Submitted:
31 December 2024
Posted:
03 January 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Antisense Oligonucleotides (ASOs)
2.1. Chemical Modifications
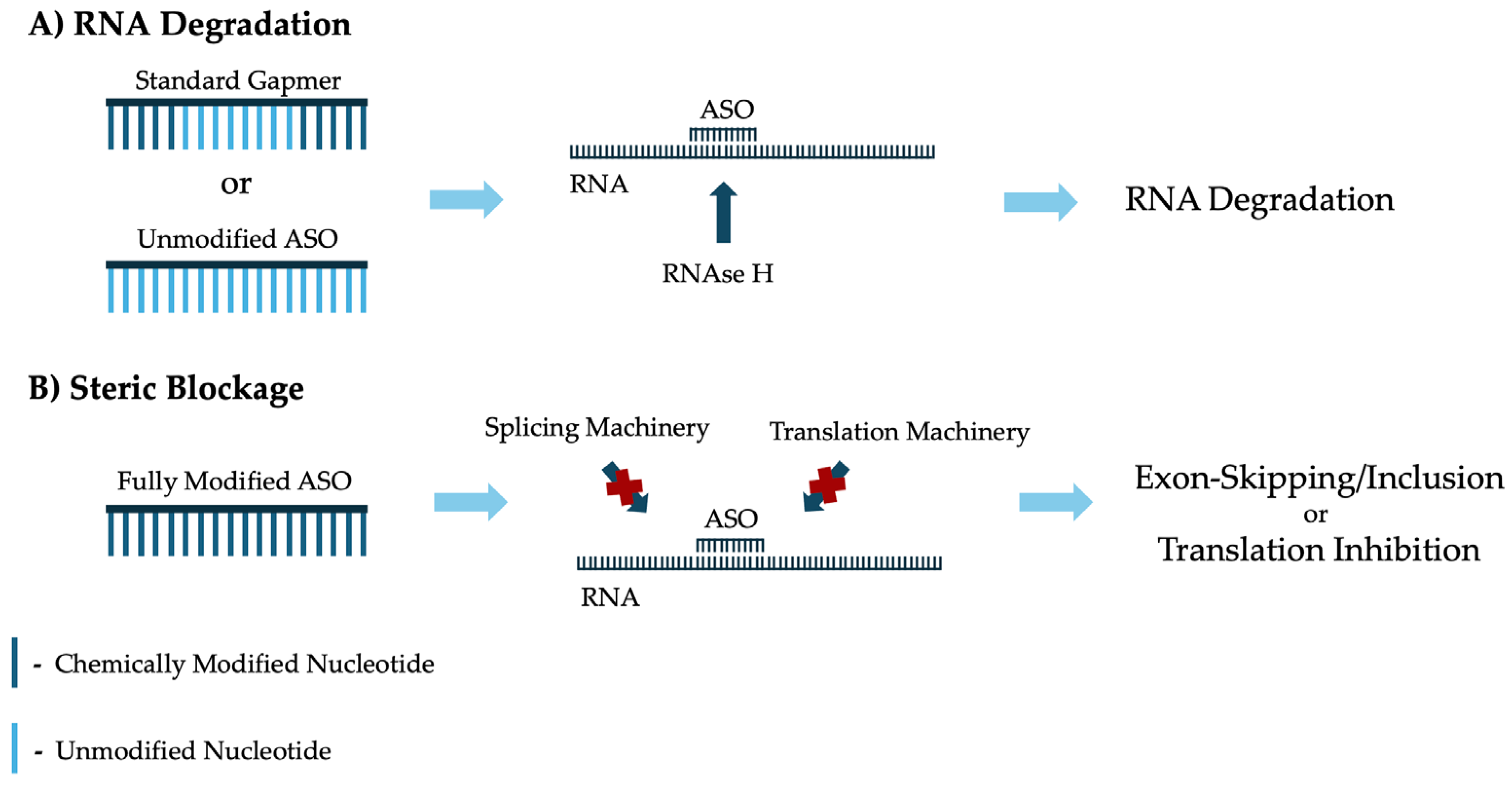
2.2. Mechanisms of Action
2.3. FDA-Approved ASOs
2.4. ASOs as N-of-1 Therapies
2.5. Challenges Associated with ASO Design
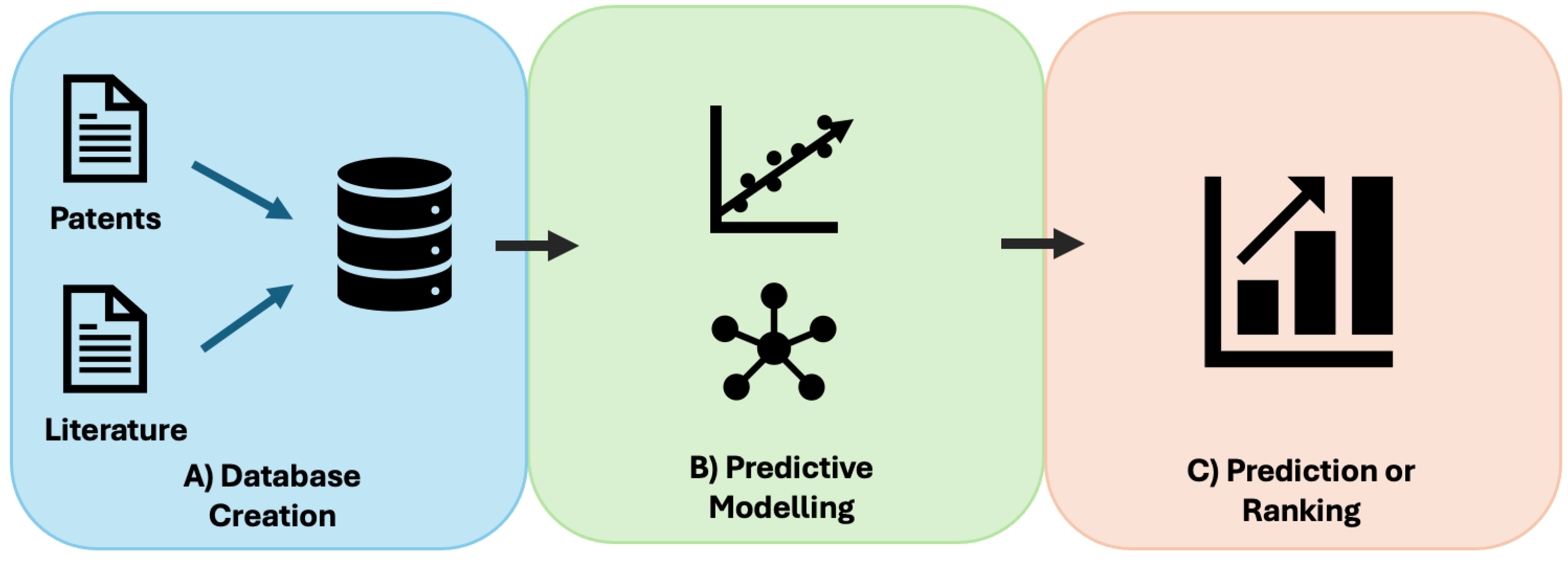
3. Machine Learning-Based Platforms to Improve Antisense Oligonucleotide Design
3.2. ASOptimizer
3.3. Limitations and Future Directions of Current Machine Learning-Based Platforms
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aronson, J. Rare Diseases, Orphan Drugs, and Orphan Diseases. BMJ 2006, 333, 127.
- Nguengang Wakap, S.; Lambert, D.M.; Olry, A.; Rodwell, C.; Gueydan, C.; Lanneau, V.; Murphy, D.; Le Cam, Y.; Rath, A. Estimating Cumulative Point Prevalence of Rare Diseases: Analysis of the Orphanet Database. Eur. J. Hum. Genet. EJHG 2020, 28, 165–173. [CrossRef]
- Haendel, M.; Vasilevsky, N.; Unni, D.; Bologa, C.; Harris, N.; Rehm, H.; Hamosh, A.; Baynam, G.; Groza, T.; McMurry, J.; et al. How Many Rare Diseases Are There? Nat. Rev. Drug Discov. 2020, 19, 77–78. [CrossRef]
- Health, T.L.G. The Landscape for Rare Diseases in 2024. Lancet Glob. Health 2024, 12, e341. [CrossRef]
- Lee, C.E.; Singleton, K.S.; Wallin, M.; Faundez, V. Rare Genetic Diseases: Nature’s Experiments on Human Development. iScience 2020, 23, 101123. [CrossRef]
- Mazzucato, M.; Visonà Dalla Pozza, L.; Minichiello, C.; Toto, E.; Vianello, A.; Facchin, P. Estimating Mortality in Rare Diseases Using a Population-Based Registry, 2002 through 2019. Orphanet J. Rare Dis. 2023, 18, 362. [CrossRef]
- Yang, G.; Cintina, I.; Pariser, A.; Oehrlein, E.; Sullivan, J.; Kennedy, A. The National Economic Burden of Rare Disease in the United States in 2019. Orphanet J. Rare Dis. 2022, 17, 163. [CrossRef]
- Tisdale, A.; Cutillo, C.M.; Nathan, R.; Russo, P.; Laraway, B.; Haendel, M.; Nowak, D.; Hasche, C.; Chan, C.-H.; Griese, E.; et al. The IDeaS Initiative: Pilot Study to Assess the Impact of Rare Diseases on Patients and Healthcare Systems. Orphanet J. Rare Dis. 2021, 16, 429. [CrossRef]
- Fehr, A.; Prütz, F. Rare Diseases: A Challenge for Medicine and Public Health. J. Health Monit. 2023, 8, 3–6. [CrossRef]
- Ng, Q.X.; Ong, C.; Chan, K.E.; Ong, T.S.K.; Lim, I.J.X.; Tang, A.S.P.; Chan, H.W.; Koh, G.C.H. Comparative Policy Analysis of National Rare Disease Funding Policies in Australia, Singapore, South Korea, the United Kingdom and the United States: A Scoping Review. Health Econ. Rev. 2024, 14, 42. [CrossRef]
- Augustine, E.F.; Adams, H.R.; Mink, J.W. Clinical Trials in Rare Disease: Challenges and Opportunities. J. Child Neurol. 2013, 28, 1142–1150. [CrossRef]
- Griggs, R.C.; Batshaw, M.; Dunkle, M.; Gopal-Srivastava, R.; Kaye, E.; Krischer, J.; Nguyen, T.; Paulus, K.; Merkel, P.A.; Rare Diseases Clinical Research Network Clinical Research for Rare Disease: Opportunities, Challenges, and Solutions. Mol. Genet. Metab. 2009, 96, 20–26. [CrossRef]
- Fermaglich, L.J.; Miller, K.L. A Comprehensive Study of the Rare Diseases and Conditions Targeted by Orphan Drug Designations and Approvals over the Forty Years of the Orphan Drug Act. Orphanet J. Rare Dis. 2023, 18, 163. [CrossRef]
