Submitted:
12 October 2024
Posted:
15 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Methods
2.1. Study Population
2.2. Selection of Cases
2.3. Data Analysis
2.4. Ethical Approval
3. Results
3.1. Significance of IDH1 Mutation
3.2. Significance of MGMT Promotor Methylation
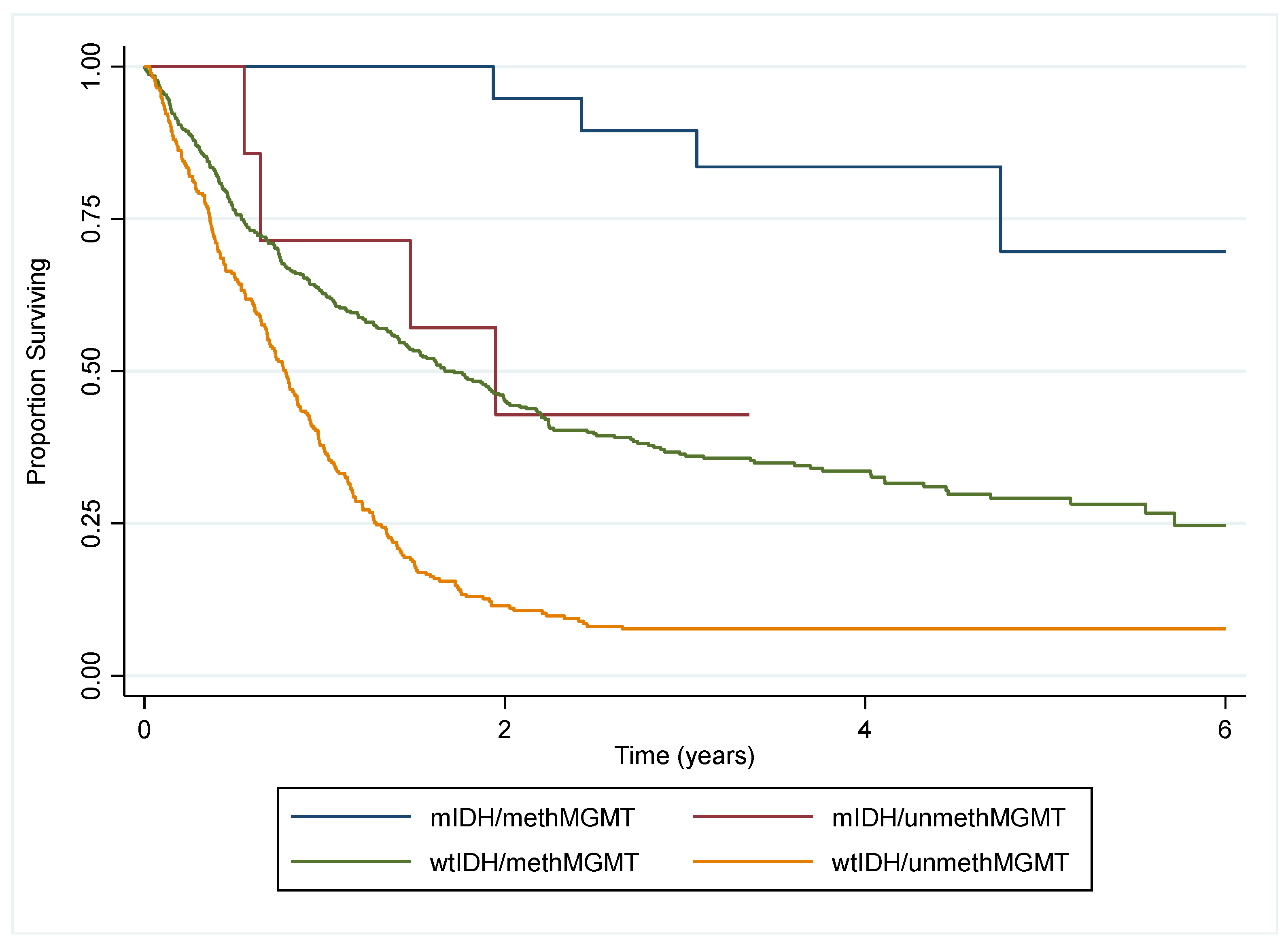
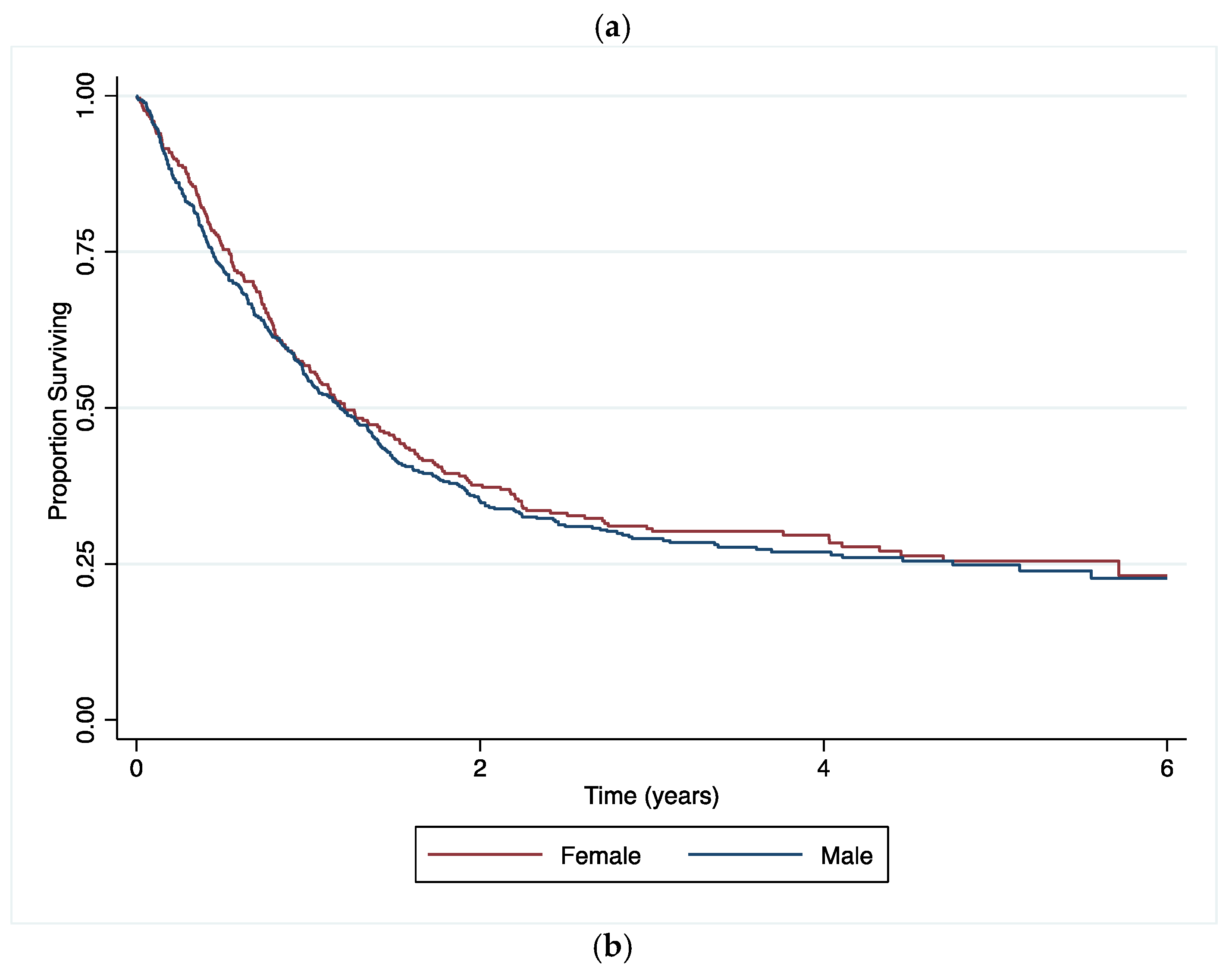
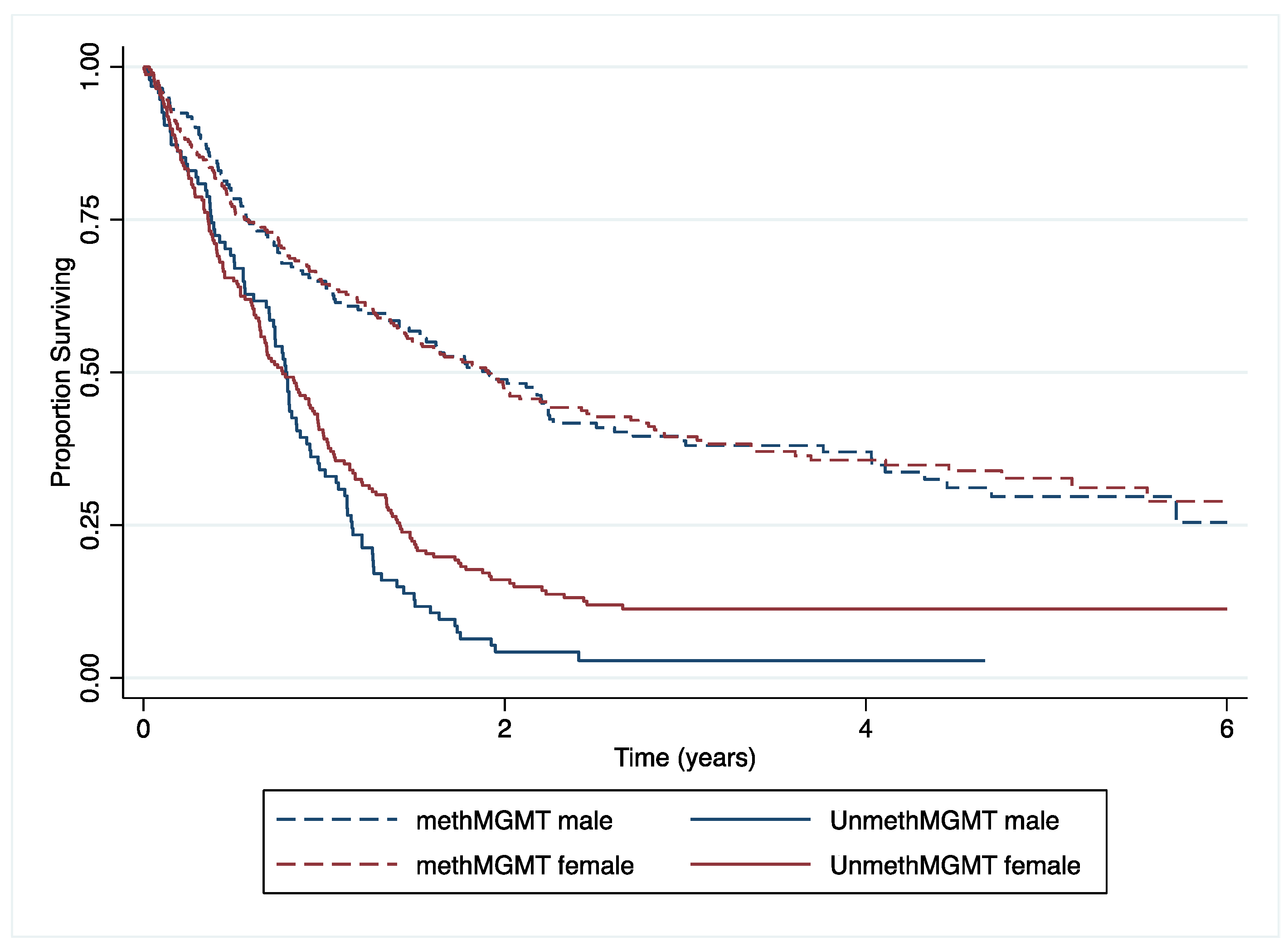
3.3. Survival Analysis
4. Discussion
4.1. Main Findings
4.2. Comparison to Other Findings
4.3. Interpretation and Implications
4.4. Strengths and Limitations
5. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgements
Conflicts of Interest
References
- Schwartzbaum, J.A.; Fisher, J.L.; Aldape, K.D.; Wrensch, M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol 2006, 2, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Larjavaara, S.; et al. Incidence of gliomas by anatomic location. Neuro Oncol 2007, 9, 319–325. [Google Scholar] [CrossRef]
- Yan, H.; et al. IDH1 and IDH2 Mutations in Gliomas. New England Journal of Medicine 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathologica 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; et al. IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br J Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef]
- Hegi, M.E.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Esteller, M.; et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000, 343, 1350–1354. [Google Scholar] [CrossRef]
- Lamborn, K.R.; Chang, S.M.; Prados, M.D. Prognostic factors for survival of patients with glioblastoma: recursive partitioning analysis. Neuro-oncology 2004, 6, 227–235. [Google Scholar] [CrossRef]
- King’s College Hospital NHS Trust. Molecular pathology test protocol. https://www.kch.nhs.uk/gps/neuropathology-guide/molecular-pathology (2021).
- Pandith, A.A.; et al. Favorable role of IDH1/2 mutations aided with MGMT promoter gene methylation in the outcome of patients with malignant glioma. Future Sci OA 2020, 7, FSO663. [Google Scholar] [CrossRef]
- Molenaar, R.J.; et al. The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma better than either IDH1 or MGMT alone. Neuro Oncol 2014, 16, 1263–1273. [Google Scholar] [CrossRef] [PubMed]
- Arora, I.; et al. MGMT gene promoter methylation and its correlation with clinicopathological parameters in glioblastomas. Neurol India 2018, 66, 1106–1114. [Google Scholar]
- Ostrom, Q.T.; et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008-2012. Neuro Oncol 2015, 17 (Suppl. 4), iv1–iv62. [Google Scholar] [CrossRef]
- Bleeker, F.E.; et al. The prognostic IDH1( R132 ) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol 2010, 119, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Sanson, M.; et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2009, 27, 4150–4154. [Google Scholar] [CrossRef]
