Submitted:
17 May 2023
Posted:
18 May 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study population
2.2. Clinical information
2.3. Sample collection for molecular biology analysis
2.4. Sequencing 324 cancer-associated genes
2.5. Mutated genes identification related with temozolomide resistance mechanisms
2.6. Bioinformatic analysis of mutations effect on the proteins
3. Results
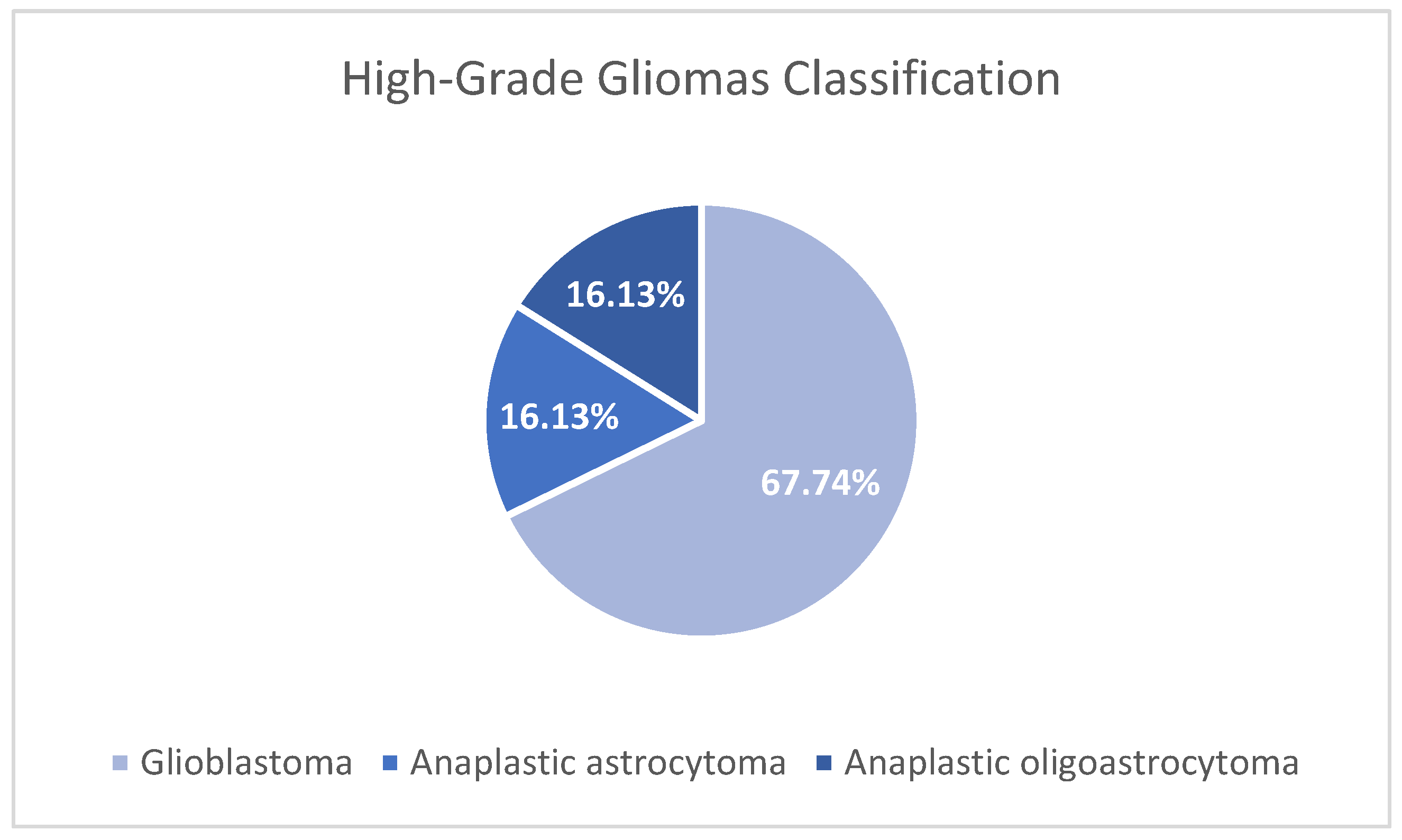
3.1. Study population
3.2. Clinical information