- Lauffer, M.C.; van Roon-Mom, W.; Aartsma-Rus, A. Possibilities and Limitations of Antisense Oligonucleotide Therapies for the Treatment of Monogenic Disorders. Commun. Med. 2024, 4, 1–11. [CrossRef]
- Dhuri, K.; Bechtold, C.; Quijano, E.; Pham, H.; Gupta, A.; Vikram, A.; Bahal, R. Antisense Oligonucleotides: An Emerging Area in Drug Discovery and Development. J. Clin. Med. 2020, 9, 2004. [CrossRef]
- Perry, C.M.; Balfour, J.A. Fomivirsen. Drugs 1999, 57, 375–380; discussion 381. [CrossRef]
- Hair, P.; Cameron, F.; McKeage, K. Mipomersen Sodium: First Global Approval. Drugs 2013, 73, 487–493. [CrossRef]
- Gupta, S.; Sharma, S.N.; Kundu, J.; Pattanayak, S.; Sinha, S. Morpholino Oligonucleotide-Mediated Exon Skipping for DMD Treatment: Past Insights, Present Challenges and Future Perspectives. J. Biosci. 2023, 48, 38.
- Aartsma-Rus, A.; Krieg, A.M. FDA Approves Eteplirsen for Duchenne Muscular Dystrophy: The Next Chapter in the Eteplirsen Saga. Nucleic Acid Ther. 2017, 27, 1–3. [CrossRef]
- Research, C. for D.E. and IND Submissions for Individualized Antisense Oligonucleotide Drug Products: Administrative and Procedural Recommendations Guidance for Sponsor-Investigators Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/ind-submissions-individualized-antisense-oligonucleotide-drug-products-administrative-and-procedural (accessed on 28 December 2024).
- Hwang, G.; Kwon, M.; Seo, D.; Kim, D.H.; Lee, D.; Lee, K.; Kim, E.; Kang, M.; Ryu, J.-H. ASOptimizer: Optimizing Antisense Oligonucleotides through Deep Learning for IDO1 Gene Regulation. Mol. Ther. Nucleic Acids 2024, 35, 102186. [CrossRef]
- Yanagihara, N.; Tadakuma, H.; Ishihama, Y.; Okabe, K.; Funatsu, T. Determination of Potent Antisense Oligonucleotides In Vitro by Semiempirical Rules. J. Biosci. Bioeng. 2007, 103, 270–277. [CrossRef]
- Aupy, P.; Echevarría, L.; Relizani, K.; Zarrouki, F.; Haeberli, A.; Komisarski, M.; Tensorer, T.; Jouvion, G.; Svinartchouk, F.; Garcia, L.; et al. Identifying and Avoiding tcDNA-ASO Sequence-Specific Toxicity for the Development of DMD Exon 51 Skipping Therapy. Mol. Ther. Nucleic Acids 2020, 19, 371–383. [CrossRef]
- Jiang, Q.-S.; Wang, S.-Q. Design and Screening of Antisense Oligodeoxynucleotides against PAI-1 mRNA in Endothelial Cells in Vitro. Acta Pharmacol. Sin. 2006, 27, 1018–1023. [CrossRef]
- Kamola, P.J.; Kitson, J.D.A.; Turner, G.; Maratou, K.; Eriksson, S.; Panjwani, A.; Warnock, L.C.; Douillard Guilloux, G.A.; Moores, K.; Koppe, E.L.; et al. In Silico and in Vitro Evaluation of Exonic and Intronic Off-Target Effects Form a Critical Element of Therapeutic ASO Gapmer Optimization. Nucleic Acids Res. 2015, 43, 8638–8650. [CrossRef]
- Lee, J.J.A.; Maruyama, R.; Duddy, W.; Sakurai, H.; Yokota, T. Identification of Novel Antisense-Mediated Exon Skipping Targets in DYSF for Therapeutic Treatment of Dysferlinopathy. Mol. Ther. Nucleic Acids 2018, 13, 596–604. [CrossRef]
- Echigoya, Y.; Lim, K.R.Q.; Trieu, N.; Bao, B.; Miskew Nichols, B.; Vila, M.C.; Novak, J.S.; Hara, Y.; Lee, J.; Touznik, A.; et al. Quantitative Antisense Screening and Optimization for Exon 51 Skipping in Duchenne Muscular Dystrophy. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 2561–2572. [CrossRef]