- Wanis, H.A.; Møller, H.; Ashkan, K.; Davies, E.A. The incidence of major subtypes of primary brain tumors in adults in England 1995-2017. Neuro-Oncology 2021, 23, 1371–1382. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009-2013. Neuro Oncol 2016, 18, v1–v75. [Google Scholar] [CrossRef]
- Sun, T.; Warrington, N.M.; Rubin, J.B. Why does Jack, and not Jill, break his crown? Sex disparity in brain tumors. Biol Sex Differ 2012, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Trifiletti, D.M.; et al. Prognostic Implications of Extent of Resection in Glioblastoma: Analysis from a Large Database. World Neurosurg 2017, 103, 330–340. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Rubin, J.B.; Lathia, J.D.; Berens, M.E.; Barnholtz-Sloan, J.S. Females have the survival advantage in glioblastoma. Neuro Oncol 2018, 20, 576–577. [Google Scholar] [CrossRef]
- Smits, A.; et al. Sex Disparities in MGMT Promoter Methylation and Survival in Glioblastoma: Further Evidence from Clinical Cohorts. J. Clin. Med. 2021, 10, 556. [Google Scholar] [CrossRef] [PubMed]
- Eoli, M.; et al. Methylation of O6-methylguanine DNA methyltransferase and loss of heterozygosity on 19q and/or 17p are overlapping features of secondary glioblastomas with prolonged survival. Clin Cancer Res 2007, 13, 2606–2613. [Google Scholar] [CrossRef] [PubMed]
- Ellingson, B.M.; et al. Anatomic localization of O6-methylguanine DNA methyltransferase (MGMT) promoter methylated and unmethylated tumors: a radiographic study in 358 de novo human glioblastomas. Neuroimage 2012, 59, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; et al. Radiogenomics of Glioblastoma: Machine Learning-based Classification of Molecular Characteristics by Using Multiparametric and Multiregional MR Imaging Features. Radiology 2016, 281, 907–918. [Google Scholar] [CrossRef]
- Paldor, I. , Drummond, K.J., Kaye, A.H. IDH1 mutation may not be prognostically favorable in glioblastoma when controlled for tumor location: A case-control study. J Clin Neurosci 2016, 34, 117–120. [Google Scholar] [CrossRef]
- Ang, S.Y.L.; et al. Incidence of biomarkers in high-grade gliomas and their impact on survival in a diverse SouthEast Asian cohort - a population-based study. BMC Cancer 2020, 20, 79. [Google Scholar] [CrossRef]
- Wanis, H.A.; Møller, H.; Ashkan, K.; Davies, E.A. The Influence of Ethnicity on Survival from Malignant Primary Brain Tumours in England: A Population-Based Cohort Study. Cancers 2023, 15, 1464. [Google Scholar] [CrossRef]
- Das, B.R.; Tangri, R.; Ahmad, F.; Roy, A.; Patole, K. Molecular investigation of isocitrate dehydrogenase gene (IDH) mutations in gliomas: first report of IDH2 mutations in Indian patients. Asian Pac J Cancer Prev 2013, 14, 7261–7264. [Google Scholar] [PubMed]
