Introduction
Machine Learning (ML) has revolutionized various industries, and healthcare is no exception. In the healthcare field, ML involves the use of algorithms and statistical models to enable computers to learn from and make predictions or decisions based on data. This has led to significant advancements in diagnosis, treatment, and patient care. One of the areas where ML has shown immense potential is in medical imaging analysis/image classification.
Image classification is a key aspect of machine learning, enabling computers to recognize patterns and objects in images. In healthcare, this involves training algorithms to identify specific features in medical images, aiding diagnosis and treatment planning. MRI, CT, and X-ray offer valuable insights into the body. ML algorithms can learn to analyze these images, leading to quicker and more precise diagnoses. A 2023 study published in the Journal of the American Medical Association (JAMA) reported that a deep learning model achieved 91% accuracy in detecting pneumonia from chest X-rays, exceeding the performance of radiologists on the same task. For glioblastoma, early and accurate diagnosis is vital. ML-driven image classification, using MRI scans, has great potential. Researchers can build models to spot subtle patterns related to MGMT promoter methylation, predicting glioblastoma subtypes and guiding treatment choices.
Several research studies have explored the use of machine learning models to analyze MRI scans for glioblastoma and predict MGMT promoter methylation status. A 2019 study published in the journal Radiology used a machine learning model to analyze MRI scans and predict MGMT methylation with an accuracy of 85%. The researchers found that their model was able to identify subtle features in the MRI scans that were associated with MGMT methylation status. These studies demonstrate the potential of machine learning for improving glioblastoma diagnosis and treatment planning.
This study investigates the training and testing of machine learning models utilizing MRI (magnetic resonance imaging) scans to detect the presence of MGMT promoter methylation. This study focuses on using advanced image classification techniques, particularly Keras, a popular deep learning library in Python. The developed model can predict the genetic subtype of glioblastoma, allowing for fewer procedures and better treatment decisions. By integrating imaging technology and data, we can provide an early diagnosis and offer individualized treatment options. Using this strategy, we can avoid unnecessary procedures, help determine the type of therapy required, and focus on improving the management, survival, and prospects of glioblastoma patients.
Methods
I utilized a Keras model implemented in Python using the TensorFlow library, to detect the presence of MGMT promoter methylation in patients with glioblastoma using MRI scans. I obtained a publicly available dataset from Kaggle
1, which consisted of MRI images of patients with glioblastoma. The dataset was divided into two classes: malignant and benign. I used this dataset to train and test my model. I split the dataset into a training set and a testing set. I used 89.8% of the data (86,884 images) for training, and the remaining 10.2% (9,882 images) for testing. Each folder contained 4 different types of scans corresponding to each of the structural multi-parametric MRI scans (In DICOM format). This is to ensure that the model's performance can be properly evaluated on unseen data. I Optimized the number of epochs. An epoch is one complete pass through the entire training dataset. By experimenting (i.e. hyperparameter tuning) with different numbers of epochs, I aimed to find the optimal number that produced the best results. In my case I used 100 epochs. I evaluated my model's performance using metrics like loss and accuracy. Loss is a measure of how well the model is performing, with lower values indicating better performance. Accuracy is a measurement for classification tasks, representing the percentage of correct samples.