- Chiba, S.; Lim, K.R.Q.; Sheri, N.; Anwar, S.; Erkut, E.; Shah, M.N.A.; Aslesh, T.; Woo, S.; Sheikh, O.; Maruyama, R.; et al. eSkip-Finder: A Machine Learning-Based Web Application and Database to Identify the Optimal Sequences of Antisense Oligonucleotides for Exon Skipping. Nucleic Acids Res. 2021, 49, W193–W198. [CrossRef]
- Lin, S.; Hong, L.; Wei, D.-Q.; Xiong, Y. Deep Learning Facilitates Efficient Optimization of Antisense Oligonucleotide Drugs. Mol. Ther. Nucleic Acids 2024, 35, 102208. [CrossRef]
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous Sarcoma Virus Replication and Cell Transformation by a Specific Oligodeoxynucleotide. Proc. Natl. Acad. Sci. U. S. A. 1978, 75, 280–284. [CrossRef]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [CrossRef]
- Dowdy, S.F. Overcoming Cellular Barriers for RNA Therapeutics. Nat. Biotechnol. 2017, 35, 222–229. [CrossRef]
- Bennett, C.F.; Baker, B.F.; Pham, N.; Swayze, E.; Geary, R.S. Pharmacology of Antisense Drugs. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 81–105. [CrossRef]
- Quemener, A.M.; Bachelot, L.; Forestier, A.; Donnou-Fournet, E.; Gilot, D.; Galibert, M.-D. The Powerful World of Antisense Oligonucleotides: From Bench to Bedside. Wiley Interdiscip. Rev. RNA 2020, 11, e1594. [CrossRef]
- Eckstein, F. Phosphorothioates, Essential Components of Therapeutic Oligonucleotides. Nucleic Acid Ther. 2014, 24, 374–387. [CrossRef]
- Geary, R.S.; Henry, S.P.; Grillone, L.R. Fomivirsen: Clinical Pharmacology and Potential Drug Interactions. Clin. Pharmacokinet. 2002, 41, 255–260. [CrossRef]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in Oligonucleotide Drug Delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [CrossRef]
- Qassem, S.; Breier, D.; Naidu, G.S.; Hazan-Halevy, I.; Peer, D. Unlocking the Therapeutic Potential of Locked Nucleic Acids through Lipid Nanoparticle Delivery. Mol. Ther. Nucleic Acids 2024, 35, 102224. [CrossRef]
- Juliano, R.L. The Delivery of Therapeutic Oligonucleotides. Nucleic Acids Res. 2016, 44, 6518–6548. [CrossRef]
- Manoharan, M. 2’-Carbohydrate Modifications in Antisense Oligonucleotide Therapy: Importance of Conformation, Configuration and Conjugation. Biochim. Biophys. Acta 1999, 1489, 117–130. [CrossRef]
- Hebb, M.O.; Robertson, H.A. End-Capped Antisense Oligodeoxynucleotides Effectively Inhibit Gene Expression in Vivo and Offer a Low-Toxicity Alternative to Fully Modified Phosphorothioate Oligodeoxynucleotides. Brain Res. Mol. Brain Res. 1997, 47, 223–228. [CrossRef]
- Summerton, J.; Weller, D. Morpholino Antisense Oligomers: Design, Preparation, and Properties. Antisense Nucleic Acid Drug Dev. 1997, 7, 187–195. [CrossRef]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-Selective Recognition of DNA by Strand Displacement with a Thymine-Substituted Polyamide. Science 1991, 254, 1497–1500. [CrossRef]
- Demidov, V.V.; Potaman, V.N.; Frank-Kamenetskii, M.D.; Egholm, M.; Buchard, O.; Sönnichsen, S.H.; Nielsen, P.E. Stability of Peptide Nucleic Acids in Human Serum and Cellular Extracts. Biochem. Pharmacol. 1994, 48, 1310–1313. [CrossRef]
- Hudziak, R.M.; Barofsky, E.; Barofsky, D.F.; Weller, D.L.; Huang, S.B.; Weller, D.D. Resistance of Morpholino Phosphorodiamidate Oligomers to Enzymatic Degradation. Antisense Nucleic Acid Drug Dev. 1996, 6, 267–272. [CrossRef]