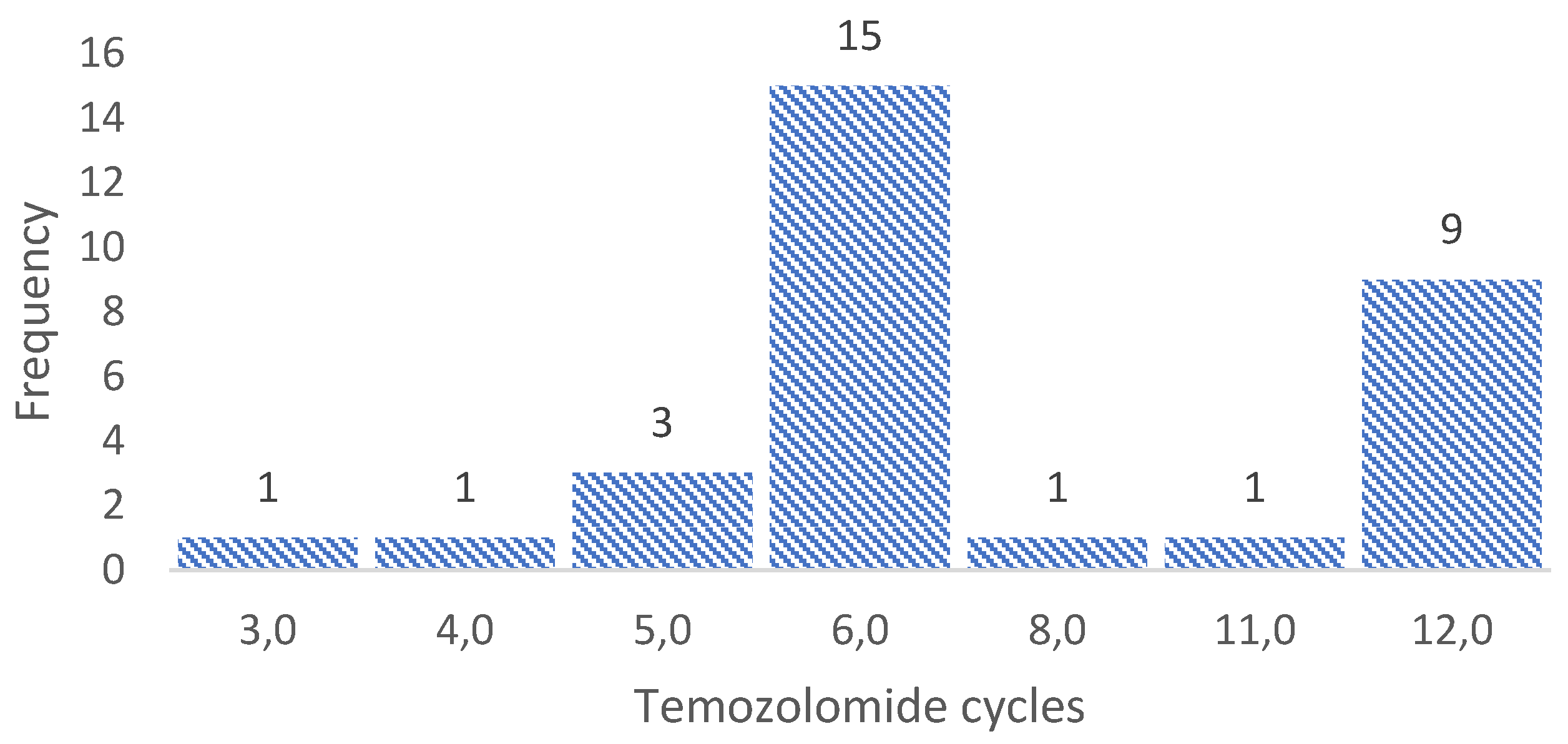
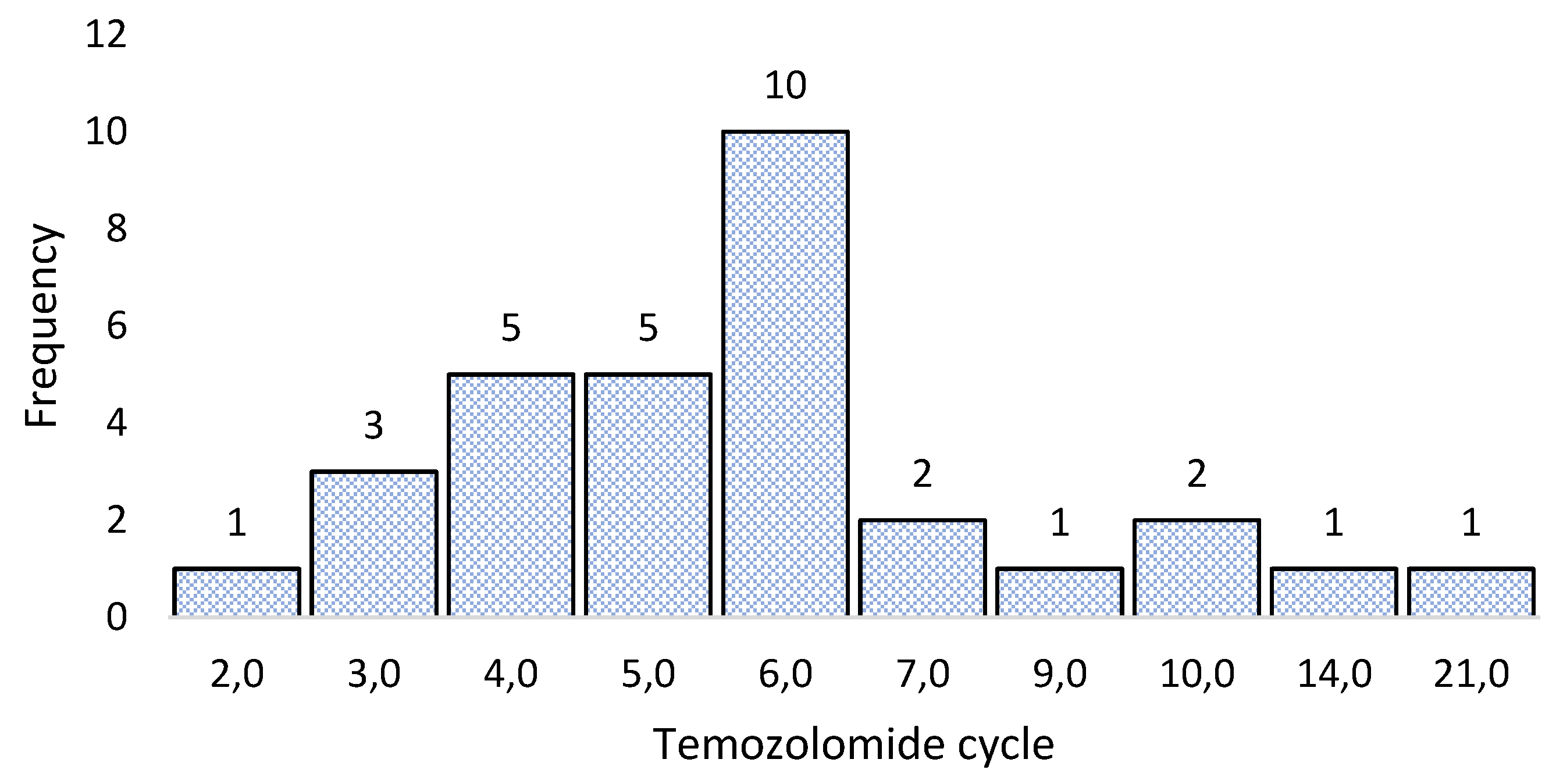
3.3. Sample collection for molecular biology analysis
3.4. Sequencing 324 cancer-associated genes
3.5. Mutated genes identification related with temozolomide resistance mechanisms
3.6. Bioinformatic analysis of mutations effect on the proteins
4. Discussion
5. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021 May;71(3):209–49. [CrossRef]
- Gómez Vega JC, Ocampo Navia MI, De Vries E, Feo Lee O. Sobrevida de los tumores cerebrales primarios en Colombia. Univ Médica [Internet]. 2020 May 7 [cited 2022 Jul 31];61(3).
- Low JT, Ostrom QT, Cioffi G, Neff C, Waite KA, Kruchko C, et al. Primary brain and other central nervous system tumors in the United States (2014-2018): A summary of the CBTRUS statistical report for clinicians. Neuro-Oncol Pract. 2022 May 13;9(3):165–82. [CrossRef]
- Jeans A, Esiri M. Brain histology. Pract Neurol. 2008 Oct 1;8(5):303–10.