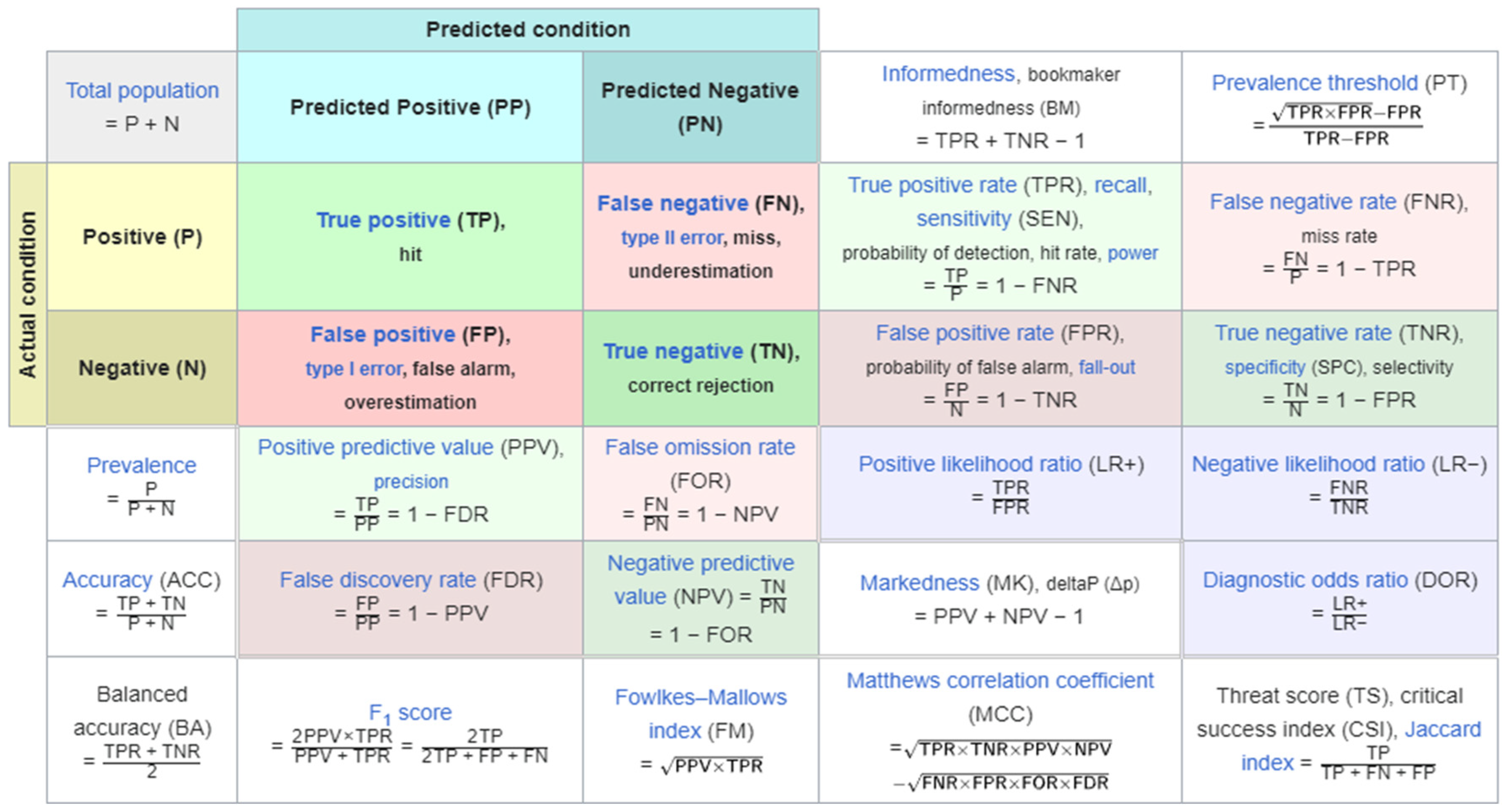
In predictive analysis, a table of confusion/confusion matrix is a table with 2 columns and 2 rows which display all the number of true positives, false negatives, false positives, and true negatives. Multi-panel figures must be submitted as a single image as demonstrated above, and each panel can be described in the legend.