- Lee, J.J.A.; Yokota, T. Antisense Therapy in Neurology. J. Pers. Med. 2013, 3, 144–176. [CrossRef]
- Thakur, S.; Sinhari, A.; Jain, P.; Jadhav, H.R. A Perspective on Oligonucleotide Therapy: Approaches to Patient Customization. Front. Pharmacol. 2022, 13. [CrossRef]
- Egli, M.; Manoharan, M. Chemistry, Structure and Function of Approved Oligonucleotide Therapeutics. Nucleic Acids Res. 2023, 51, 2529–2573. [CrossRef]
- Sumner, C.J.; Miller, T.M. The Expanding Application of Antisense Oligonucleotides to Neurodegenerative Diseases. J. Clin. Invest. 2024, 134, e186116. [CrossRef]
- Hyjek, M.; Figiel, M.; Nowotny, M. RNases H: Structure and Mechanism. DNA Repair 2019, 84, 102672. [CrossRef]
- Wu, H.; Lima, W.F.; Zhang, H.; Fan, A.; Sun, H.; Crooke, S.T. Determination of the Role of the Human RNase H1 in the Pharmacology of DNA-like Antisense Drugs. J. Biol. Chem. 2004, 279, 17181–17189. [CrossRef]
- ten Asbroek, A.L.M.A.; van Groenigen, M.; Nooij, M.; Baas, F. The Involvement of Human Ribonucleases H1 and H2 in the Variation of Response of Cells to Antisense Phosphorothioate Oligonucleotides. Eur. J. Biochem. 2002, 269, 583–592. [CrossRef]
- Khvorova, A.; Watts, J.K. The Chemical Evolution of Oligonucleotide Therapies of Clinical Utility. Nat. Biotechnol. 2017, 35, 238–248. [CrossRef]
- Kuespert, S.; Heydn, R.; Peters, S.; Wirkert, E.; Meyer, A.-L.; Siebörger, M.; Johannesen, S.; Aigner, L.; Bogdahn, U.; Bruun, T.-H. Antisense Oligonucleotide in LNA-Gapmer Design Targeting TGFBR2—A Key Single Gene Target for Safe and Effective Inhibition of TGFβ Signaling. Int. J. Mol. Sci. 2020, 21, 1952. [CrossRef]
- Kiełpiński, Ł.J.; Hagedorn, P.H.; Lindow, M.; Vinther, J. RNase H Sequence Preferences Influence Antisense Oligonucleotide Efficiency. Nucleic Acids Res. 2017, 45, 12932–12944. [CrossRef]
- Bauman, J.; Jearawiriyapaisarn, N.; Kole, R. Therapeutic Potential of Splice-Switching Oligonucleotides. Oligonucleotides 2009, 19, 1–13. [CrossRef]
- Havens, M.A.; Hastings, M.L. Splice-Switching Antisense Oligonucleotides as Therapeutic Drugs. Nucleic Acids Res. 2016, 44, 6549–6563. [CrossRef]
- Tomkiewicz, T.Z.; Suárez-Herrera, N.; Cremers, F.P.M.; Collin, R.W.J.; Garanto, A. Antisense Oligonucleotide-Based Rescue of Aberrant Splicing Defects Caused by 15 Pathogenic Variants in ABCA4. Int. J. Mol. Sci. 2021, 22, 4621. [CrossRef]
- Evers, M.M.; Tran, H.-D.; Zalachoras, I.; Pepers, B.A.; Meijer, O.C.; den Dunnen, J.T.; van Ommen, G.-J.B.; Aartsma-Rus, A.; van Roon-Mom, W.M.C. Ataxin-3 Protein Modification as a Treatment Strategy for Spinocerebellar Ataxia Type 3: Removal of the CAG Containing Exon. Neurobiol. Dis. 2013, 58, 49–56. [CrossRef]
- Han, Z.; Chen, C.; Christiansen, A.; Ji, S.; Lin, Q.; Anumonwo, C.; Liu, C.; Leiser, S.C.; Meena, null; Aznarez, I.; et al. Antisense Oligonucleotides Increase Scn1a Expression and Reduce Seizures and SUDEP Incidence in a Mouse Model of Dravet Syndrome. Sci. Transl. Med. 2020, 12, eaaz6100. [CrossRef]
- Singh, N.N.; Howell, M.D.; Androphy, E.J.; Singh, R.N. How the Discovery of ISS-N1 Led to the First Medical Therapy for Spinal Muscular Atrophy. Gene Ther. 2017, 24, 520–526. [CrossRef]
