Submitted:
08 March 2023
Posted:
09 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crooke, S.T.; Liang, X.-H.; Baker, B.F.; Crooke, R.M. Antisense technology: A review. Journal of Biological Chemistry 2021, 296. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, M.L.; Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proceedings of the National Academy of Sciences 1978, 75, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.H.; Lim, S.; Wong, W.F. Antisense oligonucleotides: from design to therapeutic application. Clinical and experimental pharmacology and physiology 2006, 33, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Quemener, A.M.; Bachelot, L.; Forestier, A.; Donnou-Fournet, E.; Gilot, D.; Galibert, M.D. The powerful world of antisense oligonucleotides: From bench to bedside. Wiley Interdisciplinary Reviews: RNA 2020, 11, e1594. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, C.; Wood, M.J. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nature Reviews Neurology 2018, 14, 9–21. [Google Scholar] [CrossRef]
- Inoue, H.; Hayase, Y.; Iwai, S.; Ohtsuka, E. Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett 1987, 215, 327–330. [Google Scholar] [CrossRef]
- Lundin, K.E.; Gissberg, O.; Smith, C.E. Oligonucleotide therapies: the past and the present. Human gene therapy 2015, 26, 475–485. [Google Scholar] [CrossRef]
- Walder, J.A.; Walder, R.Y. Nucleic acid hybridization and amplification method for detection of specific sequences in which a complementary labeled nucleic acid probe is cleaved. Google Patents: 1995.
- Lim, S.R.; Hertel, K.J. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. Journal of Biological Chemistry 2001, 276, 45476–45483. [Google Scholar] [CrossRef]
- Shirley, M. Casimersen: First Approval. Drugs 2021, 81, 875–879. [Google Scholar] [CrossRef]
- Nelson, S.F.; Miceli, M.C. FDA Approval of Eteplirsen for Muscular Dystrophy. JAMA 2017, 317, 1480. [Google Scholar] [CrossRef]
- Roshmi, R.R.; Yokota, T. Viltolarsen: From Preclinical Studies to FDA Approval. Methods Mol Biol 2023, 2587, 31–41. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Corey, D.R. The 10th Oligonucleotide Therapy Approved: Golodirsen for Duchenne Muscular Dystrophy. Nucleic Acid Ther 2020, 30, 67–70. [Google Scholar] [CrossRef]