- Goryaynov SA, Potapov AA, Ignatenko MA, Zhukov VYu, Protskiy SV, Zakharova NA, et al. Glioblastoma metastases: a literature review and a description of six clinical observations. Vopr Neirokhirurgii Im NN Burdenko. 2015;79(2):33. [CrossRef]
- Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma Subclassifications and Their Clinical Significance. Neurotherapeutics. 2017 Apr;14(2):284–97. [CrossRef]
- Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro-Oncol. 2021 Aug 2;23(8):1231–51. [CrossRef]
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N Engl J Med. 2005 Mar 10;352(10):987–96. [CrossRef]
- Martinez R, Esteller M. The DNA methylome of glioblastoma multiforme. Neurobiol Dis. 2010 Jul;39(1):40–6. [CrossRef]
- Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N Engl J Med. 2005 Mar 10;352(10):997–1003. [CrossRef]
- Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. Ha P, editor. PLOS ONE. 2020 Sep 25;15(9): e0237802. [CrossRef]
- Back, M., Rodriguez, M., Jayamanne, D., Khasraw, M., Lee, A., & Wheeler, H. (2018). Understanding the Revised Fourth Edition of the World Health Organization Classification of Tumours of the Central Nervous System (2016) for Clinical Decision-making: A Guide for Oncologists Managing Patients with Glioma. Clinical oncology, 30(9), 556–562. [CrossRef]
- Komori T. Grading of adult diffuse gliomas according to the 2021 WHO Classification of Tumors of the Central Nervous System. Lab Invest. 2022 Feb;102(2):126–33. [CrossRef]
- Mikkelsen VE, Solheim O, Salvesen Ø, Torp SH. The histological representativeness of glioblastoma tissue samples. Acta Neurochir (Wien). 2021 Jul;163(7):1911–20. [CrossRef]
- Iwanaga T, Takahashi Y, Fujita T. Immunohistochemistry of neuron-specific and glia-specific proteins. Arch Histol Cytol. 1989;52(Suppl):13–24. [CrossRef]
- Hervey-Jumper SL, Berger MS. Maximizing safe resection of low- and high-grade glioma. J Neurooncol. 2016 Nov;130(2):269–82. [CrossRef]
- Lakomy R, Kazda T, Selingerova I, Poprach A, Pospisil P, Belanova R, et al. Real-World Evidence in Glioblastoma: Stupp’s Regimen After a Decade. Front Oncol. 2020 Jul 3;10:840. [CrossRef]
- Paleologos N, Newton H, editors. Oligodendroglioma: clinical presentation, pathology, molecular biology, imaging, and treatment. San Diego: Elsevier/Academic Press; 2019. 402 p.
- Profyris C, Chen E, Young IM, Chendeb K, Ahsan SA, Briggs RG, et al. Anaplastic Oligodendroglioma – Is Adjuvant Radiotherapy Mandatory following Maximal Surgical Resection? Clin Neurol Neurosurg. 2021 Jan;200:106303. [CrossRef]
- Weller, M., Wick, W., Aldape, K., Brada, M., Berger, M., Pfister, S. M., Nishikawa, R., Rosenthal, M., Wen, P. Y., Stupp, R., & Reifenberger, G. (2015). Glioma. Nature reviews. Disease primers, 1, 15017. [CrossRef]
- Zhang J, F.G. Stevens M, D. Bradshaw T. Temozolomide: Mechanisms of Action, Repair and Resistance. Curr Mol Pharmacol. 2012 Jan 1;5(1):102–14. [CrossRef]
- Wang JYJ, Edelmann W. Mismatch repair proteins as sensors of alkylation DNA damage. Cancer Cell. 2006 Jun;9(6):417–8. [CrossRef]
- Massoud TF, Paulmurugan R, editors. Glioblastoma resistance to chemotherapy: molecular mechanisms and innovative reversal strategies. 1st ed. San Diego: Academic Press is an imprint of Elsevier; 2021.