- Rigo, F.; Seth, P.P.; Bennett, C.F. Antisense Oligonucleotide-Based Therapies for Diseases Caused by Pre-mRNA Processing Defects. Adv. Exp. Med. Biol. 2014, 825, 303–352. [CrossRef]
- Summerton, J. Morpholino Antisense Oligomers: The Case for an RNase H-Independent Structural Type. Biochim. Biophys. Acta 1999, 1489, 141–158. [CrossRef]
- Bennett, C.F.; Swayze, E.E. RNA Targeting Therapeutics: Molecular Mechanisms of Antisense Oligonucleotides as a Therapeutic Platform. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 259–293. [CrossRef]
- Dias, N.; Dheur, S.; Nielsen, P.E.; Gryaznov, S.; Van Aerschot, A.; Herdewijn, P.; Hélène, C.; Saison-Behmoaras, T.E. Antisense PNA Tridecamers Targeted to the Coding Region of Ha-Ras mRNA Arrest Polypeptide Chain Elongation. J. Mol. Biol. 1999, 294, 403–416. [CrossRef]
- Anderson, K.P.; Fox, M.C.; Brown-Driver, V.; Martin, M.J.; Azad, R.F. Inhibition of Human Cytomegalovirus Immediate-Early Gene Expression by an Antisense Oligonucleotide Complementary to Immediate-Early RNA. Antimicrob. Agents Chemother. 1996, 40, 2004–2011. [CrossRef]
- Arend, K.C.; Ziehr, B.; Vincent, H.A.; Moorman, N.J. Multiple Transcripts Encode Full-Length Human Cytomegalovirus IE1 and IE2 Proteins during Lytic Infection. J. Virol. 2016, 90, 8855–8865. [CrossRef]
- Vitravene Study Group A Randomized Controlled Clinical Trial of Intravitreous Fomivirsen for Treatment of Newly Diagnosed Peripheral Cytomegalovirus Retinitis in Patients with AIDS. Am. J. Ophthalmol. 2002, 133, 467–474. [CrossRef]
- Vitravene Study Group. Safety of Intravitreous Fomivirsen for Treatment of Cytomegalovirus Retinitis in Patients with AIDS. Am. J. Ophthalmol. 2002, 133, 484–498. [CrossRef]
- Piper, H.; Ciulla, T.A.; Danis, R.P.; Pratt, L.M. Changing Therapeutic Paradigms in CMV Retinitis in AIDS. Expert Opin. Pharmacother. 2000, 1, 1343–1352. [CrossRef]
- Alhamadani, F.; Zhang, K.; Parikh, R.; Wu, H.; Rasmussen, T.P.; Bahal, R.; Zhong, X.-B.; Manautou, J.E. Adverse Drug Reactions and Toxicity of the Food and Drug Administration-Approved Antisense Oligonucleotide Drugs. Drug Metab. Dispos. Biol. Fate Chem. 2022, 50, 879–887. [CrossRef]
- Crooke, S.T.; Geary, R.S. Clinical Pharmacological Properties of Mipomersen (Kynamro), a Second Generation Antisense Inhibitor of Apolipoprotein B. Br. J. Clin. Pharmacol. 2013, 76, 269–276. [CrossRef]
- Santos, R.D.; Duell, P.B.; East, C.; Guyton, J.R.; Moriarty, P.M.; Chin, W.; Mittleman, R.S. Long-Term Efficacy and Safety of Mipomersen in Patients with Familial Hypercholesterolaemia: 2-Year Interim Results of an Open-Label Extension. Eur. Heart J. 2015, 36, 566–575. [CrossRef]
- Gales, L. Tegsedi (Inotersen): An Antisense Oligonucleotide Approved for the Treatment of Adult Patients with Hereditary Transthyretin Amyloidosis. Pharm. Basel Switz. 2019, 12, 78. [CrossRef]
- Sekijima, Y.; Nakamura, K. Hereditary Transthyretin Amyloidosis. In GeneReviews®; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington, Seattle: Seattle (WA), 1993.
- Blair, H.A. Tofersen: First Approval. Drugs 2023, 83, 1039–1043. [CrossRef]
- Duan, D.; Goemans, N.; Takeda, S.; Mercuri, E.; Aartsma-Rus, A. Duchenne Muscular Dystrophy. Nat. Rev. Dis. Primer 2021, 7, 13. [CrossRef]