- Rajmohan, K.S.; et al. Prognostic significance of histomolecular subgroups of adult anaplastic (WHO Grade III) gliomas: applying the ‘integrated’ diagnosis approach. J Clin Pathol 2016, 69, 686–694. [Google Scholar] [CrossRef]
- Juratli, T.A.; Cahill, D.P.; McCutcheon, I.E. Determining optimal treatment strategy for diffuse glioma: the emerging role of IDH mutations. Expert Rev Anticancer Ther 2015, 15, 603–606. [Google Scholar] [CrossRef]
- Mulholland, S.; et al. MGMT CpG island is invariably methylated in adult astrocytic and oligodendroglial tumors with IDH1 or IDH2 mutations. Int J Cancer 2012, 131, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Mellai, M.; et al. MGMT promoter hypermethylation and its associations with genetic alterations in a series of 350 brain tumors. J Neurooncol 2012, 107, 617–631. [Google Scholar] [CrossRef] [PubMed]
- Ohka, F.; et al. The global DNA methylation surrogate LINE-1 methylation is correlated with MGMT promoter methylation and is a better prognostic factor for glioma. PLoS One 2011, 6, e23332. [Google Scholar] [CrossRef]
- Preusser, M.; et al. Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol 2008, 18, 520–532. [Google Scholar] [CrossRef]
- Minniti, G.; et al. IDH1 mutation and MGMT methylation status predict survival in patients with anaplastic astrocytoma treated with temozolomide-based chemoradiotherapy. J Neurooncol 2014, 118, 377–383. [Google Scholar] [CrossRef]
- Horbinski, C. What do we know about IDH1/2 mutations so far, and how do we use it? Acta Neuropathol 2013, 125, 621–636. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; et al. IDH mutation and MGMT promoter methylation in glioblastoma: results of a prospective registry. Oncotarget 2015, 6, 40896–40906. [Google Scholar] [CrossRef] [PubMed]
- McGirt, M.J.; et al. Independent association of extent of resection with survival in patients with malignant brain astrocytoma. J Neurosurg 2009, 110, 156–162. [Google Scholar] [CrossRef]
- Stummer, W.; et al. Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: a randomised controlled multicentre phase III trial. Lancet Oncol 2006, 7, 392–401. [Google Scholar] [CrossRef]
- Sanai, N.; Polley, M.-Y.; McDermott, M.W.; Parsa, A.T.; Berger, M.S. An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg 2011, 115, 3–8. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.; Li, Y.; Fang, S.; Jiang, T. Role of molecular biomarkers in glioma resection: a systematic review. Chin Neurosurg J 2020, 6, 18. [Google Scholar] [PubMed]
- Wick, W.; et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol 2009, 27, 5874–5880. [Google Scholar] [PubMed]