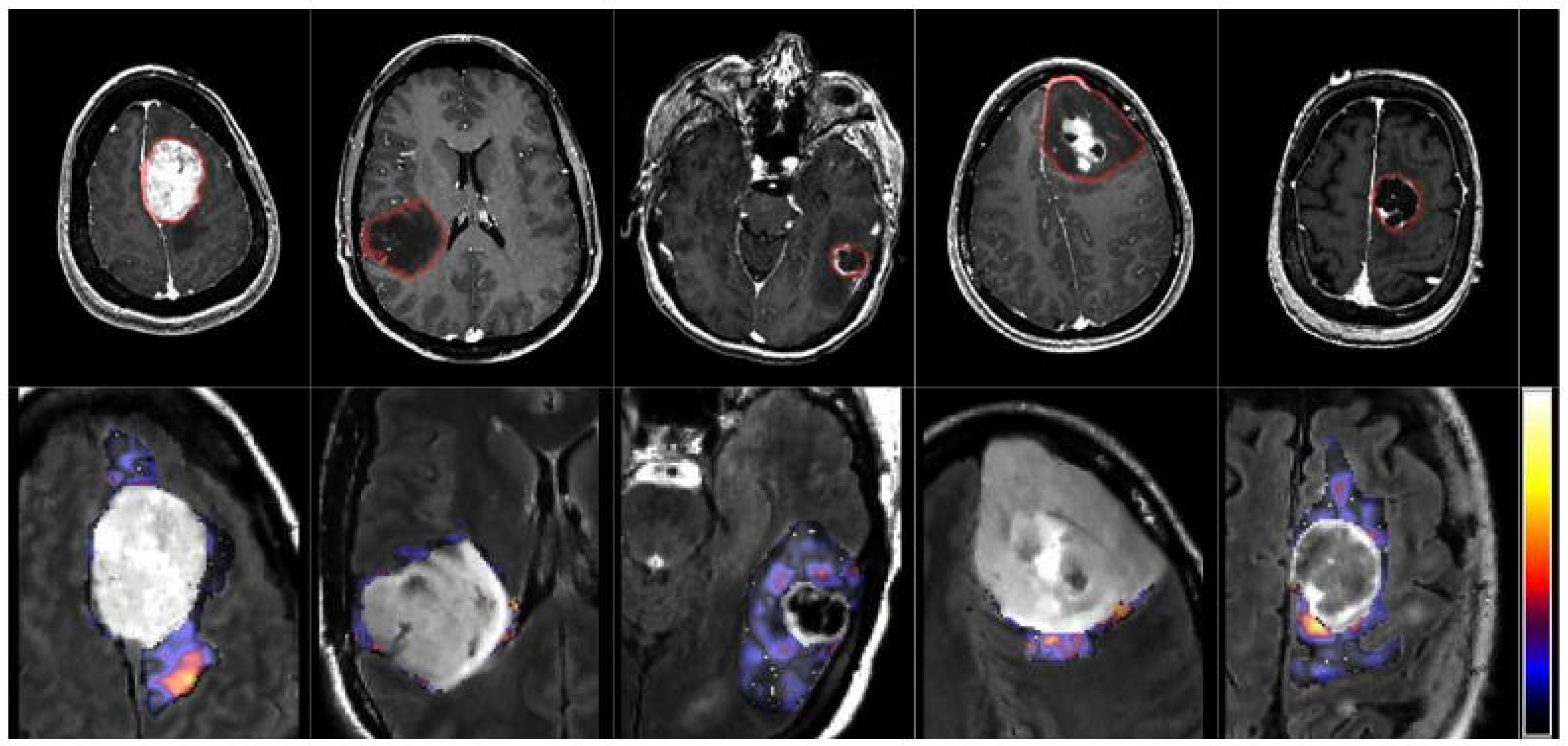
Glioblastoma multiforme represents an advanced and aggressive form of brain cancer characterized by significant tumor growth and infiltration into surrounding brain tissues. Patients typically experience symptoms such as headaches, seizures, cognitive decline, and neurological deficits. Treatment involves a combination of surgery, radiation therapy, and chemotherapy, with the goal of removing or shrinking the tumor. Despite aggressive intervention, the prognosis for glioblastoma remains poor, with limited median survival. Ongoing research focuses on developing new therapeutic approaches, including immunotherapy and targeted therapies, to improve outcomes for individuals facing this challenging diagnosis. Patients and their families are encouraged to collaborate closely with healthcare teams, explore treatment options, and consider participation in clinical trials for access to emerging therapies and advancements in glioblastoma treatment. Analyzing a random sample of MRI images is crucial for accurately diagnosing and monitoring glioblastoma multiforme, aiding healthcare professionals in determining the extent of tumor growth and guiding the most effective treatment strategies.
Figure 2.
From left to right: meningioma, glioma grade II, grade III, grade IV and metastasis.
Figure 2.
From left to right: meningioma, glioma grade II, grade III, grade IV and metastasis.