- Vengalil, S.; Preethish-Kumar, V.; Polavarapu, K.; Mahadevappa, M.; Sekar, D.; Purushottam, M.; Thomas, P.T.; Nashi, S.; Nalini, A. Duchenne Muscular Dystrophy and Becker Muscular Dystrophy Confirmed by Multiplex Ligation-Dependent Probe Amplification: Genotype-Phenotype Correlation in a Large Cohort. J. Clin. Neurol. Seoul Korea 2017, 13, 91–97. [CrossRef]
- Takeshima, Y.; Yagi, M.; Okizuka, Y.; Awano, H.; Zhang, Z.; Yamauchi, Y.; Nishio, H.; Matsuo, M. Mutation Spectrum of the Dystrophin Gene in 442 Duchenne/Becker Muscular Dystrophy Cases from One Japanese Referral Center. J. Hum. Genet. 2010, 55, 379–388. [CrossRef]
- Aartsma-Rus, A.; Janson, A.A.M.; Kaman, W.E.; Bremmer-Bout, M.; den Dunnen, J.T.; Baas, F.; van Ommen, G.-J.B.; van Deutekom, J.C.T. Therapeutic Antisense-Induced Exon Skipping in Cultured Muscle Cells from Six Different DMD Patients. Hum. Mol. Genet. 2003, 12, 907–914. [CrossRef]
- Lim, K.R.Q.; Nguyen, Q.; Yokota, T. Genotype-Phenotype Correlations in Duchenne and Becker Muscular Dystrophy Patients from the Canadian Neuromuscular Disease Registry. J. Pers. Med. 2020, 10, 241. [CrossRef]
- Bladen, C.L.; Salgado, D.; Monges, S.; Foncuberta, M.E.; Kekou, K.; Kosma, K.; Dawkins, H.; Lamont, L.; Roy, A.J.; Chamova, T.; et al. The TREAT-NMD DMD Global Database: Analysis of More than 7,000 Duchenne Muscular Dystrophy Mutations. Hum. Mutat. 2015, 36, 395–402. [CrossRef]
- Matsuo, M. Antisense Oligonucleotide-Mediated Exon-Skipping Therapies: Precision Medicine Spreading from Duchenne Muscular Dystrophy. JMA J. 2021, 4, 232–240. [CrossRef]
- Sabrina Haque, U.; Kohut, M.; Yokota, T. Comprehensive Review of Adverse Reactions and Toxicology in ASO-Based Therapies for Duchenne Muscular Dystrophy: From FDA-Approved Drugs to Peptide-Conjugated ASO. Curr. Res. Toxicol. 2024, 7, 100182. [CrossRef]
- Mercuri, E.; Sumner, C.J.; Muntoni, F.; Darras, B.T.; Finkel, R.S. Spinal Muscular Atrophy. Nat. Rev. Dis. Primer 2022, 8, 52. [CrossRef]
- Ruhno, C.; McGovern, V.L.; Avenarius, M.R.; Snyder, P.J.; Prior, T.W.; Nery, F.C.; Muhtaseb, A.; Roggenbuck, J.S.; Kissel, J.T.; Sansone, V.A.; et al. Complete Sequencing of the SMN2 Gene in SMA Patients Detects SMN Gene Deletion Junctions and Variants in SMN2 That Modify the SMA Phenotype. Hum. Genet. 2019, 138, 241–256. [CrossRef]
- Chi, X.; Gatti, P.; Papoian, T. Safety of Antisense Oligonucleotide and siRNA-Based Therapeutics. Drug Discov. Today 2017, 22, 823–833. [CrossRef]
- Kim, J.; Hu, C.; Achkar, C.M.E.; Black, L.E.; Douville, J.; Larson, A.; Pendergast, M.K.; Goldkind, S.F.; Lee, E.A.; Kuniholm, A.; et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N. Engl. J. Med. 2019, 381, 1644–1652. [CrossRef]
- McBride, J.L.; Neuringer, M.; Ferguson, B.; Kohama, S.G.; Tagge, I.J.; Zweig, R.C.; Renner, L.M.; McGill, T.J.; Stoddard, J.; Peterson, S.; et al. Discovery of a CLN7 Model of Batten Disease in Non-Human Primates. Neurobiol. Dis. 2018, 119, 65–78. [CrossRef]
- Kousi, M.; Lehesjoki, A.-E.; Mole, S.E. Update of the Mutation Spectrum and Clinical Correlations of over 360 Mutations in Eight Genes That Underlie the Neuronal Ceroid Lipofuscinoses. Hum. Mutat. 2012, 33, 42–63. [CrossRef]
- Ray, D.A.; Batzer, M.A. Reading TE Leaves: New Approaches to the Identification of Transposable Element Insertions. Genome Res. 2011, 21, 813–820. [CrossRef]