- Nuño, M.; et al. Survival and prognostic factors of anaplastic gliomas. Neurosurgery 2013, 73, 458–465. [Google Scholar]
- Panesar, S.; Tailor, J.; Bhangoo, R.; Ashkan, K. Multidisciplinary Team Management of Cerebral Metastases: Recent Trends and Future Implications. Clin Oncol (R Coll Radiol) 2016, 28, 343–344. [Google Scholar] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar]
- Brat, D.J.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 2015, 372, 2481–2498. [Google Scholar]
- Modrek, A.S.; et al. Low-Grade Astrocytoma Mutations in IDH1, P53, and ATRX Cooperate to Block Differentiation of Human Neural Stem Cells via Repression of SOX2. Cell Rep 2017, 21, 1267–1280. [Google Scholar]
| Variables | Groups | No. of patients | (%) | |
|---|---|---|---|---|
| Patient characteristics | Sex | Female | 296 | (39.5) |
| Male | 453 | (60.5) | ||
| Age at diagnosis | Mean (SD) | 56.4 (14.7) | ||
| Age group | =<39 | 119 | (15.9) | |
| 40-49 | 95 | (12.7) | ||
| 50-59 | 165 | (22.0) | ||
| 60-69 | 225 | (30.0) | ||
| >=70 | 145 | (19.4) | ||
| Year of diagnosis | 2015 | 151 | (20.2) | |
| 2016 | 136 | (18.2) | ||
| 2017 | 163 | (218) | ||
| 2018 | 147 | (19.6) | ||
| 2019 | 152 | (20.3) | ||
| Ethnic group | White British | 484 | (64.6) | |
| Asian/Asian British | 11 | (1.5) | ||
| Black/Black British | 23 | (3.1) | ||
| Mixed Ethnic Group | 10 | (1.3) | ||
| Any Other White | 30 | (4.1) | ||
| Any Other Ethnic Group | 19 | (2.5) | ||
| Not Stated/Specified | 172 | (23.0) | ||
| Performance status at referral | 0 | 312 | (41.7) | |
| 1 | 244 | (32.6) | ||
| 2 | 63 | (8.4) | ||
| 3/4 | 27 | (3.6) | ||
| Unknown | 103 | (13.8) | ||
| Vital status | Alive | 210 | (28.0) | |
| (on 30/07/2021) | Dead | 539 | (72.0) | |
| Histology | Glioblastoma | 570 | (76.1) | |
| Tumour characteristics | Astrocytoma | 98 | (13.1) | |
| Oligodendroglioma | 81 | (10.8) | ||
| WHO grade | I/II | 77 | (10.3) | |
| III/IV | 672 | (89.7) | ||
| Tumour location | Frontal Lobe | 237 | (31.6) | |
| Occipital Lobe | 27 | (3.6) | ||
| Parietal Lobe | 116 | (15.5) | ||
| Temporal Lobe | 176 | (23.5) | ||
| Overlapping Lesion of Brain | 131 | (17.5) | ||
| Cerebrum | 31 | (4.1) | ||
| Ventricle | 10 | (1.3) | ||
| Brain, other | 21 | (2.8) | ||
| IDH1 mutation | Mutated | 146 | (19.5) | |
| Wild Type | 587 | (78.4) | ||
| Not Tested/Unknown | 16 | (2.1) | ||
| MGMT promotor methylation | Methylated | 407 | (54.3) | |
| Unmethylated | 291 | (38.9) | ||
| Not Tested/Unknown | 51 | (6.8) | ||
| 1p19q co-deletion | Co-deleted | 86 | (11.5) | |
| Not Co-deleted | 50 | (6.7) | ||
| Not Tested/Unknown | 613 | (81.8) | ||
| Excision type | Total/Total Macroscopic | 149 | (19.9) | |
| Gross Subtotal/Partial | 192 | (25.6) | ||
| Extent Uncertain | 408 | (54.5) |