- Brudvig, J.J.; Weimer, J.M. On the cusp of cures: breakthroughs in Batten disease research. Current Opinion in Neurobiology 2022, 72, 48–54. [Google Scholar] [CrossRef]
- Kim, J.; Hu, C.; Moufawad El Achkar, C.; Black, L.E.; Douville, J.; Larson, A.; Pendergast, M.K.; Goldkind, S.F.; Lee, E.A.; Kuniholm, A.; et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N Engl J Med 2019, 381, 1644–1652. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; van Roon-Mom, W.; Lauffer, M.; Siezen, C.; Duijndam, B.; Coenen-de Roo, T.; Schule, R.; Synofzik, M.; Graessner, H. Development of tailored splice switching oligonucleotides for progressive brain disorders in Europe: development, regulation and implementation considerations. RNA, 2023; 10.1261/rna.079540.122. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Garanto, A.; van Roon-Mom, W.; McConnell, E.M.; Suslovitch, V.; Yan, W.X.; Watts, J.K.; Yu, T.W. Consensus Guidelines for the Design and In Vitro Preclinical Efficacy Testing N-of-1 Exon Skipping Antisense Oligonucleotides. Nucleic Acid Ther 2023, 33, 17–25. [Google Scholar] [CrossRef]
- Lemaitre, M.M. Individualized Antisense Oligonucleotide Therapies: How to Approach the Challenge of Manufacturing These Oligos from a Chemistry, Manufacturing, and Control-Regulatory Standpoint. Nucleic Acid Ther 2022, 32, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Bateman-House, A.; Kearns, L. Individualized Therapeutics Development for Rare Diseases: The Current Ethical Landscape and Policy Responses. Nucleic Acid Ther 2022, 32, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.F.; Meisler, M.H. Antisense Oligonucleotide Therapy for Neurodevelopmental Disorders. Dev Neurosci 2021, 43, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Amariles, P.; Madrigal-Cadavid, J. Ethical, Economic, Societal, Clinical, and Pharmacology Uncertainties Associated With Milasen and Other Personalized Drugs. Ann Pharmacother 2020, 54, 937–938. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Watts, J.K. The Munich Meeting: Medical Maturation, More Mechanisms, and Milasen. Nucleic Acid Ther 2019, 29, 302–304. [Google Scholar] [CrossRef]