- Shen, W., Hu, J. A., & Zheng, J. S. (2014). Mechanism of temozolomide-induced antitumour effects on glioma cells. The Journal of international medical research, 42(1), 164–172. [CrossRef]
- Chen Y, Hu F, Zhou Y, Chen W, Shao H, Zhang Y. MGMT Promoter Methylation and Glioblastoma Prognosis: A Systematic Review and Meta-analysis. Arch Med Res. 2013 May;44(4):281–90. [CrossRef]
- Nabors LB, Portnow J, Ahluwalia M, Baehring J, Brem H, Brem S, et al. Central Nervous System Cancers, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2020 Nov;18(11):1537–70. [CrossRef]
- Weller M, Stupp R, Reifenberger G, Brandes AA, van den Bent MJ, Wick W, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010 Jan;6(1):39–51. [CrossRef]
- Binabaj, M. M., Bahrami, A., ShahidSales, S., Joodi, M., Joudi Mashhad, M., Hassanian, S. M., Anvari, K., & Avan, A. (2018). The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials. Journal of cellular physiology, 233(1), 378–386. [CrossRef]
- Knobbe, C. B., & Reifenberger, G. (2003). Genetic alterations and aberrant expression of genes related to the phosphatidyl-inositol-3'-kinase/protein kinase B (Akt) signal transduction pathway in glioblastomas. Brain pathology (Zurich, Switzerland), 13(4), 507–518. [CrossRef]
- Shepherd P. R. (2005). Mechanisms regulating phosphoinositide 3-kinase signalling in insulin-sensitive tissues. Acta physiologica Scandinavica, 183(1), 3–12. [CrossRef]
- Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017 Mar 9;168(6):960-976. [CrossRef]
- Jaiswal BS, Kljavin NM, Stawiski EW, Chan E, Parikh C, Durinck S, et al. Oncogenic ERBB3 Mutations in Human Cancers. Cancer Cell. 2013 May;23(5):603–17.
- Roskoski R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014 Jan;79:34–74. [CrossRef]
- Vanajothi R, Rajamanika S, Sudha A, Srinivasan P. Structural and Functional Analysis of KIT Gene Encoding Receptor Tyrosine Kinase and its Interaction with Sunitinib and HDAC Inhibitors: An in silico Approach. Pak J Biol Sci. 2012 Jan 15;15(3):121–31. [CrossRef]
- Pathania S, Pentikäinen OT, Singh PK. A holistic view on c-Kit in cancer: Structure, signaling, pathophysiology and its inhibitors. Biochim Biophys Acta BBA - Rev Cancer. 2021 Dec;1876(2):188631. [CrossRef]
- RR, Li KKW, Zhang ZY, Chan AKY, Wang WW, Chan DTM, et al. Mismatch repair proteins PMS2 and MLH1 can further refine molecular stratification of IDH-mutant lower grade astrocytomas. Clin Neurol Neurosurg. 2021 Sep;208:106882. [CrossRef]
- Harder BG, Peng S, Sereduk CP, Sodoma AM, Kitange GJ, Loftus JC, et al. Inhibition of phosphatidylinositol 3-kinase by PX-866 suppresses temozolomide-induced autophagy and promotes apoptosis in glioblastoma cells. Mol Med. 2019 Dec;25(1):49. [CrossRef]
- Gomes AL, Reis-Filho JS, Lopes JM, Martinho O, Lambros MBK, Martins A, et al. Molecular Alterations of KIT Oncogene in Gliomas. Anal Cell Pathol. 2007 Jan 1;29(5):399–408. [CrossRef]
- Beauchamp RL, Erdin S, Witt L, Jordan JT, Plotkin SR, Gusella JF, et al. mTOR kinase inhibition disrupts neuregulin 1-ERBB3 autocrine signaling and sensitizes NF2-deficient meningioma cellular models to IGF1R inhibition. J Biol Chem. 2021 Jan;296:100157. [CrossRef]
- Botchkareva NV, Khlgatian M, Jack Longley B, Botchkarev VA, Gilchrest BA. SCF/c-kit signaling is required for cyclic regeneration of the hair pigmentation unit. FASEB J. 2001 Mar;15(3):645–58. [CrossRef]
- Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017 Mar;168(6):960–76. [CrossRef]
- Parthasarathy R, Eisenberg F. The inositol phospholipids: a stereochemical view of biological activity. Biochem J. 1986 Apr 15;235(2):313–22. [CrossRef]
- Tanaka S, Batchelor TT, Iafrate AJ, Dias-Santagata D, Borger DR, Ellisen LW, et al. PIK3CA activating mutations are associated with more disseminated disease at presentation and earlier recurrence in glioblastoma. Acta Neuropathol Commun. 2019 Dec;7(1):66. [CrossRef]
- Liggett LA, DeGregori J. Changing mutational and adaptive landscapes and the genesis of cancer. Biochim Biophys Acta BBA - Rev Cancer. 2017 Apr;1867(2):84–94. [CrossRef]
- Gozzelino L, Kochlamazashvili G, Baldassari S, Mackintosh AI, Licchetta L, Iovino E, et al. Defective lipid signalling caused by mutations in PIK3C2B underlies focal epilepsy. Brain. 2022 Jul 29;145(7):2313–31. [CrossRef]
- Knobbe CB, Reifenberger G. Genetic Alterations and Aberrant Expression of Genes Related to the Phosphatidyl-lnositol-3′-Kinase/Protein Kinase B (Akt) Signal Transduction Pathway in Glioblastomas. Brain Pathol. 2006 Apr 5;13(4):507–18. [CrossRef]
- Rao SK, Edwards J, Joshi AD, Siu IM, Riggins GJ. A survey of glioblastoma genomic amplifications and deletions. J Neurooncol. 2010 Jan;96(2):169–79. [CrossRef]
- Maini J, Ghasemi M, Yandhuri D, Thakur SS, Brahmachari V. Human PRE-PIK3C2B, an intronic cis-element with dual function of activation and repression. Biochim Biophys Acta BBA - Gene Regul Mech. 2017 Feb;1860(2):196–204. [CrossRef]
- Aung KT, Yoshioka K, Aki S, Ishimaru K, Takuwa N, Takuwa Y. The class II phosphoinositide 3-kinases PI3K-C2α and PI3K-C2β differentially regulate clathrin-dependent pinocytosis in human vascular endothelial cells. J Physiol Sci. 2019 Mar;69(2):263–80. [CrossRef]
- Wang C, Yang Y, Yin L, Wei N, Hong T, Sun Z, et al. Novel Potential Biomarkers Associated With Epithelial to Mesenchymal Transition and Bladder Cancer Prognosis Identified by Integrated Bioinformatic Analysis. Front Oncol. 2020 Jun 30;10:931. [CrossRef]
- Margaria JP, Ratto E, Gozzelino L, Li H, Hirsch E. Class II PI3Ks at the Intersection between Signal Transduction and Membrane Trafficking. Biomolecules. 2019 Mar 15;9(3):104. [CrossRef]
- Liu Z, Li X, Ma J, Li D, Ju H, Liu Y, et al. Integrative Analysis of the IQ Motif-Containing GTPase-Activating Protein Family Indicates That the IQGAP3-PIK3C2B Axis Promotes Invasion in Colon Cancer. OncoTargets Ther. 2020 Aug;Volume 13:8299–311. [CrossRef]
- Liu Z, Sun C, Zhang Y, Ji Z, Yang G. Phosphatidylinositol 3-Kinase-C2β Inhibits Cisplatin-Mediated Apoptosis via the Akt Pathway in Oesophageal Squamous Cell Carcinoma. J Int Med Res. 2011 Aug;39(4):1319–32. [CrossRef]
- Cisse O, Quraishi M, Gulluni F, Guffanti F, Mavrommati I, Suthanthirakumaran M, et al. Downregulation of class II phosphoinositide 3-kinase PI3K-C2β delays cell division and potentiates the effect of docetaxel on cancer cell growth. J Exp Clin Cancer Res. 2019 Dec;38(1):472. [CrossRef]
- Löw S, Vougioukas VI, Hielscher T, Schmidt U, Unterberg A, Halatsch ME. Pathogenetic pathways leading to glioblastoma multiforme: association between gene expressions and resistance to erlotinib. Anticancer Res. 2008 Dec;28(6A):3729–32.