| Glioblastoma N=570 (76.1%) |
Astrocytoma N=98 (13.1%) |
Oligodendroglioma N=81 (10.8%) |
Total | p-value | |||||
|---|---|---|---|---|---|---|---|---|---|
| IDH Mutation | |||||||||
| IDH1Mutated | 19 | (3.3%) | 52 | (53.1%) | 75 | (92.6%) | 146 | (19.5%) | <0.001 |
| Wildtype | 550 | (96.5%) | 31 | (31.6%) | 6 | (7.4%) | 587 | (78.4%) | |
| Not Tested/Unknown | 1 | (0.2%) | 15 | (15.3%) | 0 | (0.0%) | 16 | (2.1%) | |
| MGMT Methylation | |||||||||
| MGMT methylated | 285 | (50.0%) | 60 | (61.2%) | 62 | (76.5%) | 407 | (54.3%) | <0.001 |
| Unmethylated | 277 | (48.6%) | 12 | (12.2%) | 2 | (2.5%) | 291 | (38.9%) | |
| Not Tested/Unknown | 8 | (1.4%) | 26 | (26.5%) | 17 | (21%) | 51 | (6.8%) | |
| 1p19q Co-deletion | |||||||||
| Co-deleted | 6 | (1.0%) | 4 | (4.1%) | 76 | (93.8%) | 86 | (11.5%) | <0.001 |
| Not Co-deleted | 21 | (3.7%) | 24 | (24.5%) | 5 | (6.2%) | 50 | (6.7%) | |
| Not Tested/Unknown | 543 | (95.3%) | 70 | (71.4%) | 0 | (0.0%) | 613 | (81.8%) | |
| Unadjusted | Adjusted for age, sex, PS, ethnicity |
and tumour biomarkers | and tumour histology, tumour location | and extent of excision | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | |||||||||
| Age at diagnosis | ||||||||||||||||||
| =<39 | 5.48 | 3.17 | 9.47 | 4.86 | 2.77 | 8.54 | 6.04 | 2.48 | 14.73 | 2.18 | 0.75 | 6.39 | 1.83 | 0.61 | 5.50 | |||
| 40-49 | 2.45 | 1.35 | 4.42 | 2.34 | 1.27 | 4.30 | 3.78 | 1.42 | 10.08 | 2.63 | 0.80 | 8.60 | 2.29 | 0.70 | 7.51 | |||
| 50-59 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |||||||||||||
| 60-69 | 0.45 | 0.25 | 0.84 | 0.48 | 0.26 | 0.89 | 0.53 | 0.21 | 1.37 | 0.60 | 0.18 | 1.95 | 0.51 | 0.15 | 1.72 | |||
| >=70 | 0.20 | 0.08 | 0.51 | 0.20 | 0.08 | 0.51 | 0.46 | 0.14 | 1.53 | 0.61 | 0.16 | 2.38 | 0.65 | 0.16 | 2.60 | |||
| χ2 and p-value | χ2 (4)=103.85; p<0.001 | χ2 (4)=86.56; p<0.001 | χ2 (4)=40.42; p<0.001 | χ2 (4)=9.03; p= 0.0603 | χ2 (4)= 7.54; p=0.1098 | |||||||||||||
|
Sex |
||||||||||||||||||
| Male | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |||||||||||||
| Female | 1.11 | 0.76 | 1.60 | 0.99 | 0.65 | 1.50 | 0.66 | 0.35 | 1.24 | 0.72 | 0.33 | 1.55 | 0.72 | 0.33 | 1.58 | |||
| χ2 and p-value | χ2 (1)=0.29; p=0.5913 | χ2 (1)=0.00; p=0.9456 | χ2 (1)=1.65; p=0.1984 | χ2 (1)=0.70; p= 0.4012 | χ2 (1)=0.65; p= 0.4193 | |||||||||||||
|
Ethnic groups |
||||||||||||||||||
| White British | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |||||||||||||
| Any Other Ethnic Group | 1.70 | 0.63 | 4.58 | 1.08 | 0.36 | 3.27 | 1.50 | 0.36 | 6.19 | 3.00 | 0.57 | 15.63 | 2.61 | 0.50 | 13.58 | |||
| Any Other White | 1.84 | 0.84 | 4.06 | 1.43 | 0.58 | 3.54 | 1.75 | 0.39 | 7.90 | 1.23 | 0.18 | 8.60 | 1.38 | 0.20 | 9.50 | |||