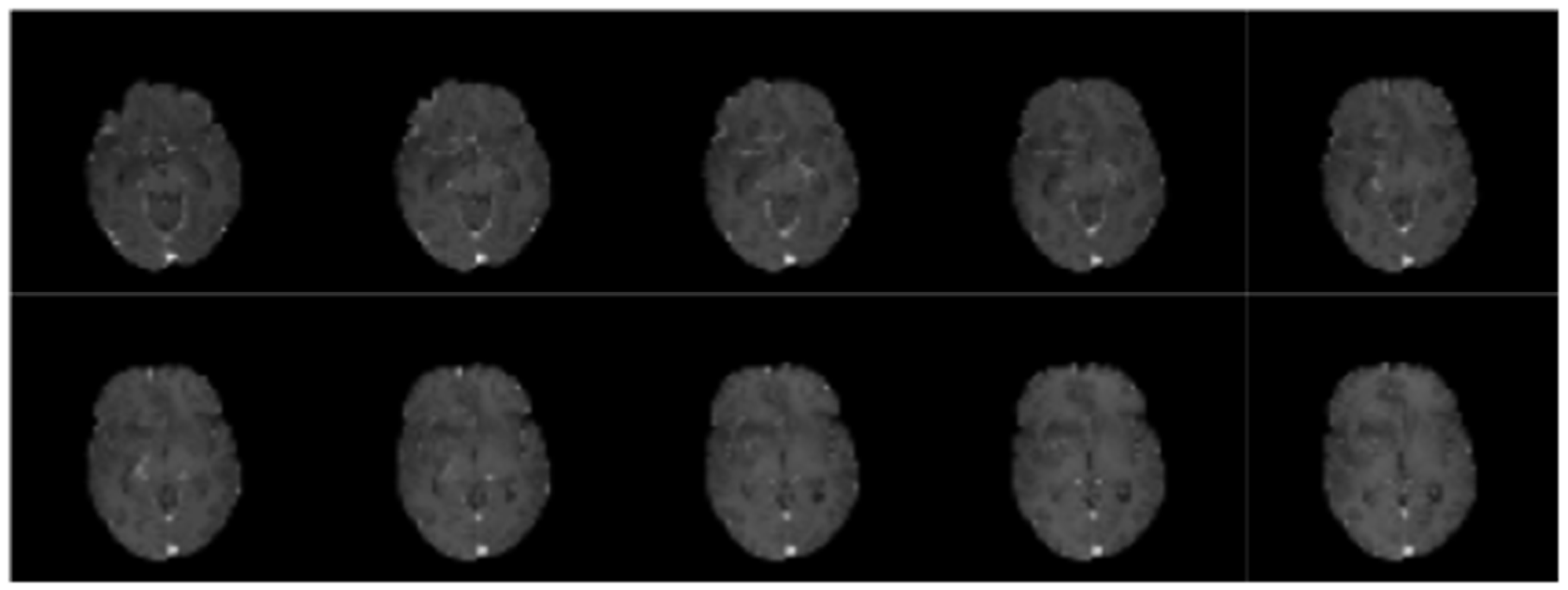
Figure 3.
Sample scans from testing dataset.
Figure 3.
Sample scans from testing dataset.
Table 1.
Confusion matrix array (from testing dataset).
Table 1.
Confusion matrix array (from testing dataset).
| |
Predicted Positive |
Predicted Negative |
| Positive |
3210 |
1608 |
| Negative |
3539 |
1525 |
1608 out of 9882 images were labeled as false negatives, which is an issue because it means that the unhealthy patient is more likely to be diagnosed as healthy and not receive any treatment. 3210 images were labeled as true positives, this is good because the image was correctly labeled as having cancer and the patient is more likely to be diagnosed as unhealthy.
Results
Each independent case has a dedicated folder identified by a five-digit number. Within each of these “case” folders, there are four sub-folders, each of them corresponding to each of the structural multi-parametric MRI (mpMRI) scans, in DICOM format. The exact mpMRI scans included are:
Fluid Attenuated Inversion Recovery (FLAIR)
T1-weighted pre-contrast (T1w)
T1-weighted post-contrast (T1wCE)
T2-weighted (T2w)
The tables in the image show the prediction results per image. The BraTS21ID column is a unique identifier for each test subject. The MGMT_value shows the probability that the MGMT gene promoter is present for a particular test subject, on a scale between 0-1.