- Kim, Y.J.; Sivetz, N.; Layne, J.; Voss, D.M.; Yang, L.; Zhang, Q.; Krainer, A.R. Exon-skipping antisense oligonucleotides for cystic fibrosis therapy. Proceedings of the National Academy of Sciences 2022, 119, e2114858118. [Google Scholar] [CrossRef] [PubMed]
- Covello, G.; Ibrahim, G.H.; Bacchi, N.; Casarosa, S.; Denti, M.A. Exon skipping through chimeric antisense U1 snRNAs to correct retinitis pigmentosa GTPase-regulator (RPGR) splice defect. nucleic acid therapeutics 2022, 32, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Cai, J.; de Gorter, D.J.; Sanchez-Duffhues, G.; Kemaladewi, D.U.; Hoogaars, W.M.; Aartsma-Rus, A.; ’t Hoen, P.A.; ten Dijke, P. Antisense-oligonucleotide mediated exon skipping in activin-receptor-like kinase 2: inhibiting the receptor that is overactive in fibrodysplasia ossificans progressiva. PloS one 2013, 8, e69096. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Yokota, T. An overview of recent advances and clinical applications of exon skipping and splice modulation for muscular dystrophy and various genetic diseases. Exon Skipping and Inclusion Therapies: Methods and Protocols, 2018; 31–55. [Google Scholar]
- Isom, L.L.; Knupp, K.G. Dravet syndrome: novel approaches for the most common genetic epilepsy. Neurotherapeutics 2021, 18, 1524–1534. [Google Scholar] [CrossRef] [PubMed]
- Barthelemy, F.; Blouin, C.; Wein, N.; Mouly, V.; Courrier, S.; Dionnet, E.; Kergourlay, V.; Mathieu, Y.; Garcia, L.; Butler-Browne, G.; et al. Exon 32 Skipping of Dysferlin Rescues Membrane Repair in Patients’ Cells. J Neuromuscul Dis 2015, 2, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.A.; Maruyama, R.; Duddy, W.; Sakurai, H.; Yokota, T. Identification of Novel Antisense-Mediated Exon Skipping Targets in DYSF for Therapeutic Treatment of Dysferlinopathy. Mol Ther Nucleic Acids 2018, 13, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, E.J.; Demonbreun, A.R.; Kim, E.Y.; Puckelwartz, M.J.; Vo, A.H.; Dellefave-Castillo, L.M.; Gao, Q.Q.; Vainzof, M.; Pavanello, R.C.M.; Zatz, M.; et al. Efficient exon skipping of SGCG mutations mediated by phosphorodiamidate morpholino oligomers. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.A.; Ashton, G.H.; Mellerio, J.E.; Salas-Alanis, J.C.; Swensson, O.; McMillan, J.R.; Eady, R.A. Moderation of phenotypic severity in dystrophic and junctional forms of epidermolysis bullosa through in-frame skipping of exons containing non-sense or frameshift mutations. J Invest Dermatol 1999, 113, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Kalbfuss, B.; Mabon, S.A.; Misteli, T. Correction of alternative splicing of tau in frontotemporal dementia and parkinsonism linked to chromosome 17. J Biol Chem 2001, 276, 42986–42993. [Google Scholar] [CrossRef]
- Renshaw, J.; Orr, R.M.; Walton, M.I.; Te Poele, R.; Williams, R.D.; Wancewicz, E.V.; Monia, B.P.; Workman, P.; Pritchard-Jones, K. Disruption of WT1 gene expression and exon 5 splicing following cytotoxic drug treatment: antisense down-regulation of exon 5 alters target gene expression and inhibits cell survival. Mol Cancer Ther 2004, 3, 1467–1484. [Google Scholar] [CrossRef]
- Yu, A.M.; Tu, M.J. Deliver the promise: RNAs as a new class of molecular entities for therapy and vaccination. Pharmacol Ther 2022, 230, 107967. [Google Scholar] [CrossRef]
- Maruyama, R.; Yokota, T. Tips to Design Effective Splice-Switching Antisense Oligonucleotides for Exon Skipping and Exon Inclusion. Methods Mol Biol 2018, 1828, 79–90. [Google Scholar] [CrossRef]
- Sciabola, S.; Xi, H.; Cruz, D.; Cao, Q.; Lawrence, C.; Zhang, T.; Rotstein, S.; Hughes, J.D.; Caffrey, D.R.; Stanton, R.V. PFRED: A computational platform for siRNA and antisense oligonucleotides design. PloS one 2021, 16, e0238753. [Google Scholar] [CrossRef] [PubMed]
- Shimo, T.; Maruyama, R.; Yokota, T. Designing effective antisense oligonucleotides for exon skipping. Duchenne Muscular Dystrophy: Methods and Protocols, 2018; 143–155. [Google Scholar]
- Chiba, S.; Lim, K.R.Q.; Sheri, N.; Anwar, S.; Erkut, E.; Shah, M.N.A.; Aslesh, T.; Woo, S.; Sheikh, O.; Maruyama, R.; et al. eSkip-Finder: a machine learning-based web application and database to identify the optimal sequences of antisense oligonucleotides for exon skipping. Nucleic Acids Res 2021, 49, W193–W198. [Google Scholar] [CrossRef]
- Bishop, C.M.; Nasrabadi, N.M. Pattern recognition and machine learning; Springer: 2006; Vol. 4.
- Chandra, A.; Yao, X. Ensemble learning using multi-objective evolutionary algorithms. Journal of Mathematical Modelling and Algorithms 2006, 5, 417–445. [Google Scholar] [CrossRef]