| Asian/Asian British | 3.68 | 1.05 | 12.97 | 2.30 | 0.49 | 10.78 | 1.56 | 0.13 | 18.47 | 1.20 | 0.03 | 54.51 | 1.11 | 0.03 | 39.17 | |||
| Black/Black British | 0.55 | 0.16 | 1.90 | 0.42 | 0.11 | 1.64 | 0.93 | 0.17 | 4.96 | 0.65 | 0.08 | 5.17 | 0.68 | 0.09 | 5.39 | |||
| Mixed Ethnic Group | 0.41 | 0.05 | 3.27 | 0.17 | 0.02 | 1.47 | 0.03 | 0.00 | 0.55 | 0.11 | 0.00 | 2.64 | 0.14 | 0.01 | 2.95 | |||
| Not Stated/Specified | 0.50 | 0.30 | 0.83 | 0.57 | 0.33 | 1.01 | 0.59 | 0.26 | 1.33 | 0.60 | 0.22 | 1.61 | 0.59 | 0.22 | 1.60 | |||
| χ2 and p-value | χ2 (6)=17.87; p=0.0066 | χ2 (6)=9.90; p=0.1288 | χ2 (6)= 8.34; p=0.2140 | χ2 (6)=5.02; p=0.5414 | χ2 (6)= 4.46; p=0.6148 | |||||||||||||
|
MGMT methylation status |
||||||||||||||||||
| Unmethylated | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |||||||||||||
| Methylated | 15.92 | 7.30 | 34.75 | 19.73 | 8.64 | 45.08 | 18.74 | 6.31 | 55.68 | 13.33 | 3.78 | 47.08 | 14.13 | 3.88 | 51.43 | |||
| χ2 and p-value | χ2 (1)= 48.33; p<0.001 | χ2 (1)= 50.18; p<0.001 | χ2 (1)=27.56; p<0.001 | χ2 (1)= 16.31; p=0.0001 | χ2 (1)= 16.21; p=0.0001 | |||||||||||||
|
Tumour location |
||||||||||||||||||
| Frontal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |||||||||||||
| Brain, other | 0.54 | 0.17 | 1.68 | 0.30 | 0.09 | 1.03 | 0.64 | 0.07 | 5.52 | 1.32 | 0.14 | 12.52 | 2.30 | 0.22 | 23.84 | |||
| Cerebrum | 0.08 | 0.01 | 0.58 | 0.05 | 0.01 | 0.40 | 0.11 | 0.01 | 1.26 | 0.06 | 0.00 | 0.85 | 0.08 | 0.00 | 1.82 | |||
| Occipital Lobe | 0.17 | 0.04 | 0.73 | 0.16 | 0.03 | 0.77 | 0.26 | 0.04 | 1.65 | 0.33 | 0.03 | 3.19 | 1.02 | 0.10 | 10.57 | |||
| Overlapping Brain Lesions | 0.48 | 0.29 | 0.81 | 0.68 | 0.37 | 1.23 | 1.12 | 0.47 | 2.67 | 1.33 | 0.47 | 3.74 | 1.69 | 0.56 | 5.08 | |||
| Parietal Lobe | 0.28 | 0.15 | 0.52 | 0.38 | 0.19 | 0.77 | 0.53 | 0.19 | 1.46 | 0.50 | 0.15 | 1.65 | 0.62 | 0.17 | 2.25 | |||
| Temporal Lobe | 0.28 | 0.17 | 0.48 | 0.35 | 0.19 | 0.63 | 0.44 | 0.18 | 1.06 | 0.41 | 0.15 | 1.17 | 0.67 | 0.23 | 1.99 | |||
| Ventricle | 0.29 | 0.03 | 2.39 | 0.20 | 0.02 | 1.92 | 0.16 | 0.01 | 3.57 | 0.09 | 0.00 | 1.83 | 0.08 | 0.00 | 2.19 | |||
| χ2 and p-value | χ2 (7)=39.34; p<0.001 | χ2 (7)=27.16; p=0.0003 | χ2 (7)=9.60; p=0.2122 | χ2 (7)=11.04; p=0.1370 | χ2 (7)=7.84; p=0.3466 | |||||||||||||
|
Tumour histology |
||||||||||||||||||
| Astrocytoma | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |||||||||||||
| Glioblastoma | 0.02 | 0.01 | 0.04 | 0.03 | 0.01 | 0.07 | 0.05 | 0.02 | 0.12 | 0.04 | 0.02 | 0.11 | 0.03 | 0.01 | 0.09 | |||
| Oligodendroglioma | 7.45 | 2.90 | 19.13 | 10.54 | 3.73 | 29.78 | 2.49 | 0.40 | 15.56 | 2.47 | 0.37 | 16.61 | 3.73 | 0.47 | 29.79 | |||
| χ2 and p-value | χ2 (2)=215.81; p<0.001 | χ2 (2)=154.87; p<0.001 | χ2 (2)=55.46; p<0.001 | χ2 (2)=55.89; p<0.001 | χ2 (2)= 53.13; p<0.001 | |||||||||||||