Discussion
The study is of significant importance because it focuses on using machine learning to improve the diagnosis and treatment of glioblastoma, a life-threatening form of brain cancer. Glioblastoma is known for its poor prognosis and limited survival rates, and early and accurate detection is crucial for improving patient outcomes. Glioblastoma has a very short survival time, and early diagnosis can make a difference in treatment results. Machine learning can assist physicians in detecting glioblastoma earlier in patients and marking such cases for review. By using machine learning to look at MRI scans, the study aimed to detect the presence of MGMT promoter methylation, a genetic subtype indicator. Early detection can lead to faster work and better treatment planning. By accurately classifying patients into malignant and benign cases using our model, the study could reduce any unnecessary surgical procedures. This not only helps patients by saving them from unnecessary problems but also reduces healthcare costs and utilizes resources better.
The paper “Improving Survival Prediction of High-Grade Glioma (HGG) via Machine Learning Techniques Based on MRI Radiomic, Genetic and Clinical Risk Factors.” looks at high grade glioblastoma prognosis by creating a model that incorporates radiomic, clinical, and genetic factors. This study investigated 147 high grade glioblastoma cases and divided them into training (112 patients) and testing (35 Patients). They gave MRI images, genetic data, and clinical information to their model. The model used Kaplan-Meier survival analysis to look between genetical, radiomic, and clinical factors. According to the article - the radiomics signature is an independent prognostic biomarker for HGG, and successfully stratified patients. Age and isocitrate dehydrogenase mutation (IDH-M) were important supplements for radiomics signature, especially for the low-risk group. A nomogram incorporating radiomics signature, IDH-M and age improved the performance of individualized OS estimation, which might be a new complement to the treatment guidelines of glioma for clinical use.
Methodology from paper:
Patient Cohorts: Studied 147 HGG cases, dividing them into training (112 patients) and independent test cohorts (35 patients).
Data Collection: Used MRI images, genetic data, and clinical information.
Radiomics Analysis: Extracted features from tumor and peritumoral edema areas on MRI images (CE-T1WI and T2 FLAIR).
Analysis Methods: Employed Kaplan-Meier survival analysis, log-rank test, and multivariate Cox regression to explore associations between radiomics, genetic, clinical factors, and OS.
Nomogram Construction: Developed a predictive model integrating radiomics, genetic (IDH mutation), and clinical (age) factors.
Table 2.
For each BraTS21ID in the test set, probability for the target MGMT_value is predicted above.
Table 2.
For each BraTS21ID in the test set, probability for the target MGMT_value is predicted above.
| |
BraTS21ID |
MGMT_value |
|
BraTS21ID |
MGMT_value |
|
BraTS21ID |
MGMT_value |
| 0 |
1 |
0.550388 |
24 |
208 |
0.661458 |
48 |
460 |
0.799065 |
| 1 |
13 |
0.627778 |
25 |
213 |
0.614583 |
49 |
462 |
0.508333 |
| 2 |
15 |
0.875000 |
26 |
229 |
0.489583 |
50 |
463 |
0.900568 |
| 3 |
27 |
0.984496 |
27 |
252 |
0.828125 |
51 |
467 |
0.665000 |
| 4 |
37 |
0.992248 |
28 |
256 |
0.536458 |
52 |
474 |
0.412500 |
| 5 |
47 |
0.961240 |
29 |
264 |
0.744792 |
53 |
489 |
0.928571 |
| 6 |
79 |
0.466667 |
30 |
287 |
0.539683 |
54 |
492 |
0.888889 |
| 7 |
80 |
0.638889 |
31 |
307 |
0.696429 |
55 |
503 |
0.740741 |
| 8 |
82 |
0.650000 |
32 |
323 |
0.789720 |
56 |
521 |
0.666667 |
| 9 |
91 |
0.550000 |
33 |
333 |
0.843434 |
57 |
535 |
0.468750 |
| 10 |
114 |
0.697917 |
34 |
335 |
0.742424 |
58 |
553 |
0.521739 |
| 11 |
119 |
0.652174 |
35 |
337 |
0.767442 |
|
|
|
| 12 |
125 |
0.770833 |
36 |
355 |
0.329268 |
|
|
|
| 13 |
129 |
0.791667 |
37 |
372 |
0.750000 |
|
|
|
| 14 |
135 |
0.661458 |
38 |
381 |
0.676136 |
|
|
|
| 15 |
145 |
0.406250 |
39 |
384 |