- Wilton-Clark, H.; Yan, E.; Yokota, T. Preparing for Patient-Customized N-of-1 Antisense Oligonucleotide Therapy to Treat Rare Diseases. Genes 2024, 15, 821. [CrossRef]
- Kim, J.; Woo, S.; de Gusmao, C.M.; Zhao, B.; Chin, D.H.; DiDonato, R.L.; Nguyen, M.A.; Nakayama, T.; Hu, C.A.; Soucy, A.; et al. A Framework for Individualized Splice-Switching Oligonucleotide Therapy. Nature 2023, 619, 828–836. [CrossRef]
- Shiloh, Y. ATM and Related Protein Kinases: Safeguarding Genome Integrity. Nat. Rev. Cancer 2003, 3, 155–168. [CrossRef]
- Verhagen, M.M.M.; Abdo, W.F.; Willemsen, M. a. a. P.; Hogervorst, F.B.L.; Smeets, D.F.C.M.; Hiel, J. a. P.; Brunt, E.R.; van Rijn, M.A.; Majoor Krakauer, D.; Oldenburg, R.A.; et al. Clinical Spectrum of Ataxia-Telangiectasia in Adulthood. Neurology 2009, 73, 430–437. [CrossRef]
- Mendell, J.R.; Goemans, N.; Lowes, L.P.; Alfano, L.N.; Berry, K.; Shao, J.; Kaye, E.M.; Mercuri, E.; Eteplirsen Study Group and Telethon Foundation DMD Italian Network Longitudinal Effect of Eteplirsen versus Historical Control on Ambulation in Duchenne Muscular Dystrophy. Ann. Neurol. 2016, 79, 257–271. [CrossRef]
- Sheikh, O.; Yokota, T. Pharmacology and Toxicology of Eteplirsen and SRP-5051 for DMD Exon 51 Skipping: An Update. Arch. Toxicol. 2022, 96, 1–9. [CrossRef]
- Aartsma-Rus, A.; Krieg, A.M. FDA Approves Eteplirsen for Duchenne Muscular Dystrophy: The Next Chapter in the Eteplirsen Saga. Nucleic Acid Ther. 2017, 27, 1–3. [CrossRef]
- Chan, J.H.P.; Lim, S.; Wong, W.S.F. Antisense Oligonucleotides: From Design to Therapeutic Application. Clin. Exp. Pharmacol. Physiol. 2006, 33, 533–540. [CrossRef]
- Gagliardi, M.; Ashizawa, A.T. The Challenges and Strategies of Antisense Oligonucleotide Drug Delivery. Biomedicines 2021, 9, 433. [CrossRef]
- Vázquez-Domínguez, I.; Anido, A.A.; Duijkers, L.; Hoppenbrouwers, T.; Hoogendoorn, A.D.M.; Koster, C.; Collin, R.W.J.; Garanto, A. Efficacy, Biodistribution and Safety Comparison of Chemically Modified Antisense Oligonucleotides in the Retina. Nucleic Acids Res. 2024, 52, 10447–10463. [CrossRef]
- Seth, P.P.; Tanowitz, M.; Bennett, C.F. Selective Tissue Targeting of Synthetic Nucleic Acid Drugs. J. Clin. Invest. 129, 915–925. [CrossRef]
- Echigoya, Y.; Mouly, V.; Garcia, L.; Yokota, T.; Duddy, W. In Silico Screening Based on Predictive Algorithms as a Design Tool for Exon Skipping Oligonucleotides in Duchenne Muscular Dystrophy. PloS One 2015, 10, e0120058. [CrossRef]
- Echigoya, Y.; Lim, K.R.Q.; Trieu, N.; Bao, B.; Miskew Nichols, B.; Vila, M.C.; Novak, J.S.; Hara, Y.; Lee, J.; Touznik, A.; et al. Quantitative Antisense Screening and Optimization for Exon 51 Skipping in Duchenne Muscular Dystrophy. Mol. Ther. 2017, 25, 2561–2572. [CrossRef]
- Ding, Y.; Chan, C.Y.; Lawrence, C.E. Sfold Web Server for Statistical Folding and Rational Design of Nucleic Acids. Nucleic Acids Res. 2004, 32, W135-141. [CrossRef]
- Shao, Y.; Wu, Y.; Chan, C.Y.; McDonough, K.; Ding, Y. Rational Design and Rapid Screening of Antisense Oligonucleotides for Prokaryotic Gene Modulation. Nucleic Acids Res. 2006, 34, 5660–5669. [CrossRef]