|
Extent of excision |
||||||||||||||||||
| Extent Uncertain | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |||||||||||||
| Gross Total/Total Macroscopic | 1.95 | 1.24 | 3.08 | 1.87 | 1.11 | 3.17 | 1.92 | 0.88 | 4.20 | 2.73 | 1.01 | 7.33 | 2.73 | 1.01 | 7.33 | |||
| Subtotal/Partial | 1.13 | 0.71 | 1.80 | 1.33 | 0.78 | 2.26 | 1.09 | 0.49 | 2.42 | 1.77 | 0.63 | 5.00 | 1.77 | 0.63 | 5.00 | |||
| χ2 and p-value | χ2 (2)=8.59; p=0.0136 | χ2 (2)=5.50; p=0.0638 | χ2 (2)=2.86; p=0.2387 | χ2 (2)=4.08; p=0.1299 | χ2 (2)=4.08; p=0.1299 | |||||||||||||
| Unadjusted | Adjusted for age, sex, PS, ethnicity | and tumour biomarkers | and tumour histology, tumour location | and extent of excision | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | ||||||
| Age at diagnosis | |||||||||||||||
| =<39 | 2.56 | 1.48 | 4.43 | 2.54 | 1.45 | 4.45 | 1.42 | 0.73 | 2.77 | 1.15 | 0.54 | 2.45 | 1.20 | 0.56 | 2.56 |
| 40-49 | 1.32 | 0.77 | 2.27 | 1.29 | 0.75 | 2.24 | 0.83 | 0.44 | 1.57 | 0.82 | 0.42 | 1.57 | 0.82 | 0.43 | 1.59 |
| 50-59 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||||
| 60-69 | 1.35 | 0.90 | 2.05 | 1.45 | 0.95 | 2.21 | 1.69 | 1.08 | 2.67 | 1.73 | 1.09 | 2.73 | 1.76 | 1.11 | 2.78 |
| >=70 | 1.11 | 0.70 | 1.75 | 1.18 | 0.74 | 1.88 | 1.56 | 0.95 | 2.55 | 1.56 | 0.95 | 2.57 | 1.53 | 0.93 | 2.53 |
| χ2 and p-value | χ2 (4)=12.41; p=0.0145 | χ2 (4)=11.42; p=0.0223 | χ2 (4)=9.01; p=0.0608 | χ2 (4)=9.26; p=0.0550 | χ2 (4)=9.28; p=0.0544 | ||||||||||
|
Sex |
|||||||||||||||
| Male | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||||
| Female | 1.52 | 1.11 | 2.08 | 1.54 | 1.11 | 2.12 | 1.72 | 1.22 | 2.44 | 1.72 | 1.20 | 2.46 | 1.75 | 1.22 | 2.51 |
| χ2 and p-value | χ2 (1)=6.76; p=0.0093 | χ2 (1)= 6.77; p=0.0093 | χ2 (1)=9.39; p= 0.0022 | χ2 (1)=8.88; p=0.0029 | χ2 (1)=9.38; p= 0.0022 | ||||||||||
|
Ethnic groups |
|||||||||||||||
| White British | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||||
| Any Other Ethnic Group | 0.94 | 0.37 | 2.39 | 0.78 | 0.30 | 2.03 | 0.82 | 0.27 | 2.48 | 0.90 | 0.29 | 2.76 | 0.92 | 0.30 | 2.82 |
| Any Other White | 0.86 | 0.39 | 1.87 | 0.73 | 0.33 | 1.63 | 0.59 | 0.24 | 1.48 | 0.52 | 0.20 | 1.36 | 0.49 | 0.19 | 1.28 |
| Asian/Asian British | 0.69 | 0.17 | 2.78 | 0.62 | 0.14 | 2.66 | 0.38 | 0.06 | 2.37 | 0.47 | 0.07 | 2.90 | 0.44 | 0.07 | 2.78 |
| Black/Black British | 0.69 | 0.28 | 1.68 | 0.69 | 0.27 | 1.74 | 0.79 | 0.30 | 2.10 | 0.76 | 0.28 | 2.04 | 0.75 | 0.28 | 2.01 |
| Mixed Ethnic Group | 1.60 | 0.41 | 6.27 | 1.17 | 0.29 | 4.76 | 1.53 | 0.36 | 6.50 | 1.36 | 0.32 | 5.84 | 1.37 | 0.32 | 5.90 |
| Not Stated/Specified | 0.90 | 0.63 | 1.29 | 0.93 | 0.64 | 1.35 | 1.01 | 0.68 | 1.50 | 1.00 | 0.67 | 1.50 | 1.00 | 0.67 | 1.48 |
