dynamics, and Function Using NetworkX. in (2008). [CrossRef]
- QGIS Development Team. QGIS Geographic Information System. (QGIS Association).
- Kimi, an AI Assistant by Moonshot AI.
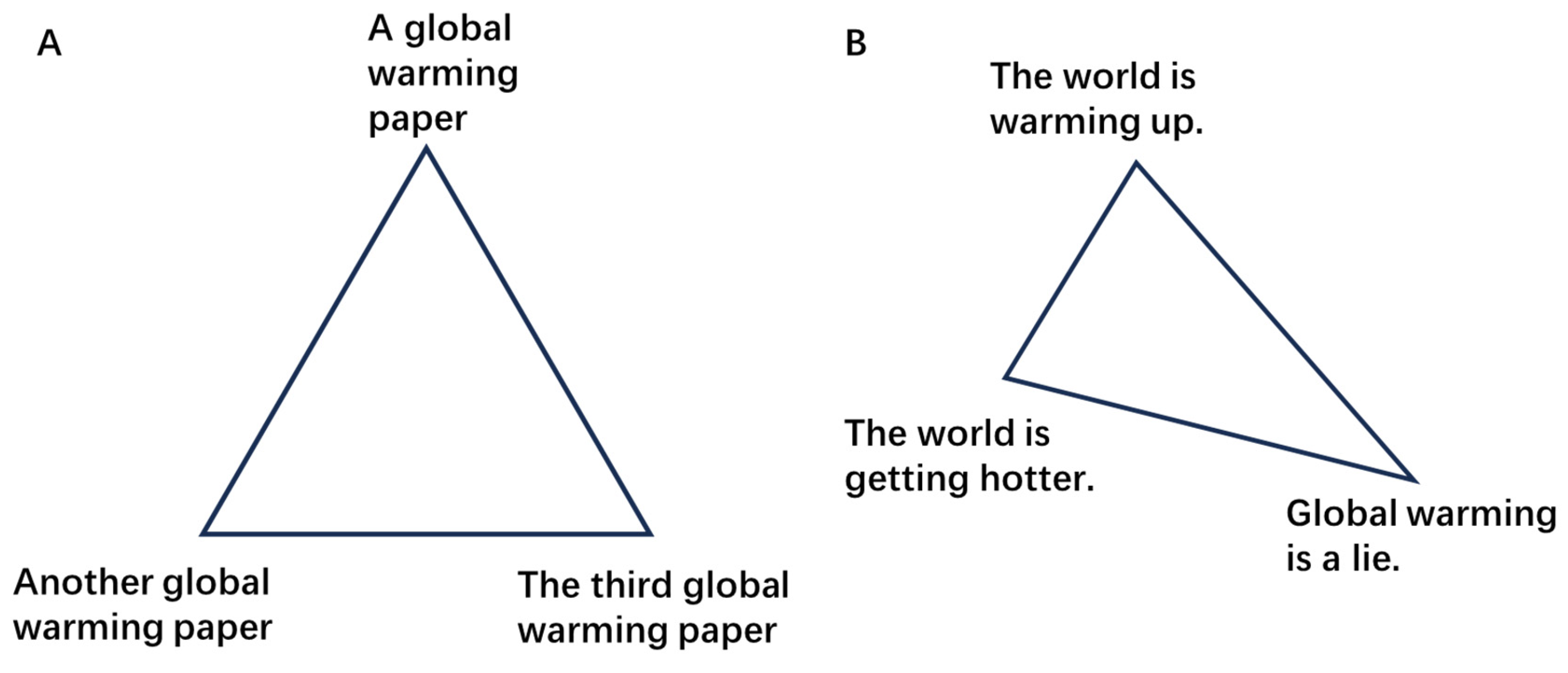
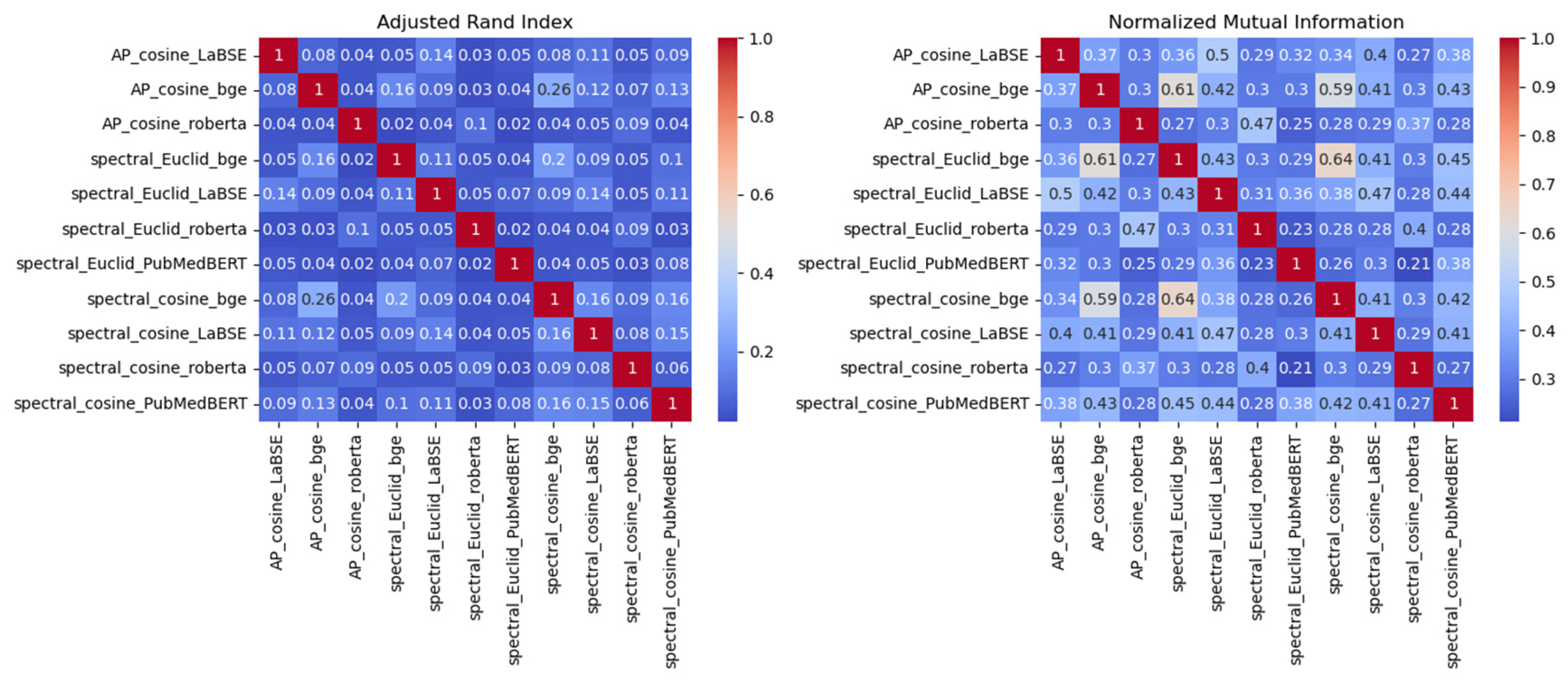

| Embedding-clustering pair | # of same Cluster | # of different Cluster | Same Cluster Ratio |
| AP_cosine_LaBSE | 97 | 229 | 0.30 |
| AP_cosine_bge | 189 | 137 | 0.58 |
| AP_cosine_roberta | 55 | 271 | 0.17 |
| spectral_Euclid_bge | 209 | 117 | 0.64 |
| spectral_Euclid_LaBSE | 99 | 227 | 0.30 |
| spectral_Euclid_roberta | 71 | 255 | 0.22 |
| spectral_Euclid_PubMedBERT | 35 | 291 | 0.11 |
| spectral_cosine_bge | 172 | 154 | 0.53 |
| spectral_cosine_LaBSE | 101 | 225 | 0.31 |
| spectral_cosine_roberta | 84 | 242 | 0.26 |
| spectral_cosine_PubMedBERT | 138 | 188 | 0.42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