- Sciabola, S.; Xi, H.; Cruz, D.; Cao, Q.; Lawrence, C.; Zhang, T.; Rotstein, S.; Hughes, J.D.; Caffrey, D.R.; Stanton, R.V. PFRED: A Computational Platform for siRNA and Antisense Oligonucleotides Design. PLoS ONE 2021, 16, e0238753. [CrossRef]
- Altmann, A.; Toloşi, L.; Sander, O.; Lengauer, T. Permutation Importance: A Corrected Feature Importance Measure. Bioinforma. Oxf. Engl. 2010, 26, 1340–1347. [CrossRef]
- Stadler, M.B.; Shomron, N.; Yeo, G.W.; Schneider, A.; Xiao, X.; Burge, C.B. Inference of Splicing Regulatory Activities by Sequence Neighborhood Analysis. PLoS Genet. 2006, 2, e191. [CrossRef]
- Ham, K.A.; Aung-Htut, M.T.; Fletcher, S.; Wilton, S.D. Nonsequential Splicing Events Alter Antisense-Mediated Exon Skipping Outcome in COL7A1. Int. J. Mol. Sci. 2020, 21, 7705. [CrossRef]
- Reiser, P.; Neubert, M.; Eberhard, A.; Torresi, L.; Zhou, C.; Shao, C.; Metni, H.; van Hoesel, C.; Schopmans, H.; Sommer, T.; et al. Graph Neural Networks for Materials Science and Chemistry. Commun. Mater. 2022, 3, 1–18. [CrossRef]
- Zhai, L.; Ladomersky, E.; Lenzen, A.; Nguyen, B.; Patel, R.; Lauing, K.L.; Wu, M.; Wainwright, D.A. IDO1 in Cancer: A Gemini of Immune Checkpoints. Cell. Mol. Immunol. 2018, 15, 447–457. [CrossRef]
- Akoglu, H. User’s Guide to Correlation Coefficients. Turk. J. Emerg. Med. 2018, 18, 91–93. [CrossRef]
- McNally, E.M.; Wyatt, E.J. Welcome to the Splice Age: Antisense Oligonucleotide–Mediated Exon Skipping Gains Wider Applicability. J. Clin. Invest. 126, 1236–1238. [CrossRef]
- McClorey, G.; Banerjee, S. Cell-Penetrating Peptides to Enhance Delivery of Oligonucleotide-Based Therapeutics. Biomedicines 2018, 6, 51. [CrossRef]
- Yang, L.; Ma, F.; Liu, F.; Chen, J.; Zhao, X.; Xu, Q. Efficient Delivery of Antisense Oligonucleotides Using Bioreducible Lipid Nanoparticles In Vitro and In Vivo. Mol. Ther. Nucleic Acids 2020, 19, 1357–1367. [CrossRef]
- Huang, S.; Hao, X.-Y.; Li, Y.-J.; Wu, J.; Xiang, D.-X.; Luo, S. Nonviral Delivery Systems for Antisense Oligonucleotide Therapeutics. Biomater. Res. 2022, 26, 49. [CrossRef]
- Zhu, A.; Chiba, S.; Shimizu, Y.; Kunitake, K.; Okuno, Y.; Aoki, Y.; Yokota, T. Ensemble-Learning and Feature Selection Techniques for Enhanced Antisense Oligonucleotide Efficacy Prediction in Exon Skipping. Pharmaceutics 2023, 15, 1808. [CrossRef]
| Therapy Name | Disease | Target RNA | Mechanism of Action | Chemical Modifications |
|---|---|---|---|---|
| Fomiversen | CMV retinitis | MIE | RNA degradation | PS |
| Mipomersen | HoFH | APOB | RNA degradation | PS and 2’-MOE |
| Eteplirsen | DMD | DMD (exon 51) | Splice switching | PMO |
| Nusinersen | SMA | SMN2 (exon 7) | Splice switching | PS and 2’-MOE |
| Inotersen | hATTR | TTR | RNA degradation | PS and 2’-MOE |
| Golodirsen | DMD | DMD (exon 53) | Splice switching | PMO |
| Viltolersen | DMD | DMD (exon 53) | Splice switching | PMO |
| Casimersen | DMD | DMD (exon 45) | Splice switching | PMO |
| Tofersen | ALS | SOD1 | RNA degradation | PS and 2’-MOE |
| Therapy Name | Disease | Target | Mechanism of Action | Chemical Modifications |
|---|---|---|---|---|
| Milasen | Batten Disease | CLN7 (intron 6) | Splice Switching | PS and 2’-MOE |
| Atipeksen | HoFH | ATM (exon 53) | Splice Switching | PS and 2’-MOE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