| χ2 and p-value | χ2 (6)=1.79; p= 0.9379 | χ2 (6)=1.80; p= 0.9373 | χ2 (6)=3.01; p= 0.8070 | χ2 (6)=2.95; p= 0.8156 | χ2 (6)= 3.36; p= 0.7631 | ||||||||||
|
IDH1 mutation |
|||||||||||||||
| Unmutated | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||||
| Mutated | 15.92 | 7.30 | 34.75 | 19.88 | 8.72 | 45.33 | 16.71 | 6.15 | 45.43 | 12.61 | 4.14 | 38.41 | 15.54 | 4.73 | 51.05 |
| χ2 and p-value | χ2 (1)=48.33; p<0.001 | χ2 (1)=50.56; p<0.001 | χ2 (1)=30.46; p<0.001 | χ2 (1)=19.88; p<0.001 | χ2 (1)=20.42; p<0.001 | ||||||||||
|
Tumour location |
|||||||||||||||
| Frontal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||||
| Brain, other | 0.46 | 0.17 | 1.23 | 0.38 | 0.14 | 1.07 | 0.62 | 0.21 | 1.82 | 0.65 | 0.22 | 1.94 | 0.60 | 0.20 | 1.82 |
| Cerebrum | 0.34 | 0.15 | 0.80 | 0.28 | 0.12 | 0.67 | 0.43 | 0.17 | 1.07 | 0.43 | 0.17 | 1.07 | 0.40 | 0.16 | 1.01 |
| Occipital Lobe | 0.61 | 0.26 | 1.42 | 0.66 | 0.27 | 1.62 | 0.88 | 0.34 | 2.27 | 0.86 | 0.33 | 2.25 | 0.75 | 0.28 | 2.02 |
| Overlapping Brain Lesions | 0.81 | 0.52 | 1.28 | 0.83 | 0.51 | 1.33 | 0.93 | 0.56 | 1.55 | 0.95 | 0.57 | 1.58 | 0.93 | 0.55 | 1.58 |
| Parietal Lobe | 0.62 | 0.39 | 0.98 | 0.67 | 0.41 | 1.08 | 0.88 | 0.53 | 1.48 | 0.90 | 0.53 | 1.52 | 0.87 | 0.51 | 1.47 |
| Temporal Lobe | 0.55 | 0.36 | 0.82 | 0.56 | 0.37 | 0.87 | 0.71 | 0.45 | 1.13 | 0.71 | 0.45 | 1.13 | 0.68 | 0.42 | 1.09 |
| Ventricle | 3.08 | 0.36 | 26.06 | 1.80 | 0.20 | 15.93 | 3.40 | 0.38 | 30.69 | 3.08 | 0.34 | 27.96 | 3.03 | 0.33 | 27.55 |
| χ2 and p-value | χ2 (7)=15.90; p=0.0261 | χ2 (7)=15.37; p=0.0316 | χ2 (7)=6.80; p=0.4499 | χ2 (7)=6.59; p=0.4732 | χ2 (7)=7.48; p=0.3805 | ||||||||||
|
Tumour histology |
|||||||||||||||
| Astrocytoma | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||||
| Glioblastoma | 0.21 | 0.11 | 0.39 | 0.19 | 0.09 | 0.39 | 0.45 | 0.19 | 1.08 | 0.46 | 0.19 | 1.12 | 0.44 | 0.18 | 1.09 |
| Oligodendroglioma | 6.20 | 1.33 | 28.88 | 7.01 | 1.45 | 33.91 | 0.21 | 0.02 | 2.45 | 0.22 | 0.02 | 2.51 | 0.10 | 0.01 | 1.80 |
| χ2 and p-value | χ2 (2)=44.17; p<0.001 | χ2 (2)=40.62; p<0.001 | χ2 (2)=4.34; p=0.1141 | χ2 (2)=4.05; p=0.1318 | χ2 (2)=5.03; p=0.0808 | ||||||||||
|
Extent of excision |
|||||||||||||||
| Extent Uncertain | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||||||||
| Gross Total/Total Macroscopic | 0.95 | 0.64 | 1.40 | 0.84 | 0.56 | 1.26 | 0.70 | 0.44 | 1.10 | 0.68 | 0.43 | 1.08 | 0.68 | 0.43 | 1.08 |
| Subtotal/Partial | 0.95 | 0.66 | 1.37 | 0.97 | 0.66 | 1.41 | 0.93 | 0.62 | 1.40 | 0.93 | 0.61 | 1.40 | 0.93 | 0.61 | 1.40 |
| χ2 and p-value | χ2 (2)=0.11; p=0.9450 | χ2 (2)=0.72; p=0.6994 | χ2 (2)=2.47; p=0.2903 | χ2 (2)=0.11; p=0.9450 | χ2 (2)=0.11; p=0.9450 | ||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