- Ortiz R, Perazzoli G, Cabeza L, Jiménez-Luna C, Luque R, Prados J, et al. Temozolomide: An Updated Overview of Resistance Mechanisms, Nanotechnology Advances and Clinical Applications. Curr Neuropharmacol. 2021 Apr;19(4):513–37. [CrossRef]
- Indraccolo S, Lombardi G, Fassan M, Pasqualini L, Giunco S, Marcato R, et al. Genetic, Epigenetic, and Immunologic Profiling of MMR-Deficient Relapsed Glioblastoma. Clin Cancer Res. 2019 Mar 15;25(6):1828–37. [CrossRef]
- Felsberg J, Thon N, Eigenbrod S, Hentschel B, Sabel MC, Westphal M, et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer. 2011 Aug 1;129(3):659–70. [CrossRef]
- Higuchi F, Nagashima H, Ning J, Koerner MVA, Wakimoto H, Cahill DP. Restoration of Temozolomide Sensitivity by PARP Inhibitors in Mismatch Repair Deficient Glioblastoma is Independent of Base Excision Repair. Clin Cancer Res. 2020 Apr 1;26(7):1690–9. [CrossRef]
- Yang B, Han N, Sun J, Jiang H, Xu HY. CtIP contributes to non-homologous end joining formation through interacting with ligase IV and promotion of TMZ resistance in glioma cells. Eur Rev Med Pharmacol Sci. 2019 Mar;23(5):2092–102. [CrossRef]
- Zhang P, Wang H, Chen Y, Lodhi AF, Sun C, Sun F, et al. DR5 related autophagy can promote apoptosis in gliomas after irradiation. Biochem Biophys Res Commun. 2020 Feb;522(4):910–6. [CrossRef]

| Gene | Gene presence | P-value | Hazard Ratio (CI 95%) | |
|---|---|---|---|---|
| Si | Not | |||
| PIK3C2B | 2(11,7) | 15(88,2) | 0,004 | 13,81 (2,258 - 84,45) |
| NOTCH3 | 4(23,5) | 13(76,4) | 0,005 | 6,026 (1,742 - 20,84) |
| KIT | 4(23,5) | 13(76,4) | 0,024 | 3,985 (1,204 - 13,18) |
| ERBB3 | 3(17,6) | 14(82,3) | 0,04 | 3,871 (1,066 - 14,04) |
| MLH1 | 3(17,6) | 14(82,3) | 0,06 | 3,528 (0,950 - 13,09) |
| Gene | P-value | HR Raw value (CI 95%) | P-value | HR Adjusted value (CI 95%) | |
|---|---|---|---|---|---|
| PIK3C2B | 0,004 | 13,81 (2,258 - 84,45) | 0,000 | 82,37 (8,36 - 811,67) | |
| KIT | 0,024 | 3,985 (1,204 - 13,18) | 0,002 | 10,24 (2,42 - 43,34) | |
| ERBB3 | 0,04 | 3,871 (1,066 - 14,04) | 0,001 | 13,20 (2,77 - 62,77) | |
| MLH1 | 0,06 | 3,528 (0,950 - 13,09) | 0,006 | 8,50 (1,83 - 39,45) |
| Protein | Domains | Biological processes | Molecular function | Cellular component |
|---|---|---|---|---|
| PIK3C2B | -R458Q mutation affects domain (PI3K_Ras-bd_dom) - Amplification - Rearrangement |
-Phosphatidylinositol-mediated signaling - Phosphatidylinositol phosphatase biosynthetic process |
-Phosphatidylinositol binding -kinase activity |
None |
| ERBB3 | - R164K mutation affects domain (Rcp_L) - L1177l the mutation does not affect any functional domain |
-Membrane receptor protein tyrosine kinase signaling pathway. -Protein phosphorylation |
-Protein kinase activity -Protein tyrosine kinase activity -ATP binding |
Membrane |
| KIT | - Amplification - Wrong amplification |
-Protein phosphorylation -Transmembrane receptor protein tyrosine kinase signaling pathway -Kit signaling pathway -Fc receptor signaling pathway |
-ATP binding -Transmembrane receptor -Protein tyrosine kinase activity -Protein kinase activity -Cytokine binding -Protein tyrosine kinase activity |