0.748120 |
|
|
|
| 16 |
153 |
0.468750 |
40 |
393 |
0.760331 |
|
|
|
| 17 |
161 |
0.389423 |
41 |
422 |
0.696809 |
|
|
|
| 18 |
163 |
0.218750 |
42 |
428 |
0.666667 |
|
|
|
| 19 |
174 |
0.864583 |
43 |
434 |
0.664634 |
|
|
|
| 20 |
181 |
0.421875 |
44 |
438 |
0.785714 |
|
|
|
| 21 |
182 |
0.338542 |
45 |
447 |
0.811688 |
|
|
|
| 22 |
190 |
0.822917 |
46 |
450 |
0.897590 |
|
|
|
| 23 |
200 |
0.458333 |
47 |
458 |
0.520548 |
|
|
|
This model from above performs better because of the increased dataset available to the researchers along with real-time data from patients. This model, like mine, includes MRI images from patients to help predict HGG. But the researchers were able to connect with real patients - the model also has access to genetic information and clinical records which allows it to make much more accurate and real predictions which is like a likely future possibility of my work.
Conclusions
Overall, this project explores training and testing of machine learning models using MRI (magnetic resonance imaging) scans to detect for the presence of MGMT promoter methylation. The resulting models can predict the genetic subtype of glioblastoma, leading to fewer surgeries and better treatment decisions. By combining imaging technology with data, we can come up with an early diagnosis and provide personalized treatment plans. By using this method we can cut unnecessary surgeries, help decide the type of therapy needed, and overall focus on improving the management, survival, and prospects of patients with glioblastoma. I utilized a CNN model, the code was written in python using the TensorFlow library. I evaluated the model using loss and accuracy and optimized for loss. The results can contribute to early and accurate detection of brain tumors in patients.
Limitations
The drawbacks of AI include job displacement, ethical concerns about bias and privacy, security risks from hacking, a lack of human-like creativity and empathy. In the realm of utilizing machine learning for predicting glioblastoma based on MRI scans, there exist limitations for innovation. The size of available datasets can be a significant limitation. Larger datasets are often needed to train more complex models effectively. The scarcity of comprehensive and diverse datasets might restrict the model's generalizability. The dataset used for training the model might have biases related to demographics, geographic location, or race. This could lead to a lack of representation and generalization issues for populations not adequately included in the dataset. Focusing solely on glioblastoma might limit the model's application in diagnosing or predicting other types of brain tumors. The model's generalizability could be enhanced by considering various tumor types and subtypes. Glioblastoma is a complex disease with various genetic and molecular subtypes. Current models might not capture the full spectrum of these subtypes, limiting their predictive accuracy and treatment implications.
Acknowledgements
I would like to thank RSNA for publishing the huge dataset along with giving researchers across the world an opportunity to work on this very interesting ML project.
References
- European Journal of Radiology, Elsevier. (2019). Improving Survival Prediction of High-Grade Glioma via Machine Learning Techniques Based on MRI Radiomic, Genetic and Clinical Risk Factors. https://www.sciencedirect.com/science/article/abs/pii/S0720048X19302505.
- Havaei, M., Davy, A., Warde-Farley, D., Biard, A., Courville, A., Bengio, Y., ... & Pal, C. (2017). Brain tumor segmentation with Deep Neural Networks. https://www.sciencedirect.com/science/article/abs/pii/S1361841516300330\.
- Mobadersany, P., Yousefi, S., Amgad, M., Gutman, D. A., Barnholtz-Sloan, J. S., Velázquez Vega, J. E., & Brat, D. J. (2018). Predicting cancer outcomes from histology and genomics using convolutional networks. Proceedings of the National Academy of Sciences. https://www.pnas.org/doi/10.1073/pnas.1717139115.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).