- Sahin, E.K. Assessing the predictive capability of ensemble tree methods for landslide susceptibility mapping using XGBoost, gradient boosting machine, and random forest. SN Applied Sciences 2020, 2, 1308. [Google Scholar] [CrossRef]
- Malueka, R.G.; Takaoka, Y.; Yagi, M.; Awano, H.; Lee, T.; Dwianingsih, E.K.; Nishida, A.; Takeshima, Y.; Matsuo, M. Categorization of 77 dystrophin exons into 5 groups by a decision tree using indexes of splicing regulatory factors as decision markers. BMC Genet 2012, 13, 23. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: machine learning in Python. J Mach Learn Res.
- Ramraj, S.; Uzir, N.; Sunil, R.; Banerjee, S. Experimenting XGBoost algorithm for prediction and classification of different datasets. International Journal of Control Theory and Applications 2016, 9, 651–662. [Google Scholar]
- Iuchi, H.; Matsutani, T.; Yamada, K.; Iwano, N.; Sumi, S.; Hosoda, S.; Zhao, S.; Fukunaga, T.; Hamada, M. Representation learning applications in biological sequence analysis. Computational and Structural Biotechnology Journal 2021, 19, 3198–3208. [Google Scholar] [CrossRef] [PubMed]
- Echigoya, Y.; Mouly, V.; Garcia, L.; Yokota, T.; Duddy, W. In silico screening based on predictive algorithms as a design tool for exon skipping oligonucleotides in Duchenne muscular dystrophy. PLoS One 2015, 10, e0120058. [Google Scholar] [CrossRef]
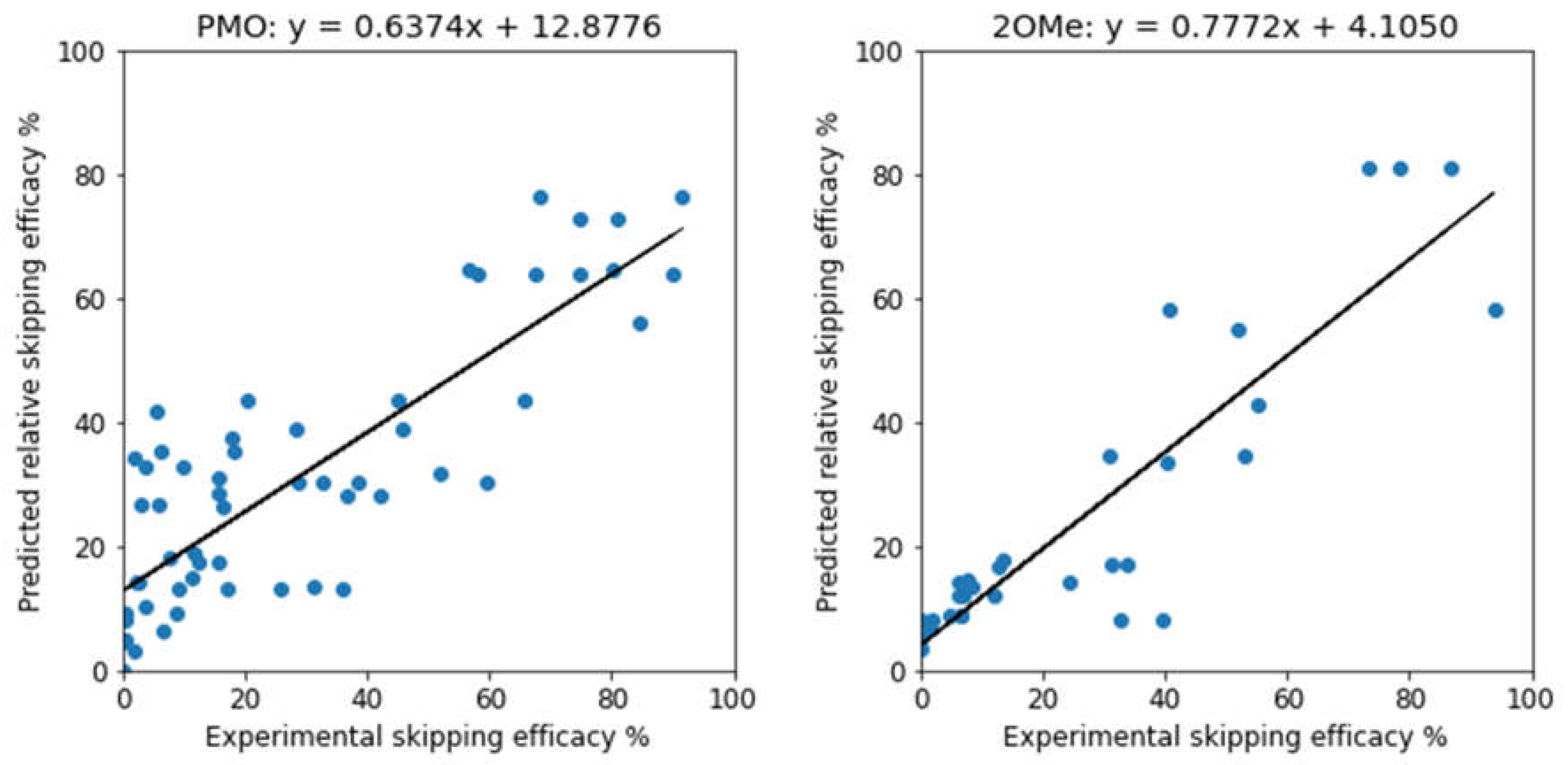
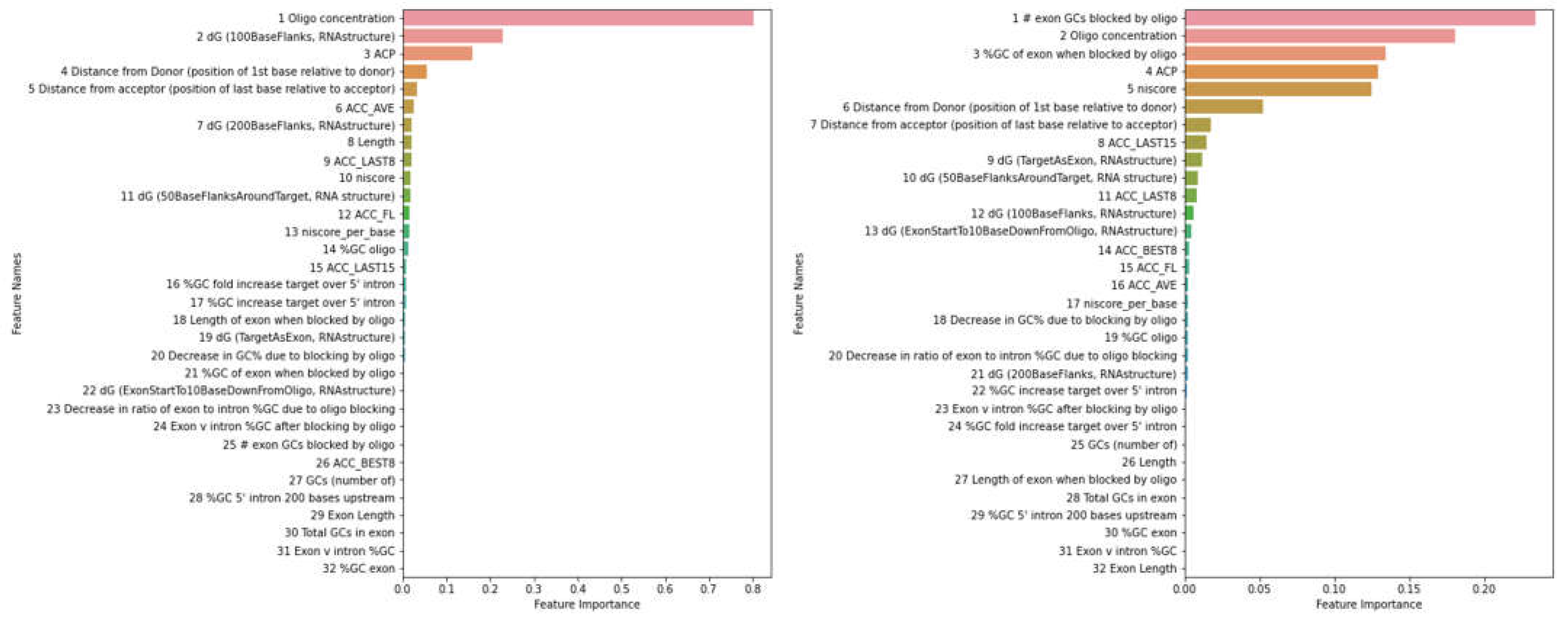
| Methods | PMO | 2OMe | ||
| R2 | MAE | R2 | MAE | |
| Support Vector | 0.138 ± 0.076 | 22.06 ± 4.02 | 0.558 ± 0.093 | 17.70 ± 5.32 |
| Random Forest | 0.555 ± 0.247 | 15.39 ± 4.84 | 0.729 ± 0.169 | 10.59 ± 3.31 |
| Gradient Boosting | 0.564 ± 0.234 | 14.97 ± 4.58 | 0.721 ± 0.152 | 10.13 ± 2.77 |
| XGBoost | 0.530 ± 0.214 | 15.58 ± 3.87 | 0.717 ± 0.164 | 10.56 ± 3.49 |
| 3-way Voting | 0.576 ± 0.244 | 14.87 ± 4.63 | 0.740 ± 0.157 | 10.07 ± 3.29 |
| ASO Name | Voting predicted | eSkip predicted | Experimental [14] |
| H73A(+16+40) | 63% (ranked #1) | 60% (ranked #1) | 100% (ranked #1) |
| H73A(+16+35) | 37% (ranked #3) | 23% (ranked #3) | 40% (ranked #3) |
| H73A(+21+40) | 42% (ranked #2) | 48% (ranked #2) | 85% (ranked #2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