None |
| MLH1 | -(F261fs*7) mutation affects domain (DNA_mismatch_repair_N and DNA_mismatch_S5_2-like) | DNA mismatch repair | -Mismatched DNA binding -ATP binding -ATP-dependent DNA damage Sensor activity -ATP hydrolysis activity |
Mismatch Repair Complex |
| Protein | protein-protein interactions | Metabolic routes |
|---|---|---|
| PIK3C2B | RICTOR, EGFR, GRB2, ANKRD32, SBF2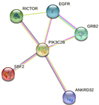
|
-DNA damage response (only ATM dependent) -Focal adhesion: PI3K-Akt-mTOR-signaling pathway -Glioblastoma signaling pathways -Phosphoinositides metabolism -Regulation of actin cytoskeleton |
| ERBB3 | ERBB2, EGFR, NNRG1, GRB2, SHC1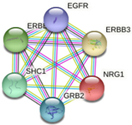
|
-Apoptosis-related network due to altered Notch3 in ovarian cancer -EGFR tyrosine kinase inhibitor resistance -ErbB signaling pathway -Glioblastoma signaling pathways -Heart development |
| KIT | PIK3R1, PLCG1, KITLG, GRB2, CRK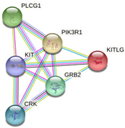
|
-Breast cancer pathway -Cardiac progenitor differentiation -Focal adhesion: PI3K-Akt-mTOR-signaling pathway -Gastrin signaling pathway -Hippo signaling regulation pathways |
| MLH1 | MSH2, PMS2, PMS1, BLM, EXO1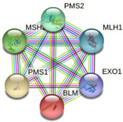
|
-Chromosomal and microsatellite instability in colorectal cancer -DNA IR-damage and cellular response via ATR -DNA mismatch repair -DNA repair pathways, full network -Ovarian infertility |
| Gene | Normal protein | Mutated protein |
|---|---|---|
|
PIK3C2B R458Q Mutation |
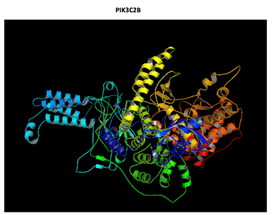 |
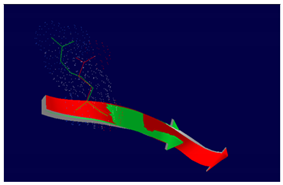 The effect of the mutation on the structure of the protein leads to the shortening of the radical, reducing the Van der Waals forces. At structural level, the mutation is located be-tween the amino acids (457-464) that make up a beta sheet, generating an elongation of the beta sheet in the mutated protein. |
|
ERBB3 R164K Mutation |
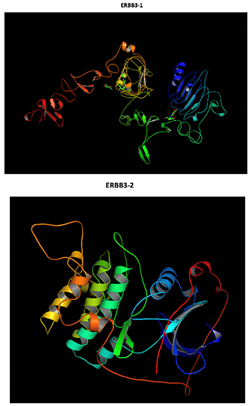 Two-part patterned protein |
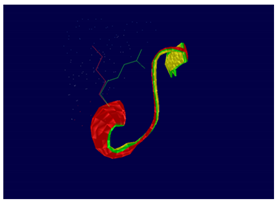 At structural level, an amino acid change is observed which leads to an alteration in the orientation of the radical group that alters the spatial distribution of Van der Waals forces. |
|
MLH1 F261fs*31 Mutation |
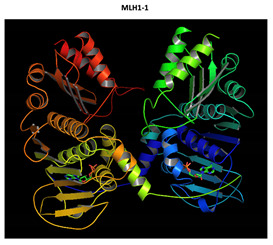 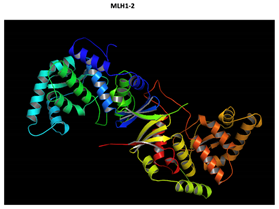 Two-part patterned protein. |
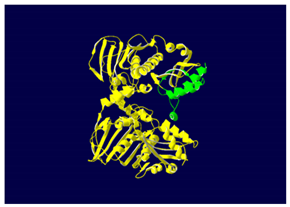 The yellow color shows the common region between the wild-type and mutated protein, the green color shows the region that would be lost in the mutated protein by the occurrence of a premature stop codon. |
| Gene | Associated diseases |
|---|---|
| PIK3C2B | -High grade glioma -Maffucci -Hepatitis C |
| ERBB3 | -High grade glioma -Breast cancer -Frontal myelination -Enteric nervous system -Diabetes type 1 |
| KIT | -High grade glioma -Piebaldism -GIST -Mastocytosis -LMA -Melanoma - Germ cell tumors |
| MLH1 | -High grade glioma -Hereditary nonpolyposis colorectal cancer (Lynch) -Turcot type I |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





